Abstract
Small animal fluorescence molecular imaging (FMI) can be a powerful tool for preclinical drug discovery and development studies1. However, light absorption by tissue chromophores (e.g., hemoglobin, water, lipids, melanin) typically limits optical signal propagation through thicknesses larger than a few millimeters2. Compared to other visible wavelengths, tissue absorption for red and near-infrared (near-IR) light absorption dramatically decreases and non-elastic scattering becomes the dominant light-tissue interaction mechanism. The relatively recent development of fluorescent agents that absorb and emit light in the near-IR range (600-1000 nm), has driven the development of imaging systems and light propagation models that can achieve whole body three-dimensional imaging in small animals3.
Despite great strides in this area, the ill-posed nature of diffuse fluorescence tomography remains a significant problem for the stability, contrast recovery and spatial resolution of image reconstruction techniques and the optimal approach to FMI in small animals has yet to be agreed on. The majority of research groups have invested in charge-coupled device (CCD)-based systems that provide abundant tissue-sampling but suboptimal sensitivity4-9, while our group and a few others10-13 have pursued systems based on very high sensitivity detectors, that at this time allow dense tissue sampling to be achieved only at the cost of low imaging throughput. Here we demonstrate the methodology for applying single-photon detection technology in a fluorescence tomography system to localize a cancerous brain lesion in a mouse model.
The fluorescence tomography (FT) system employed single photon counting using photomultiplier tubes (PMT) and information-rich time-domain light detection in a non-contact conformation11. This provides a simultaneous collection of transmitted excitation and emission light, and includes automatic fluorescence excitation exposure control14, laser referencing, and co-registration with a small animal computed tomography (microCT) system15. A nude mouse model was used for imaging. The animal was inoculated orthotopically with a human glioma cell line (U251) in the left cerebral hemisphere and imaged 2 weeks later. The tumor was made to fluoresce by injecting a fluorescent tracer, IRDye 800CW-EGF (LI-COR Biosciences, Lincoln, NE) targeted to epidermal growth factor receptor, a cell membrane protein known to be overexpressed in the U251 tumor line and many other cancers18. A second, untargeted fluorescent tracer, Alexa Fluor 647 (Life Technologies, Grand Island, NY) was also injected to account for non-receptor mediated effects on the uptake of the targeted tracers to provide a means of quantifying tracer binding and receptor availability/density27. A CT-guided, time-domain algorithm was used to reconstruct the location of both fluorescent tracers (i.e., the location of the tumor) in the mouse brain and their ability to localize the tumor was verified by contrast-enhanced magnetic resonance imaging.
Though demonstrated for fluorescence imaging in a glioma mouse model, the methodology presented in this video can be extended to different tumor models in various small animal models potentially up to the size of a rat17.
Keywords: Cancer Biology, Issue 65, Medicine, Physics, Molecular Biology, fluorescence, glioma, light transport, tomography, CT, molecular imaging, epidermal growth factor receptor, biomarker
Protocol
1. Animal Preparation
Anesthetize nude mouse (Charles River, Wilmington, MA) with intra-peritoneal injection of ketamine-xylazine (100 mg/kg:10 mg/kg i.p.).
Place mouse in stereotactic frame, make an incision into the scalp on the left-side of the skull and, using an 18-gauge needle, create a 1 mm-diameter hole into the skull 2 mm from the central line and 2 mm behind the bregma.
Inject 5×105 U251 human neuronal glioblastoma cells (kindly provided by Dr. Mark Israel at Dartmouth College, Hanover, NH) in 5 μl of phosphate buffer solution into the left cerebral hemisphere at a depth of approximately 2 mm below the surface of the brain. Use a Hamilton micro-syringe18 and a blunt ended 27-gauge needle for cell implantation and insert the tip of the needle 3 mm from the outer surface of the skull and then withdraw 1 mm to create a pocket for the cells.
Suture incision site and allow recovery from surgery.
Wait ~14 days to allow the tumor to grow before imaging.
2. Fluorescence Tomography System Calibration
On the day of mouse imaging, initiate system and allow lasers and light detectors to warm-up for approximately 20 minutes to avoid drifts in system sensitivity.
