Abstract
Summary: The FSelector package contains a comprehensive list of feature selection algorithms for supporting bioinformatics and machine learning research. FSelector primarily collects and implements the filter type of feature selection techniques, which are computationally efficient for mining large datasets. In particular, FSelector allows ensemble feature selection that takes advantage of multiple feature selection algorithms to yield more robust results. FSelector also provides many useful auxiliary tools, including normalization, discretization and missing data imputation.
Availability: FSelector, written in the Ruby programming language, is free and open-source software that runs on all Ruby supporting platforms, including Windows, Linux and Mac OS X. FSelector is available from https://rubygems.org/gems/fselector and can be installed like a breeze via the command gem install fselector. The source code is available (https://github.com/need47/fselector) and is fully documented (http://rubydoc.info/gems/fselector/frames).
Contact: ywang@ncbi.nlm.nih.gov or bryant@ncbi.nlm.nih.gov
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Feature selection is of great importance for building statistical models when mining large datasets of high dimension, such as those generated from microarray and mass spectra analysis (Saeys et al., 2007). It proves to be effective in the data mining and bioinformatics fields for reducing dimensionality, selecting relevant and removing redundant features, increasing predictive accuracy and improving model interpretability (Guyon and Elisseeff, 2003).
Depending on how they interact with the learning method, various feature selection techniques roughly fall into three categories: filters, wrappers and embedded methods (Guyon, 2006). Filters investigate only the intrinsic characteristics of a given dataset and have the advantage of being fast as well as being independent of learning method. Basically, there are two types of filters: filter-by-feature-weighting and filter-by-feature-searching. The former measures independently the relevance of each feature to the target problem according to a certain evaluation criterion. It provides a weight or ranking list as output, and features are usually selected based on a given threshold. The latter also takes inter-feature correlation into account and generates a subset of the original feature set that tends to be both relevant and non-redundant. Wrappers wrap around a specific learning method and conduct a search in the space of feature subset for optimal model performance. They often report superior results than filters, but coupled with increased computational load (Inza et al., 2004). Embedded methods seek a trade-off between performance and computational cost, by use of the internal information of a learning method. In many applications, specifically in bioinformatics, the datasets are often huge with numerous samples and/or very high-dimension features. It is thus more practical to apply filters for feature selection because of their computational efficiency. To this end, we primarily implemented filters in FSelector.
A wide variety of feature selection algorithms have been proposed in the past decades (Guyon, 2006); however, most of them are scattered in literature and not readily available to researchers. Feature selection algorithms that come in a collection can be found in several machine learning software such as PyML (http://pyml.sourceforge.net/), written in Python and Weka (http://www.cs.waikato.ac.nz/ml/weka), written in Java, or serve as additional libraries for statistical platform including R (http://cran.r-project.org/web/packages/FSelector/index.html) and Matlab (http://featureselection.asu.edu/software.php).
Ruby is a high-level dynamic scripting language with a focus on simplicity and productivity, which has become increasingly popular in bioinformatics and other scientific research fields (Aerts and Law, 2009; Dahl and Crawford, 2008; Goto et al., 2010). To our best knowledge, the only Ruby package related to feature selection is the feature_selection gem that implements only three algorithms (https://rubygems.org/gems/feature_selection). In this work, we presented FSelector, providing a substantially larger collection of filters with 40+ feature selection algorithms implemented in Ruby.
2 FEATURES OF FSelector
FSelector works at the intermediate layer between data and machine learning approaches, such as random forest and support vector machines. It simply takes a local or remote dataset in CSV, ARFF or LibSVM file format as input and generates a reduced dataset with only selected features. The output dataset is interchangeable among different file formats and is compatible with several popular machine learning software, including Weka and LibSVM (http://www.csie.ntu.edu.tw/∼cjlin/libsvm/).
