Summary
Recently we demonstrated that the miRNA regulates human Mesenchymal Stem Cells (hMSCs) differentiation. To determine the role of the miRNA pathway in hMSCs proliferation, Drosha and Dicer knockdown hMSCs were generated using a lentiviral based tetracycline inducible shRNA. hMSCs with reduced Drosha expression had a significantly reduced proliferation rate, while hMSCs with reduced Dicer expression displayed a proliferation rate similar to untransduced cells. Cell cycle analysis identified that unlike Dicer knockdown, Drosha knockdown hMSCs contained an increased number of G1 phase cells, with a reduced level of cells in S phase, compared to controls. ELISAs of hMSCs revealed decreased levels of pRB and stable levels of total RB with Drosha knockdown. Two key regulators of the G1/S phase transition, cyclin dependent kinase inhibitor 2A (p16) and cyclin dependent kinase inhibitor 2B (p15), were increased in Drosha knockdown cells but not in Dicer knockdown. Transcripts of 28S and 18S rRNA were significantly reduced in Drosha knockdown hMSCs, with no change in rRNA levels in Dicer knockdown hMSCs. 45S pre-rRNA transcripts were not significantly different in either knockdown model. The above results indicate that Drosha modifies hMSCs proliferation through a miRNA independent mechanism, potentially by regulating rRNA processing.
Introduction
Human multipotent stromal cells from bone marrow (hMSCs) have demonstrated significant therapeutic capability in a variety of disease processes (Kocher et al., 2001, Kinnaird et al., 2004, Kurokawa et al., 2005, Al-Khaldi et al., 2003, Lee et al., 2006, Gnecchi et al., 2005, Chopp and Li, 2002, Wu et al., 2007, Spees et al., 2008, Ohtaki et al., 2008). MSCs were first defined as fibroblastoid colony forming units (CFU-Fs), then as mesenchymal stem/progenitor cells, and recently as multipotent mesenchymal stromal cells (Dominici et al., 2006). The cells are easily isolated from bone marrow aspirates and can be rapidly expanded in vitro, producing large quantities of potentially remedial cells (Prockop, 1997, Phinney and Prockop, 2007, Prockop and Olson, 2007, Owen and Friedenstein, 1988, Dominici et al., 2006). hMSCs appear to have the ability to undergo up to 75 population doublings without losing their differentiation potential (Sekiya et al., 2002, Ylostalo et al., 2008, Whitney et al., 2009, Larson et al., 2008). These cells may regulate key biological activities of endogenous tissue regeneration and wound healing (Sacchetti et al., 2007, Lama et al., 2007, Sasaki et al., 2008, Wu et al., 2007). In addition, hMSCs can recover from growth arrest inducing conditions such as serum deprivation, hypoxia without losing their plasticity or wound healing properties (Pochampally et al., 2004, Hung et al., 2007, Sessarego et al., 2008, Shoji et al., 2011, Oskowitz et al., 2011). Recent studies implicate MSCs as supportive cells for tumorigenesis and metastasis (Sanchez et al., 2011, Goldstein et al., 2010). Furthermore, the use of hMSCs as cellular therapeutic vectors in being investigated in various disease models (Pochampally et al., 2005, Awad et al., 2007, Nixon et al., 2007, Ozawa et al., 2008, Liu et al., 2008). Understanding the molecular mechanisms by which the cells regulate self-renewal and replication is essential in order to maximize the potential of hMSCs therapeutic activity.
MicroRNAs (miRNAs) are endogenous, small, non-coding transcripts capable of rapidly regulating cellular gene expression (Bartel, 2004). The functional forms of these molecules are generated by post-transcriptional processing enzymes, including Dicer and Drosha (Hammond, 2005). Global disruption of miRNAs through manipulation of Dicer and Drosha has been shown to alter a variety of cellular mechanisms including differentiation, plasticity, cell growth and division, and recently self-renewal of stem cells (Oskowitz et al., 2008, Murchison et al., 2005, Zhang et al., 2006, Hatfield et al., 2005, Kanellopoulou et al., 2005, Muljo et al., 2005, Wienholds et al., 2003, Cobb et al., 2005). Previous studies have demonstrated that disruption of the enzymes needed to produce mature miRNAs results in rapid proliferation of cancerous cell lines, as well as increased tumorigenicity of cancer cells (Kumar et al., 2007, Ventura and Jacks, 2009). Dicer deficient mice are also more likely to develop tumours in a lung cancer model (Kumar et al., 2007). Interestingly, in germ line and embryonic stem cells, disruption of these enzymes decreases cell proliferation (Hatfield et al., 2005, Murchison et al., 2005, Wang et al., 2007). Individual miRNAs have been implicated in molecular regulation of these same processes. Several studies have identified specific miRNAs that act as positive or negative regulators of the cell cycle (Carleton et al., 2007, Linsley et al., 2007, Gillies and Lorimer, 2007, Liu et al., 2008, Galardi et al., 2007, le et al., 2007). Individual miRNAs have also been shown to function as both tumour suppressors and oncogenes (Chen et al., 2005, Esquela-Kerscher and Slack, 2006, Pallante et al., 2006, Galardi et al., 2007, Visone et al., 2007, Kumar et al., 2007, Zhang et al., 2007, Bonci et al., 2008, Calin et al., 2008, Chung et al., 2008).
Our previous work demonstrated that global miRNA disruption through Drosha and Dicer knockdown results in significantly reduced differentiation potential of hMSCs (Oskowitz et al., 2008). Both Drosha and Dicer knockdown models showed similar reduction in differentiation potential. As stem cell differentiation and proliferation are closely related, we extended our work to analyze the role of the miRNA pathway in hMSCs proliferation and present it in the current study. We used a similar approach as described in Oskowitz et al. 2008 to generate hMSCs that could be induced to knockdown Drosha or Dicer when treated with doxycycline (Oskowitz et al., 2008). After confirming that both models had a direct and comparable impact on global expression of mature miRNA transcripts, we analyzed the proliferation and cell cycle status of these cells. Surprisingly we found significant differences between the two models. Therefore, we focused on identifying enzyme specific, molecular mechanisms that regulate hMSCs proliferation and cell cycle status. Our results indicate that Drosha but not Dicer regulates cell cycle progression through a miRNA independent mechanism.