Place a 100°-by-4° engineered line diffusor (Thorlabs, Newton, NJ) at the direct center of the imaging gantry, normal to the excitation laser: a picosecond-pulsed 80-MHz multimode 635 nm laser diode (PicoQuant Photonics North America Inc., Westfield, MA). Adjust the angle of the diffusor to maximize the amount of signal detected by all five light collection channels. A complete description of the imaging geometry is provided elsewhere11,14,15.
Place OD 2 neutral density filters (Thorlabs, Newton, NJ) in front of all fluorescence detection photomultiplier tubes (PMT) and OD 1 neutral density filters (Thorlabs, Newton, NJ) in front of all transmittance detection PMTs. Collect 100 temporal pulse spread profiles (TPSF) of the laser, each with a 1-s integration time.
Normalize each TPSF by the laser reference, correct for temporal drift in the laser reference, and average over all iterations for each detector. These averaged TPSFs are the detector-specific instrument response functions (IRF) used in the optical image reconstruction.
3. Imaging Protocol
Anesthetize the mouse with 2% isoflurane in oxygen (1 L/min).
Inject 1 nanomole of IRDye 800CW-EGF and 1 nanomole of Alexa Fluor 647 in 100 μl of phosphate buffer solution, intraperitoneally, 12 h prior to imaging to target epidermal growth factor receptor overexpression in the tumor.
Place the mouse onto the fiberglass supports of the imaging bed, arranging the mouse so that its nose remains in a cone delivering isoflurane anesthesia.
Ensure that the mouse is positioned appropriately on the bed: i.e., that when the bed is secured into the fluorescence tomography system the mouse is at the approximate center of the imaging gantry. This positioning can be guided by rotating the excitation laser 180° about the mouse, ensuring that the focal point of the laser illuminates a point roughly on the center of the mouse from the perspective of the laser at all angles.
Once positioned, carefully transfer the imaging bed and mouse to the microCT (eXplore Locus, GE Healthcare, London, ON) scanner and collect anatomical information at a resolution of 93-μm isotropic for the whole head of the mouse.
Visualize the CT image stack and choose the slice(s) to be imaged with the fluorescence tomography system.
Carefully transfer the imaging bed and mouse back to the fluorescence tomography system. Choose the number of source positions to collect data about the mouse for each imaging slice (32), the integration time for each TPSF measurement (1 s), the number of iterations for each source position (10), and the position and number of desired imaging slices from the CT image stack from Step 3.6. The numbers in parentheses are typical values for each imaging parameter yielding ~ 5 minutes of data acquisition per imaging slice.
Place triple notch filters (Chroma Technology Corp., Bellows Falls, VT) in front of the fluorescence detection PMTs, to restrict any laser light from reaching the fluorescence detectors, and OD 2 neutral density filters in front of the transmittance detection PMTs to avoid saturation of those detectors.
Run the data acquisition software, collecting fluorescence and transmittance TPSFs at each defined source detector position and for each excitation wavelength (635 nm and 755 nm to excite the Alexa Fluor 647 and IRDye 800CW-EGF tracers, respectively). For every set of TPSFs collected, monitor and record the laser intensity with a reference PMT channel.
4. Image Reconstruction
Determine the outer surface of the mouse and the location of the imaging bed support rods from the CT images and create masks that covers the confines of the mouse and the imaging rods separately.
Use the mouse mask to produce a finite-element mesh of the animal using the NIRFAST software19.
Localize the source and detector positions from the fluorescence tomography system on the surface of the mesh based on microCT and fluorescence spatial registration co-ordinates20.
Remove optical data points associated with source or detector positions that interact with the location of the imaging bed support rods.
Normalize data collected at each source detector position by the laser reference, correct for temporal drift in the laser reference, and correct for filter sensitivities, which were determined by experimental testing at time of purchase15.
Take the Born Ratio of the data (fluorescence divided by transmittance) for each source-detector position and multiply with a forward model simulation of transmittance based on the finite-element animal mesh for uniform optical properties. This is done to mitigate errors associated with source- or detector-tissue coupling21, to calibrate the data to the model22, and to adjust data for other aspects of model-data mismatch23,24.
Construct a data vector composed of the scaled difference of the born ratio data collected at both wavelengths. The scaling factor is chosen to maximize EGFR binding contrast. Perform time-domain image reconstruction with the calibrated difference data using the TPSF for each detection channel as an input, and create fluorescence maps of the contrast-enhanced targeted tracer15.