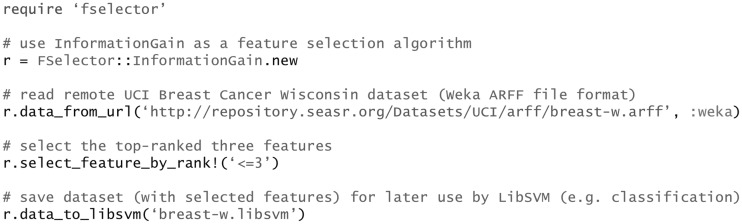
FSelector gathers and implements a rich list of feature selection algorithms (Supplementary Table S1) that have been put into practice by researchers for a wide variety of applications. It is very straightforward to get started with FSelector using a single algorithm: choose a desired algorithm, read in a data file and perform feature selection. For different algorithms, FSelector maintains a consistent interface for feature selection, depending on the algorithm type (i.e. filter-by-feature-weighting or filter-by-feature-searching). As an example, Fig. 1 shows the codes that use information gain as criterion to select the top three informative features.
Fig. 1.
Example of feature selection by using information gain as the feature selection algorithm. Note that line contents following ‘#’ are comments in the Ruby programming language
FSelector supports ensemble feature selection that takes advantage of multiple feature selection algorithms to yield more robust results (Saeys et al., 2008). It follows the same procedure as using a single algorithm and shares the same feature selection interface as well. Components in ensemble can be different algorithms of the same type (Supplementary Fig. S1A) or same algorithms with sampled data created by instance perturbation (Supplementary Fig. S1B). Depending on the type of component algorithm, results from individual algorithms are combined using various strategies (Supplementary Fig. S1).
FSelector also offers several data pre-processing techniques related to feature selection, such as normalization, discretization and missing data imputation. Normalization techniques may be useful to clean and standardize raw data. Real datasets often consist of continuous features, while many feature selection algorithms expect feature to be discrete. Discretization techniques are thus necessary prior to the use of such algorithms. Likewise, for feature selection algorithms that work on complete datasets, missing data imputation techniques are helpful to replace missing values with desired ones.
FSelector has on-line tutorials and code examples for each feature selection algorithm and for auxiliary normalization, discretization and missing data imputation techniques as well. FSelector is hosted at the largest and most popular Ruby gem repository (http://rubygems.org/), with source code available under the Git version control. FSelector is 100% documented including summaries and references, with intuitive layout generated by using the YARD tool (http://yardoc.org/). This support can lower the barrier for end users to write their own feature selection algorithms based on FSelector.
New features and updates will be added to FSelector in future. We hope the release of source code of FSelector into the public domain will encourage the community to contribute to the development and help to improve FSelector.
3 CONCLUSION
FSelector is a valuable Ruby gem that offers easy and public access to 40+ prevalent feature selection algorithms through a consistent interface. We hope the rich collection of algorithms together with other utilities (including various file formats support, auxiliary data pre-processing techniques and comprehensive help documentation) will make FSelector a useful tool for supporting various bioinformatics research, such as text mining, microarray analysis and mass spectra analysis.
Supplementary Material
ACKNOWLEDGEMENTS
Funding: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of Interest: None declared.
REFERENCES
- Aerts J, Law A. An introduction to scripting in Ruby for biologists. BMC Bioinf. 2009;10:221. doi: 10.1186/1471-2105-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl DB, Crawford S. Rinruby: accessing the r interpreter from pure ruby. J. Stat. Softw. 2008;29:1–18. [Google Scholar]
- Goto N, et al. BioRuby: bioinformatics software for the Ruby programming language. Bioinformatics. 2010;26:2617–2619. doi: 10.1093/bioinformatics/btq475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I. Feature Extraction: Foundations and Applications. Springer Verlag; 2006. [Google Scholar]
- Guyon I, Elisseeff A. An introduction to variable and feature selection. JMLR. 2003;3:1157–1182. [Google Scholar]
- Inza I, et al. Filter versus wrapper gene selection approaches in DNA microarray domains. Artif. Intell. Med. 2004;31:91–103. doi: 10.1016/j.artmed.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Saeys,Y. et al. (2008) Robust feature selection using ensemble feature selection techniques. In Proceedings of the European conference on Machine Learning and Knowledge Discovery in Databases-Part II. Springer-Verlag, Antwerp, Belgium, pp. 313–325.
- Saeys Y, et al. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.