Results
Generation of hMSCs with Global Disruption of miRNA Processing through Dicer or Drosha Knockdown
In order to develop a model to study the influence of the miRNA pathway on hMSCs proliferation, we transduced hMSCs with lentiviral, tetracycline inducible vector, containing a short hairpin RNA (shRNA) targeting either Dicer or Drosha. We have previously demonstrated (Oskowitz et al., 2008) that these transduced cells showed a specific knockdown of Dicer or Drosha at the protein level when cultured with doxycycline, as compared to transduced hMSCs cultured without doxycycline in the media. Donor matched control cells showed no variation in the expression of these enzymes when cultured in the presence or absence of doxycycline (Oskowitz et al., 2008). In order to quantify the degree of knockdown, we preformed quantitative RT-PCR on transduced hMSCs cultured with doxycycline and compared the levels of Dicer and Drosha transcripts to transduced cells cultured without doxycycline. In cell populations from two separate donors we observed a greater than five-fold reduction (p<0.001) of Dicer and Drosha transcript levels when doxycycline was added to the cell culture media (Table 1 and Supplemental Figure 1a). Furthermore, for both Dicer and Drosha, the magnitude of transcript level reduction was similar in the two donors. We then verified that the reduction of Dicer and Drosha transcripts directly affected the production of mature miRNAs. We performed quantitative RT-PCR for RNU44 endogenous controls for miRNA real-time PCR assays (Chen et al., 2005) and 5 miRNAs expressed at high levels in hMSCs (Oskowitz et al., 2008). In both the Dicer and Drosha knockdown models, we observed significant reductions (p<0.001) in the level all of these mature miRNAs in transduced cells cultured with doxycycline, as compared to transduced cells cultured without doxycycline (Table 2 and Supplemental Figure 1b). For each individual miRNA the magnitude of reduction was similar in both Dicer and Drosha knockdown models.
Table 1.
Doxycycline induces knockdown of Drosha and Dicer. Relative Cts of Dicer and Drosha transcripts from quantitative RT-PCR assays. The data are from two different hMSCs donors (one and two) transduced with a shRNA targeting Drosha or Dicer and grown in the presence (+) or absence (-) of doxycycline. For each reaction 10ng of total RNA was used and GAPDH was used as a loading control. The average of 4 sample replicates with SD is shown.
| Doxy | Donor 1 | Donor 2 | ||
|---|---|---|---|---|
| - | + | - | + | |
| Drosha | 30.7 (+/- 0.1) | 32.5 (+/- 0.2) | 31.2 (+/- 0.2) | 33.0 (+/- 0.1) |
| Dicer | 25.5 (+/- 0.1) | 27.8 (+/- 0.2) | 25.7 (+/- 0.08) | 27.9 (+/- 0.2) |
Table 2.
Knockdown of Dicer and Drosha results in reduced expression of mature miRNA. Relative Cts of specific miRNAs and GAPDH transcripts from quantitative RT-PCR assays. The data are from hMSCs transduced with a shRNA targeting Drosha or Dicer and grown in the presence (+) or absence (-) of doxycycline. The average of 4 sample replicates with SD is shown.
| Drosha | Dicer | |||
|---|---|---|---|---|
| - | + | - | + | |
| 26 | 24.7 (+/- 0.1) | 27.8 (+/- 0.2) | 24.2 (+/- 0.1) | 27.8 (+/- 0.07) |
| 23 | 24.0 (+/- 0.1) | 25.9 (+/- 0.2) | 24.3 (+/- 0.1) | 26.4 (+/- 0.09) |
| Let 7a | 19.7 (+/- 0.1) | 22.8 (+/- 0.07) | 19.9 (+/- 0.08) | 22.3 (+/- 0.01) |
| 199 | 31.3 (+/- 0.1) | 37.7 (+/- 0.6) | 31.1 (+/- 0.2) | 37.0 (+/- 0.7) |
| 125 | 25.5 (+/- 0.2) | 28.2 (+/- 0.3) | 25.7 (+/- 0.1) | 27.8 (+/- 0.2) |
| RNU 44 | 25.3 (+/- 0.08) | 25.2 (+/- 0.07) | 25.8 (+/-0.07) | 27.3 (+/- 0.01) |
| GAPDH | 21.6 (+/- 0.1) | 21.5 (+/- 0.07) | 21.3 (+/- 0.09) | 21.6 (+/- 0.2) |
Drosha but not Dicer knockdown inhibits hMSCs cell proliferation
We then tested the effect of impaired miRNA processing and global miRNA knockdowns of Dicer and Drosha on the ability of hMSCs to proliferate in culture. hMSCs transduced with a shRNA targeting Dicer or Drosha were plated at equal density in cell culture media (CCM), in the presence or absence of doxycycline. Passage matched hMSCs, from the same donor and preparation as the transduced cells, were cultured under the same conditions. Cell number was tracked over six days. Surprisingly, the hMSCs transduced with a shRNA targeting Drosha and grown in the presence of doxycycline, showed a significant growth inhibition when compared to all other cell populations. This included cells with a shRNA targeting Dicer grown the presence of doxycycline. All cell populations, except Drosha knockdown cells, grew at the same rate with the same kinetics. This included donor matched control cells grown in the presence or absence of doxycycline (Figure 1a; and Supplemental Figure 2a). We then assayed the growth rate of hMSCs during long-term passage. We observed significantly reduced numbers of population doublings per day in hMSCs with a shRNA targeting Drosha grown the presence of doxycycline when compared to all other cell populations (Figure 1b; left panel). The rate of growth for the Drosha knockdown cells continued to decline until the fourth passage, when the level began to reach a plateau. By contrast, all other cell populations grew at consistent rates during the first 5 passages (Figure 1b; left panel). To test the reversibility of this effect, we cultured a population of hMSCs transduced with a shRNA targeting Drosha in the presence of doxycycline and then removed the doxycycline following the 3rd passage. In these cells, after the third passage, we observed a gradual and significant increase in the rate of growth compared to cells grown in media with continued doxycycline supplementation (Figure 1b; left panel). Two passages after doxycycline had been removed we observed population doublings approaching hMSCs transduced with a shRNA targeting Drosha that had never been exposed to doxycycline. When the same experiment was performed using the Dicer knockdown model, no significant difference in population doubling rates were observed (Figure 1b; right panel).