5. Representative Results
An example of a fluorescence reconstruction overlaid with a co-registered CT anatomical image from the head of a mouse with a U251 orthotopic glioma tumor is presented in Figure 1b. The center of mass of the glioma determined by the fluorescent reconstruction (Figure 1b) was within 1 mm of the tumor center of mass determined by contrast-enhance magnetic resonance imaging (Figure 1a). The CT and MRI images were co-registered based on a mutual-information transformation.
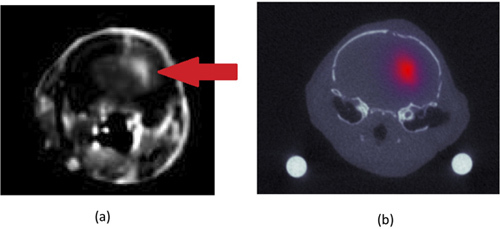 Figure 1. Contrast-enhanced (Gadolinium) magnetic resonance image of mouse head (a). The mouse was inoculated orthotopically with a U251 human glioma cell line. The location of the tumor, which absorbs more contrast agent than the normal brain, can be seen in the left cerebral hemisphere (right in image) and indicated by the white arrow. The corresponding computed tomography image (from the same location on the mouse head) is depicted in (b) with the epidermal growth factor targeted fluorescence minus the untargeted fluorescence reconstruction overlaid. The units of fluorescence are in inverse mm and relate to the absorption coefficient of bound targeted fluorescence multiplied by its quantum efficiency and by its concentration.
Figure 1. Contrast-enhanced (Gadolinium) magnetic resonance image of mouse head (a). The mouse was inoculated orthotopically with a U251 human glioma cell line. The location of the tumor, which absorbs more contrast agent than the normal brain, can be seen in the left cerebral hemisphere (right in image) and indicated by the white arrow. The corresponding computed tomography image (from the same location on the mouse head) is depicted in (b) with the epidermal growth factor targeted fluorescence minus the untargeted fluorescence reconstruction overlaid. The units of fluorescence are in inverse mm and relate to the absorption coefficient of bound targeted fluorescence multiplied by its quantum efficiency and by its concentration.
Discussion
Fluorescence tomography (FT) is a sensitive, ionizing radiation free molecular imaging modality based on visible and near-infrared light transport through biological tissue. Most of the interest in FT has been focused on its potential to expedite drug discovery and development in small animal experimental models1 and one key area of research has been the study of cancer biomarker expression and response to molecular therapies26. At present, there are two competing approaches to FT system design. The most common design is based on cooled charge-coupled device (CCD) cameras for fluorescence detection4-9. This design provides a high density of measurements, maximizing tissue sampling since each pixel in the CCD camera can detect light that has traveled a unique path through the tissue. However, CCD cameras have a limited dynamic range and read-out noise limits their ultimate sensitivity. The second design avoids the potential limitations of CCD camera detection by employing highly sensitive single-photon counting technology based on the use of such detectors as photomultiplier tubes or avalanche photodiodes10-13. The drawback of these more sensitive detection methods is that each detector can only collect light at a single point; therefore, to achieve dense tissue sampling, either many detectors have to be used (which is very expensive), or many projections have to be imaged with the same detector (which can be time consuming). While the optimal level of tissue sampling for small animal FT has not been agreed upon, and may vary on a case-by-case basis, it is agreed that single-photon counting instrumentation is better suited to explore the sensitivity limits of FT in terms of its ability to detect low concentrations of molecular markers. In this study, we provide a methodology for carrying out FT using single-photon counting detection instrumentation to localize tumors in mice.