Figure 1.
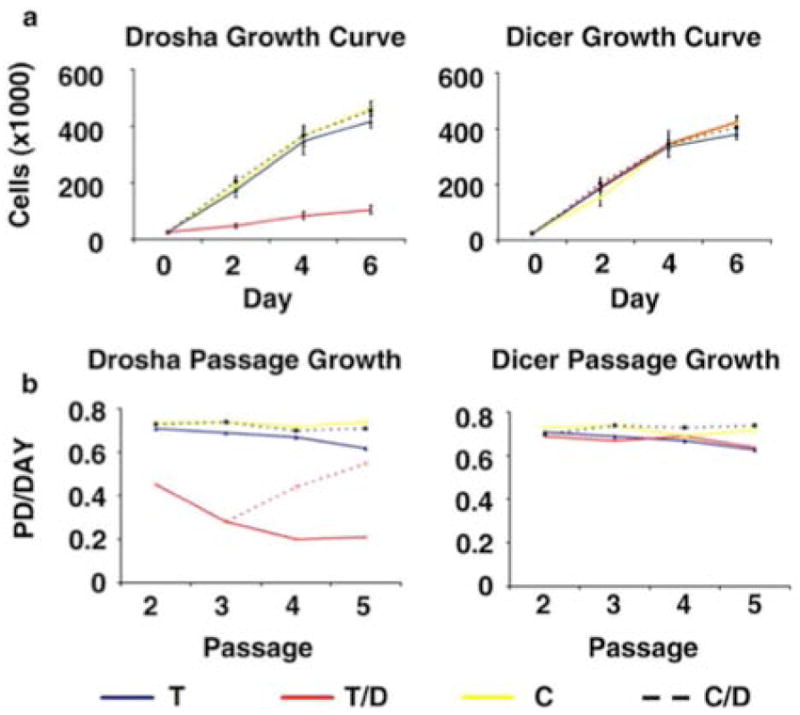
Drosha knockdown results in decreased proliferation of hMSCs. (a) Growth curve of hMSCs transduced (T) with a shRNA targeting Drosha (left panel) or Dicer (right panel), as well as control cells (C), grown in the presence and absence of doxycycline (D). (b) The number of population doublings (PD) per day for hMSCs transduced with a shRNA targeting Drosha (left panel) or Dicer (right panel), as well as control cells, grown in the presence and absence of doxycycline. The red dotted line represents cells in which doxycycline were removed from the media following passage 3. For all data the average of 3 culture replicates with SD is shown.
Drosha Knockdown Cells Do Not Demonstrate Increased Levels of Apoptosis or Changes in Culture Environment
In order to identify if apoptosis or cell death could account for the differences in cell proliferation, we assayed for DNA fragmentation using terminal deoxynucleotidyl transferase dUTP nick end-labelling (TUNEL) microscopy. Transduced hMSCs grown in the presence or absence of doxycycline showed no significant apoptosis (Supplemental Figure 2b). We then performed a mixing study to assess whether changes in culture conditions could account for the delay in growth seen in Drosha knockdown cells. hMSCs transfected with a tetracycline inducible shRNA targeting Drosha were combined with donor matched untransduced control cells. The transduced cells all had a constitutive Green Fluorescent Protein (GFP) marker while the control cells were not fluorescent (Figure 2a). Since both cell populations were grown in the same media any difference in proliferation rate would be due to intrinsic differences in cell populations and not changes in cell culture microenvironment. We observed that there was a significant decline in the percentage of GFP positive cells when doxycycline was added to the media, indicating a reduced percentage of transduced cells in culture. This effect was not seen in cultures without doxycycline supplementation (Figure 2b and 2c).
Figure 2.
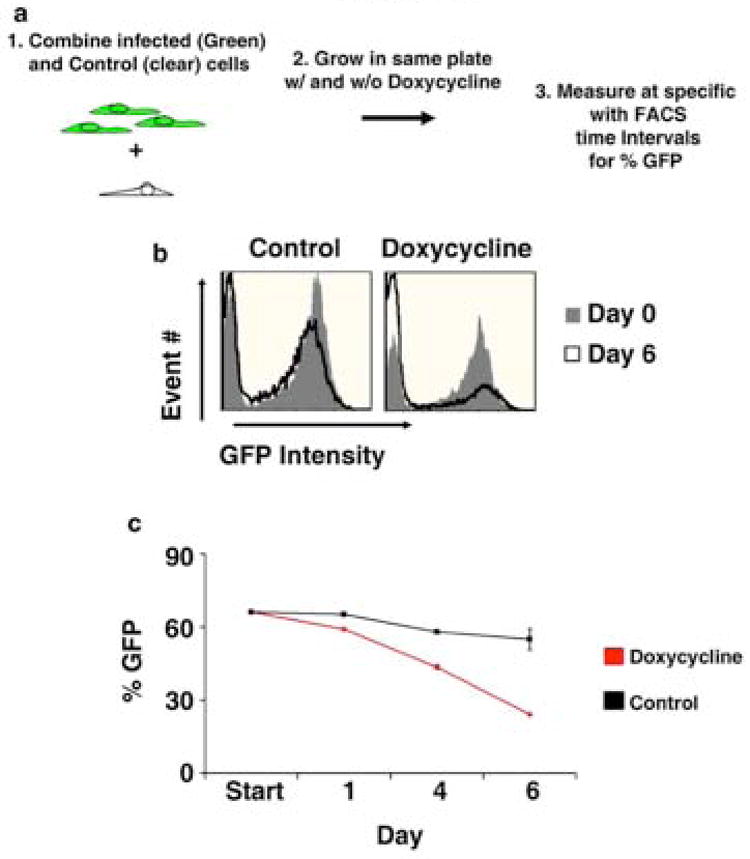
Mixing study of hMSCs transduced with a tetracycline inducible shRNA targeting Drosha and donor matched control hMSCs. A mixture of hMSCs infected with a tetracycline inducible shRNA targeting Drosha and control cells (uninfected donor matched cells) were grown in the presence (Doxycycline) or absence (Control) of doxycycline, and the percentage of GFP positive cells was measured over a six day time course. (a) Schematic of the experiment (b) Representative overlay histograms of both cell populations on day 0 and 6 of cell culture (c) Time course of GFP intensity during culture. The average of 3 culture replicates with SD is shown. The percentage of GFP positive cells is significantly different in cultures grown in the presence of doxycycline compared to cultures grown in the absence of doxycycline on Days 1, 4 and 6 (p <.001).
Drosha Knockdown Cells Have Reduced Cell Cycle Activity
To detect the basis for the reduced growth rate of Drosha knockdown cells, we analyzed the cell cycle status of hMSCs transduced with a shRNA targeting Drosha or Dicer. We observed a significant increase in the percentage of hMSCs in the G1/G0 phase of the cell cycle in Drosha knockdown cells, compared to control cells. There was also a reciprocal and significant decrease in the number of Drosha knockdown cells in the S phase of the cell cycle, compared to control cells (Figure 3a; top panel). Dicer knockdown cells showed no variation in cell cycle status compared to control cells (Figure 3a; bottom panel). To further elucidate the molecular pathways regulating the change of cell cycle status in Drosha knockdown cells, we analyzed nuclear expression of total and phosphorylated retinoblastoma (RB) protein. Enzyme linked immune sandwich assays (ELISAs) demonstrated significantly reduced levels of phosphorylated RB in Drosha knockdown cells compared to transduced control cells (Figure 3b). There was no significant difference in total RB observed between cell populations (Figure 3b). Western Blot analysis identified increased levels of phosphorylated cyclin dependent kinase 4 (pCDK4) in Drosha knockdown cells compared to transduced control cells, with similar levels of unphosphorylated CDK4 in Drosha knockdown and control cells (Figure 3c).
Figure 3.
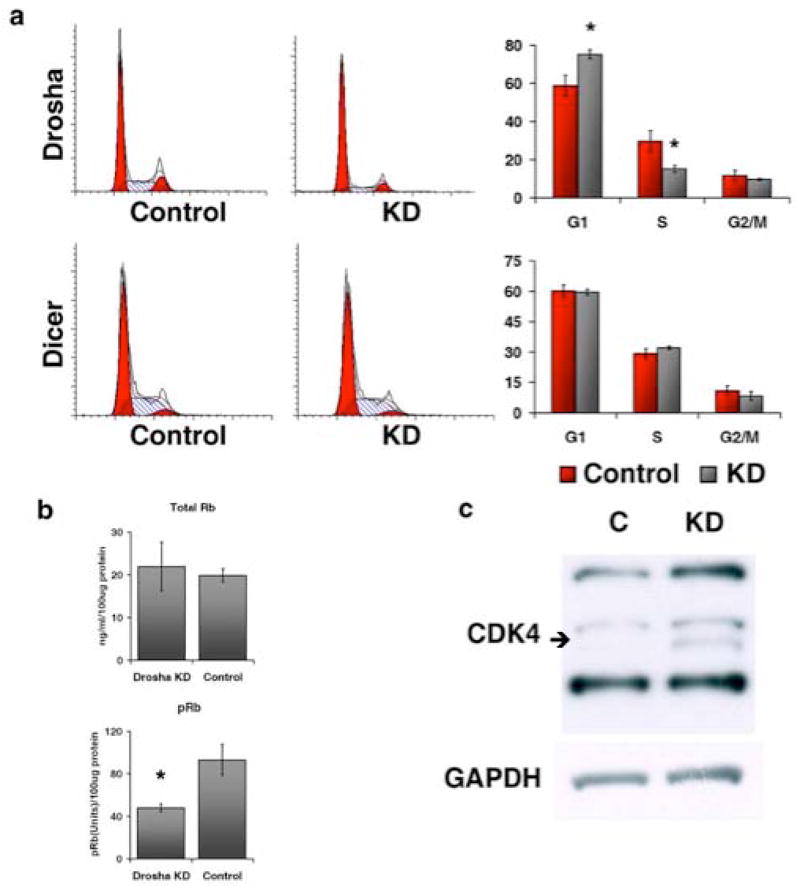
Drosha knockdown results in reduced G1/S transition. (a) Cell cycle distribution of hMSCs transduced with a shRNA targeting Drosha (top panel) or Dicer (bottom panel) grown in the presence (KD) or absence (Control) of doxycycline. Representative histograms and computer analysis of DNA content are displayed. The graphs display the average of 3 culture replicates with SD. (b) Relative levels of total RB and pRB in hMSCs transduced with an shRNA targeting Drosha and grown in the presence (KD) or absence (Control) of doxycycline. The average of 3 replicates with SD is shown. (c) Representative immunoblots of CDK4 and GAPDH from hMSCs transduced with a shRNA targeting Drosha grown in the presence (KD) or absence (C) of doxycycline. For CDK4 the bottom lines represent unphosphorylated protein and the top two lines represent phosphorylated forms of the protein. For all * indicates statistical significance (p<.001)
Drosha Knockdown Cells Have Increased Levels of G1 Phase Cell Cycle Inhibitors
Next, to understand the molecular mechanisms that were differentially expressed in Drosha knockdown cells compared to Dicer knockdown cells, we performed a quantitative RT-PCR array on 84 transcripts known to regulate cell cycle progression. Using a stringent exclusion criteria we identified two transcripts that were significantly upregulated in Drosha knockdown cells compared to transduced control cells, cyclin dependent kinase inhibitor 2A (CDKN2A) and cyclin dependent kinase inhibitor 2B (CDKN2B), also known as p16 and p15, respectively (Figure 4a and Supplemental Table 1). These two transcripts were not expressed at significantly different levels in Dicer knockdown cells compared to transduced controls (Figure 4b). The increase in transcript level in Drosha knockdown cells was confirmed by performing individual quantitative RT-PCR analysis on CDKN2A and CDKN2B (Supplemental Figure 3). We also confirmed that mRNA levels of RB and CDK4 were not significantly different in Drosha knockdown cells, compared to controls (Supplemental Figure 3). Immunofluorescent microscopy established that CDKN2A and CDKN2B were expressed at relatively higher levels within the nucleus in Drosha knockdown cells compared to control cells (Figure 4c and Supplemental Figure 4).