There are four critical steps involved to produce robust datasets with time-correlated single-photon counting FT. The first is the application of a suitable and straightforward calibration procedure. In the presented methodology, the respective sensitivities of each detection channel are accounted for by collecting a baseline measurement of excitation light transmitted through a line-diffusor designed to direct equal fractions of light to each detector15. Furthermore, the detected light during an experiment is continuously calibrated to the laser reference, in terms of both intensity and mean-time, which could fluctuate over time, by the operation of a laser reference channel11,15. The second critical step is the accurate collection and co-registration of anatomical imaging for guided fluorescence reconstructions. The FT data alone offers no anatomical information; therefore, in order to create a model of light transport that can be used to reconstruct the location of fluorescent sources within a specimen from the detected fluorescence at the surface of the specimen, the anatomy of the specimen in relation to the FT system must be accurately known. In our system, the anatomical information is acquired by a micro-computed tomography system with spatial coordinates that have been spatially registered with those of the FT system15,20. The third critical step involves ensuring that an optimal exposure (i.e., total photon detection time for each laser projection) is employed at every source-detector position. This is important for two reasons: first, to ensure that there is adequate signal-to-noise at each detection position and second to avoid detector saturation, which could damage the detection units. In order to achieve optimal exposure at each detector position, an automatic exposure control is employed, which essentially triangulates the optimum exposure from two, low-signal exposures14. The fourth critical step of the methodology is referencing the collected fluorescence data to the amount of transmitted excitation light. This referencing is often called the Born ratio, and provides many benefits for FT, with the main one being a mitigation of model-data mismatch errors23,24. The presented system was designed to detect both fluorescence and transmitted excitation light simultaneously by channeling the light in each detection channel into 2 separate photomultiplier tubes. By doing this, we avoid any effects of motion on the accuracy of the Born ratio.
With a robust dataset it hand, image reconstruction of time-domain data involves solving the inverse problem of the finite element mesh having the expression:
d=Jx
where d is a vector with n x m elements for n source-detector projections and m TPSF time gates; J is an n x m-by-l sensitivity matrix (or Jacobian), for l nodes in the mesh; and x is the vector of fluorescence optical properties in each node, having size l. d is the calibrated data collected during the experiment and J is simulated using the finite element solution to the time domain diffusion approximation of fluorescence transport25. The time-dimension of J is also convolved with the detector specific instrument response functions. x is a representation of the fluorescence map of interest and is solved for using a Levenberg-Marqardt non-negative least squares approach with Tikhonov regularization15.
The methodology presented here, which describes a procedure capable of localizing fluorescently labeled tumors in mice using highly-sensitive photon counting fluorescence detection, has the potential to push the limits of FT. In a previous study, the potential of employing this approach in larger-than-mice animals models, such as rats, as well as improved sensitivity over existing system designs in mouse-sized specimens, was demonstrated17. The immediate application of this approach would be for the monitoring of biomarker expression in vivo in small animal tumor models to assess drug efficacy in a high-throughput means. The ability of the system to excite and detect fluorescence at multiple wavelengths allows the simultaneous detection of multiple fluorescent markers. Additional fluorescent markers provide a means of interrogating multiple aspects of a pathology, simultaneously, or could be used, as in this study, to employ more quantitative imaging approaches such as dual-reporter methods of measuring in vivo binding potential, a marker of receptor density26,27.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work has been funded by National Cancer Institute's grants R01 CA120368, R01 CA109558 (KMT, RWH, FEG, BWP), RO1 CA132750 (MJ, BWP) and K25 CA138578 (FL), and Canadian Institutes of Health Research postdoctoral fellowship award (KMT). The development of the fluorescence tomography system was partially funded by Advanced Research Technologies (Montreal, QC).
References
- Rudin M, Weissleder R. Molecular imaging in drug discovery and development. Nat. Rev. Drug Discov. 2003;2:123–131. doi: 10.1038/nrd1007. [DOI] [PubMed] [Google Scholar]
- Arridge S. Optical tomography in medical imaging. Inverse Problems. 1999;15:R41–R93. [Google Scholar]