Figure 4.
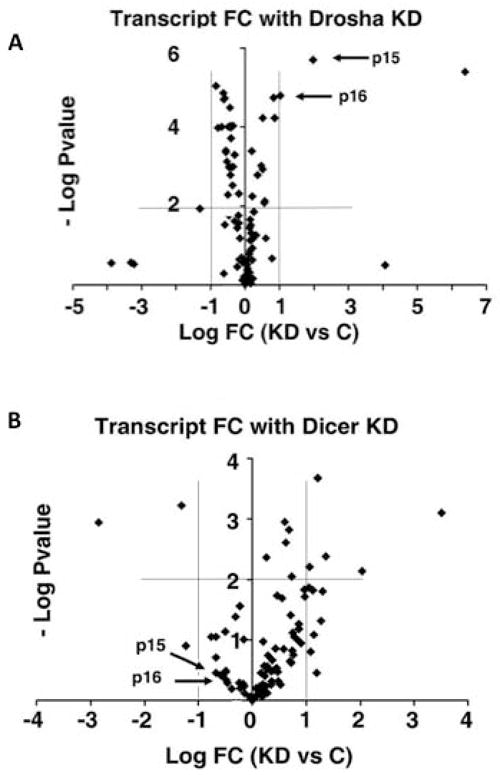
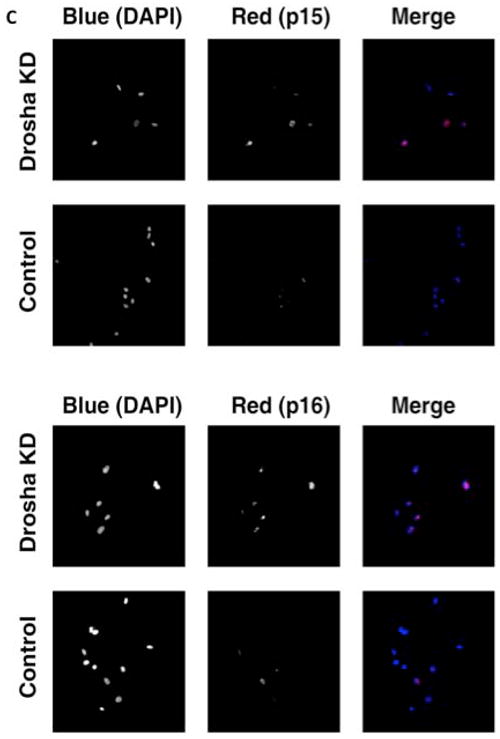
CDKN2A and CDKN2B transcripts are upregulated in Drosha knockdown hMSCs. Scatter plots of 84 cell cycle mRNAs and 5 endogenous controls. Each data point represents a specific mRNA. The x-axis represents the log fold change (FC) between transcript levels in knockdown cells (KD) and control cells (C). The y-axis represents the −log p-value for the fold change. (a) The top panel represents a comparison of transcripts from Drosha knockdown hMSCs and control cells (transduced hMSCs without doxycycline in the media) and (b) the bottom panel represents a comparison of transcripts from Dicer knockdown hMSCs and control cells (transduced hMSCs without doxycycline in the media). The horizontal thin lines identifies the p-values considered statistically significant (p<.001). The vertical lines identify the fold change considered statistically significant (FC > 2 or <-2). Two key cell cycle regulators CDKN2A (p15) and CDKN2B (p16) are identified. (c) Drosha knockdown hMSCs express increased nuclear CDKN2A and CDKN2B protein. Representative immunofluorescent images of CDKN2B (p15; top panel) and CDKN2A (p16; bottom panel) in Drosha knockdown (KD) and control hMSCs (transduced hMSCs without doxycycline in the media). The first two columns are greyscale images of blue and red channels, respectively. The last column is a merged colour image. Magnifications are 200x.
Drosha Knockdown Cells Have Reduced Levels of Mature rRNA
A previous study implicated Drosha in pre-ribosomal RNA processing (Wu et al., 2000). Furthermore, the level of cellular ribosomal RNA (rRNA) has been shown to regulate cell cycle status (Moss, 2004, Derenzini et al., 2005, Montanaro et al., 2007). Therefore, we tested the effects of Drosha knockdown on rRNA expression using quantitative RT-PCR assays. These assays demonstrated that Drosha knockdown cells contained significantly less 28S and 18S rRNA than control cells, and that the amount of cellular 28S and 18S rRNA declined in Drosha knockdown cells over time in culture (Figure 5 two bottom panels). The level of 45S rRNA, a precursor to 28S and 18S rRNA was consistent between Drosha knockdown cells and controls, with no variation over time in culture (Figure 5 top right panel). Dicer knockdown cells showed no variation in levels of 45S, 28S or 18S rRNA compared to control cells. In the Dicer knockdown model, rRNA levels did not vary over time in culture (Figure 5)
Figure 5.
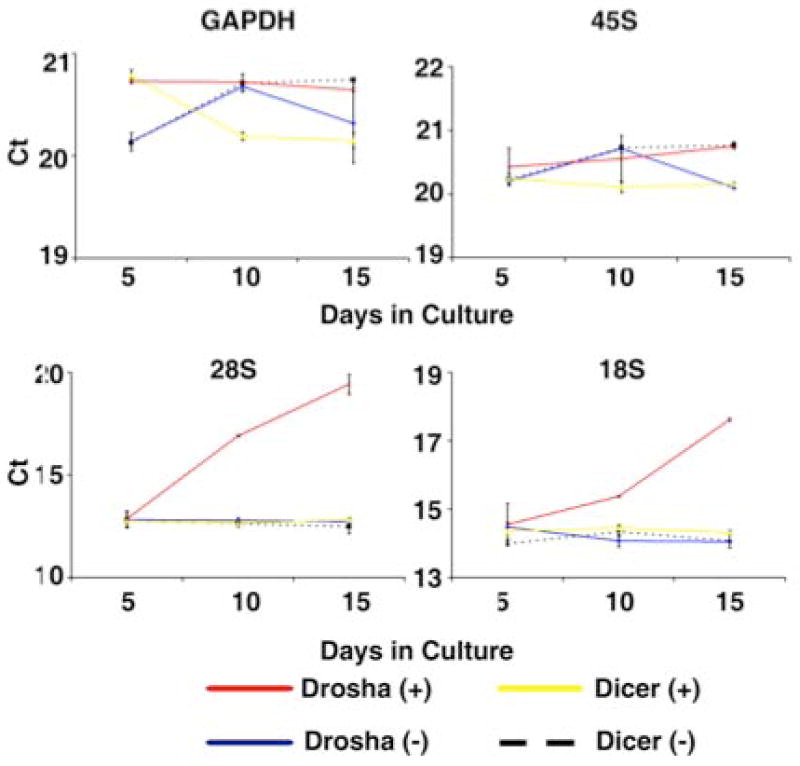
Drosha knockdown results in reduced levels of 28S and 18S rRNA. Relative Cts from real-time RT-PCR for 45S, 28S and18S rRNA as well as GAPDH over time in culture (Day 5, 10, 15). The data are from hMSCs donors transduced with a tetracycline inducible shRNA targeting Drosha or Dicer and grown in the presence (+) or absence (-) of doxycycline. For each reaction 10ng of total RNA was used. The average of 4 sample replicates with SD is shown. * indicates statistical significance (p<.001) from all other samples at that specific time point.
Discussion
We initiated this study in order to analyze the function of miRNAs in the proliferation and self-renewal of hMSCs. In our early experiments we observed that Drosha knockdown decreased the propagation of hMSCs but Dicer knockdown did not (Oskowitz et al., 2008). The result was surprising since knockout of Dicer was previously reported to decrease propagation in germ line stem cells (Hatfield et al., 2005), mouse embryonic stem cells (Murchison et al., 2005) and even primary cells (Mudhasani et al., 2008). Global disruption of miRNA processing using a DGCR8 knockout model also resulted in reduced proliferation of mouse embryonic cells (Wang et al., 2007). By contrast, in a study of transformed cell lines, global disruption of miRNA processing using Dicer, Drosha and DGCR8 knockdown models, resulted in increased cell growth and enhanced tumorigenesis (Kumar et al., 2007). Further analysis of the data in this study indicates that tumour growth in the Drosha knockdown model was considerably slower than Dicer and DGCR8 models, although no statistical analysis was presented in the manuscript. Clinical studies have shown that reduced Dicer expression is associated with poor prognosis in lung cancer (Karube et al., 2005), while increased Drosha levels are associated with poor outcomes, as well as enhanced cell growth in oesophageal cancer (Sugito et al., 2006). A number of studies have also demonstrated increased levels of Drosha in cervical cancer compared to non-cancerous tissue (Muralidhar et al., 2007, Scotto et al., 2008). Taken together these studies indicate that the role of traditional miRNA processing enzymes may be more complex than originally thought. Our data that Drosha disruption results in marked reduction of hMSCs proliferation and reduced cell cycle activity are consistent with studies demonstrating increased Drosha levels in neoplastic cells.
The finding that Drosha has a substantial effect on hMSCs rRNA levels is consistent with a previous report, in which cells treated with anti-sense to Drosha show a defect in rRNA processing and that this defect is downstream of 45S pre-ribosomal RNA production (Wu et al., 2000). Cellular rRNA appears to be a key component of cellular growth and proliferation (Moss, 2004). Disruption of rRNA synthesis has been demonstrated to significantly reduce cell cycle progression through the G1/S transition regulation. Furthermore levels of cellular rRNA appear to vary with cell cycle progression, with peak levels at the G1 end (Montanaro et al., 2007, Derenzini et al., 2005). These data indicate that rRNA levels potentially regulate cell cycle progression at the G1/S checkpoint. Certain intronic microRNA precursors called miRtrons have been shown to bypass Drosha processing in Drosophila, C. elegans, viruses and plants (Berezikov et al., 2007, Martin et al., 2009, Shapiro et al., 2010).