- Leblond F, Davis SC, Valdes PA, Pogue BW. Pre-clinical whole-body fluorescence imaging: Review of instruments, methods and applications. J. Photochem. Photobiol. B. 2010;98:77–94. doi: 10.1016/j.jphotobiol.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Moosman A, Johnson VE. A Bayesian chi-squared goodness-of-fit test for censored data models. Biometrics. 2010;66:426–434. doi: 10.1111/j.1541-0420.2009.01294.x. [DOI] [PubMed] [Google Scholar]
- Da Silva A. Optical calibration protocol for an x-ray and optical multimodality tomography system dedicated to small-animal examination. Appl. Optics. 2009;48:151–162. doi: 10.1364/ao.48.00d151. [DOI] [PubMed] [Google Scholar]
- Deliolanis N. Free-space fluorescence molecular tomography utilizing 360 degrees geometry projections. Opt. Lett. 2007;32:382–384. doi: 10.1364/ol.32.000382. [DOI] [PubMed] [Google Scholar]
- Guo X. A combined fluorescence and microcomputed tomography system for small animal imaging. IEEE Trans. Biomed. Eng. 2010;57:2876–2883. doi: 10.1109/TBME.2010.2073468. [DOI] [PubMed] [Google Scholar]
- Lin Y. Quantitative fluorescence tomography using a combined tri-modality FT/DOT/XCT system. Opt. Express. 2010;18:7835–7850. doi: 10.1364/OE.18.007835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. High-resolution reconstruction of fluorescent inclusion in mouse thorax using anatomically guided sampling and parallel Monte Carlo computing. Biomedical Optics Express. 2011;2:2449–2460. doi: 10.1364/BOE.2.002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez JB, Berube-Lauziere Y. Diffuse light propagation in biological media by a time-domain parabolic simplified spherical harmonics approximation with ray-divergence effects. Appl. Optics. 2010;49:1414–1429. doi: 10.1364/AO.49.001414. [DOI] [PubMed] [Google Scholar]
- Kepshire D. A microcomputed tomography guided fluorescence tomography system for small animal molecular imaging. Rev. Sci. Instrum. 2009;80:043701. doi: 10.1063/1.3109903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. A photo-multiplier tube-based hybrid MRI and frequency domain fluorescence tomography system for small animal imaging. Phys. Med. Biol. 2011;56:4731–4747. doi: 10.1088/0031-9155/56/15/007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedre MJ. Early photon tomography allows fluorescence detection of lung carcinomas and disease progression in mice in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19126–19131. doi: 10.1073/pnas.0804798105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepshire DL, Dehghani H, Leblond F, Pogue BW. Automatic exposure control and estimation of effective system noise in diffuse fluorescence tomography. Opt. Express. 2009;17:23272–23283. doi: 10.1364/OE.17.023272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichauer KM. Imaging workflow and calibration for CT-guided time-domain fluorescence tomography. Biomedical Optics Express. 2011;2:3021–3036. doi: 10.1364/BOE.2.003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy JC, Pottier RH. Endogenous protoporphyrin IX, a clinically useful photosensitizer for photodynamic therapy. Journal of photochemistry and photobiology. 1992;14:275–292. doi: 10.1016/1011-1344(92)85108-7. [DOI] [PubMed] [Google Scholar]
- Leblond F, Tichauer KM, Holt R, El-Ghussein F, Pogue BW. Towards whole-body optical imaging of rats using single-photon counting fluorescence tomography. Opt. Lett. 2011;36:3723–3725. doi: 10.1364/OL.36.003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs-Strauss SL. Noninvasive fluorescence monitoring for functional assessment of murine glioma treatment [dissertation] Dartmouth College; 2008. [Google Scholar]
- Dehghani H. Near infrared optical tomography using NIRFAST: Algorithm for numerical model and image reconstruction. Commun. Numer. Meth. En. 2009;25:711–732. doi: 10.1002/cnm.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R, El-Ghussein F, Tichauer KM, Leblond F, Pogue BW. Proceedings of SPIE. 2011. p. 789213.
- Ntziachristos V, Weissleder R. Experimental three-dimensional fluorescence reconstruction of diffuse media by use of a normalized Born approximation. Opt. Lett. 2001;26:893–895. doi: 10.1364/ol.26.000893. [DOI] [PubMed] [Google Scholar]
- Davis SC. Magnetic resonance-coupled fluorescence tomography scanner for molecular imaging of tissue. The Review of scientific instruments. 2008;79:064302. doi: 10.1063/1.2919131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond F, Tichauer KM, Pogue BW. Singular value decomposition metrics show limitations of detector design in diffuse fluorescence tomography. Biomedical Optics Express. 2010;1:1514–1531. doi: 10.1364/BOE.1.001514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubret A, Ripoll J, Ntziachristos V. Accuracy of fluorescent tomography in the presence of heterogeneities: Study of the normalized born ratio. Ieee T. Med. Imaging. 2005;24:1377–1386. doi: 10.1109/tmi.2005.857213. [DOI] [PubMed] [Google Scholar]
- Zhu Q. A three-dimensional finite element model and image reconstruction algorithm for time-domain fluorescence imaging in highly scattering media. Phys. Med. Biol. 2011;56:7419–7434. doi: 10.1088/0031-9155/56/23/006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichauer KM. In vivo quantification of tumor receptor binding potential with dual-reporter molecular imaging. Mol. Imag. Biol. 2011. Forthcoming. [DOI] [PMC free article] [PubMed]