Accordingly, we propose a model in which Drosha regulates cell cycle progression through a miRNA independent mechanism. It is very unlikely that differences in miRNA function could account for these changes, as the effect was absent in the Dicer model and robust in the Drosha model. Further, the magnitude of miRNA reduction was similar in both models. Drosha disruption and cell cycle abnormalities were also strongly and temporally associated with reduced rRNA levels. Our cell cycle analysis and findings that pRB is decreased while pCDK4 is increased in Drosha knockdown cells corroborates the theory that decreased cell cycle activity in Drosha knockdown hMSCs occurs at the G1/S transition. Our data, that both CDKN2A and CDKN2B are upregulated with Drosha knockdown and not Dicer knockdown, are consistent with this mechanism of regulation. CDKN2A and CDKN2B have been shown to regulate G1/S transition in by preventing the pCDK4/cyclin D complex from phosphorylating RB (Sherr, 2001). Further study is needed to evaluate the potential mechanism that links rRNA levels and cell cycle transition via CDKN2A and CDK2NB, in human MSCs. In addition, the inherent variability in the physiology of human MSCs to rodent MSCs makes the data as presented relevant to human MSCs (Chamberlain et al., 2008, Mosna et al., 2010, Parekkadan and Milwid, 2010).
The proposed model is corroborated by recent studies that highlight the alternate and paradoxical functions of miRNA machinery. Chong et al. report that Drosha or Dicer knockout mice did not always present identical phenotypes and show that Drosha can regulate the expression of mRNAs containing secondary stem–loop structures by direct cleavage albeit inefficiently in some systems (Chong et al., 2010). Furthermore, while Dicer is known to have a role in the biogenesis of multiple classes of small RNAs including siRNA, the function of Drosha is limited to generation of miRNA and miRTRONS (Okamura et al., 2008, Xiao and Rajewsky, 2009, Jinek and Doudna, 2009, Merritt et al., 2010). In another study using human diploid fibroblasts, Srikantan et al. highlighted the paradoxical effects of Drosha and Dicer downregulation on the translation in a replicative senescence model in which reduction in Dicer levels markedly enhanced cellular senescence, while reduction of Drosha levels had very little effect on senescence (Srikantan et al., 2011). Interestingly, the data presented here adds to the known functions of Drosha that are not exclusive to miRNA processing.
Materials and Methods
Isolation and Culture of hMSCs
hMSCs from bone marrow aspirates were obtained from the NIH funded National Center for Research Resources (NCRR) Tulane Center for the Preparation and Distribution of Adult Stem Cells. The cells were obtained as frozen vials of passage 1 cells that were shown to be multipotent for differentiation. The cells were negative for hematopoietic markers (CD34, CD36, CD117 and CD45), and positive for CD29 (95%), CD44 (>93%), CD49c (99%), CD49f (>70%), CD59 (>99%), CD90 (>99%), CD105 (>99%) and CD166 (>99%). The hMSCs (about 1 million per vial) were thawed, plated in a 15 cm diameter dish and incubated overnight to recover adherent, viable cells. The cells were then lifted with trypsin/EDTA (0.25 % Trypsin/1mM EDTA; GIBCO/BRL) and re-plated at 500 cells/cm2. All cultures were incubated in complete culture medium (CCM): α-MEM (GIBCO/BRL; Carlsbad, CA) containing 17 % (v/v) FBS (lot-selected for rapid growth of MSCs; Atlanta Biologicals, Lawrenceville, GA) and 2 or 4 mM L-glutamine (GIBCO/BRL) at 37 °C with 5 % humidified CO2, unless otherwise noted.
For most of the experiments medium was changed every three days until cells were 70% confluent. The cells were then lifted with trypsin/EDTA for 5 min at 37 °C. Trypsin was inactivated by adding an equal volume of FBS and the cells were concentrated by centrifugation at 500 g for 5 min. Cells were then re-suspended in CCM, and re-plated at approximately 500 cells/cm2 in a 150 mm diameter dish (Nunc; Rochester, NY), unless otherwise noted.
Cell number quantification was performed using Flow-Count Fluorospheres™ (Beckman Coulter; Fullerton, CA) according to the manufacturer’s protocol. Briefly, cells were lifted with trypsin/EDTA for 5 min at 37 °C. Trypsin was inactivated by adding an equal volume of FBS and the cells were concentrated by centrifugation at 500 g for 5 min. Cells were then re-suspended in 1000 μl PBS and aliquots of 100 μl were removed and mixed with 100 μl of fluorescent microsphere reagent (Beckman Coulter). The cell and fluorescent microsphere solution was then analyzed by fluorescent activated cell scanning (Cytomics FC 500, Beckman Coulter), using CPX2.2 software.
hMSCs were re-sorted for GFP positive cells following each passage to determine the percentage of GFP positive cells, using Fluorescence Activated Cell Sorting (FACS) (FacVantage with FACSDiva option, Becton Dickson; Franklin Lakes, NJ), to account for the significant reduction in GFP cells observed in the Drosha knockdown model.
Generation of Plasmids, Virus and Infection
The pPrime vector, consisting of a lentiviral backbone with a tetracycline inducible shRNA construct, (Stegmeier et al., 2005) was a gift of the Elledge laboratory (Cambridge, MA). We purchased a pSM2 vector with a mir-30 shRNA targeting Dicer with the following hairpin sequence TGCTGTTGACAGTGAGCGAGGCACCCATCTCTAATTATATTAGTGAAGCCACAGATGTAA TATAATTAGAGATGGGTGCCCTGCCTACTGCCTCGGA (Open Biosystems; Huntsville, AL). Both vectors were digested with XhoI and EcoRI (Invitrogen; Carlsbad, CA) and the resulting DNA was electrophoretically separated in an agarose gel. The 97bp band from the pSM2 vector, containing the mir-30 shRNA targeting Dicer and the lentiviral backbone of the pPrime vector were then isolated using QIAquick gel purification columns (Qiagen; Valencia, CA). The mir-30 shRNA targeting Dicer was ligated into the pPrime backbone in a 3:1 ratio (M/M) using T3 DNA ligase (Invitrogen). The pPrime vector containing the mir-30 shRNA targeting Dicer was then transfected into One Shot® MAX Efficiency® DH5™ T1 Phage-Resistant cells (Invitrogen). Chloramphenicol resistant colonies were selected from agar plates and grown in Circlegrow® Bacterial growth media (Qbiogene; Solon, OH) overnight, after which plasmid DNA was isolated using PureLink HiPure Maxiprep (Invitrogen). The sequences were confirmed by sequencing with the following primer 5’-GAAGTGATCTTCCGTCACAGG-3’. The construct targeting Drosha (Aagaard et al., 2007) containing the tetracycline activator and the tetracycline-response element with an shRNA targeting Drosha, as well as a constitutively expressed GFP construct, was a gift of the Rossi Lab (Duarte, CA). For both vectors, virus was generated by the lentiviral vector core of the Louisiana Cancer Research Consortium (Hasanuzzaman et al., 2007). For the construct targeting Dicer, approximately 105 passage 0 hMSCs were transduced with both the tetracycline activator and the pPrime tetracycline-response element containing mir-30 shRNA targeting Dicer (Open Biosystems; Huntsville, AL), at a multiplicity of infection (MOI) of 20. Following transduction, cells were grown out to 70% confluence in CCM containing doxycycline and GFP positive cells were selected using FACS (FacVantage with FACSDiva option, Becton Dickson). For the construct targeting Drosha, approximately 105 cells of passage 0 hMSCs were transduced with the single construct, containing both the tetracycline activator and the tetracycline-response element, at a MOI of 20. Following transduction cells were grown out to 70% confluence in CCM and GFP positive cells were selected using FACS.
Real Time RT-PCR Analysis
Cell pellets were prepared by harvesting cells with trypsin /EDTA. Total RNA was then extracted using TRIzol (Invitrogen) and RNA concentration was determined by measuring absorbance at 260 nm (SmartSpec 3000, BioRad; Hercules, CA). For miRNA analysis, quantitative RT-PCR was performed according to the manufacturer’s protocol using Taqman miRNA Assays (Applied Biosystems; Foster City, CA). The process is described in detail by Chen and colleagues (Chen et al., 2005). Briefly, 10 ng of total RNA was used to perform a reverse transcription reaction using a stem loop primer. Quantitative PCR was then performed using a miRNA specific primer/probe. Similarly, quantitative RT-PCR for mRNA was performed using Taqman gene expression assays (Applied Biosystems). For array-based assays 500 ng of total RNA was converted into cDNA with the RT2 First Strand Kit (SuperArray Bioscience Corp.; Frederick, MD). Quantitative PCR was performed using the Human Cycle RT2 Profiler™ PCR Array with the RT2 SYBR Green Master Mix (both SuperArray Bioscience Corp.). Samples were analyzed for statistical significance using the template provided by SuperArray, using 5 endogenous controls (β-2-microglobulin, β-actin, GAPDH, ribosomal protein L13a, hypoxanthine phosphoribosyltransferase-1). Samples amplifying at a Ct value greater than 35 were considered below the detectable range and the value of 35 was used for fold change calculations.
18S and 28S rRNA levels were analyzed using quantitative RT-PCR with the Taqman Human endogenous 18S control (Applied Biosystems) and the 28S endogenous control kit (Eurogentec; San Diego, CA), respectively. A custom primer/probe (primer 1: CAATTCACATTAATTCTCGCAGCTAG; primer 2:GCCAAATCGACCTCGTAGACTC; probe: CATCGACGCACGAGCCGAGTGATCC), that was designed to detect only pre-rRNA 45S transcripts (Supplemental Figure 5) based on pre-rRNA post-transcriptional processing (Wu et al., 2000), was produced by Eurogentec (AnaSpec, Fremont, CA). RNU44 reverse transcriptase and PCR primers were obtained from Applied Biosystems (catalog ID# 1094).
All quantitative RT-PCR analysis was performed using an ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems), using the SDS 2.2 program. Unless otherwise noted samples were normalized to GAPDH mRNA levels. Fold change was calculated using the ΔΔCt method of relative quantification.
Cell-Cycle Analysis
Approximately 200,000 hMSCs from day 3 of cell culture were lifted with trypsin/EDTA for 5 min at 37 °C. Trypsin was then inactivated by adding an equal volume of FBS and the cells were concentrated by centrifugation at 500 × g for 5 min. The cells were then resuspended in 100μl of PBS and mixed with 100 μl of DNA-Prep LPR (Beckman Coulter). Samples were gently vortexed for 30 seconds, and then incubated for 12 hours at 4°C in the dark in DNA-Prep Stain Reagent (Beckman Coulter). Samples were then assayed by fluorescent activated cell scanning (Cytomics FC 500; Beckman Coulter) and cell cycle status was calculated by using ModFit LT software (Verity Software House, Topsham, ME).
Western Blot Analysis
Cells pellets were prepared by harvesting cells with trypsin/EDTA and then lysed using RIPA Lysis Buffer (Santa Cruz Biotechnology; Santa Cruz, CA) according to the manufacturer’s protocol. Lysates protein concentrations were determined using the BCA protein assay kit (Pierce; Rockford, IL) according to the manufacturer’s protocol. The cell lysate was fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis using NuPage 4-12% bis tris gels (Invitrogen). The sample was transferred to a filter (Immobilon P; Millipore, Bedford, MA) by electro-blotting (Immunoblotting Apparatus; Invitrogen). The membranes were incubated for 2 hours in a blocking buffer, consisting of 5% (w/v) non-fat dry milk (Sigma-Aldrich) in PBS containing 0.1% (v/v) Tween 20 (Fischer; Pittsburgh, PA). Membranes were then incubated overnight at 4°C with the appropriate primary antibody, diluted in blocking buffer. The following primary antibody dilutions were used for Western Blotting: mouse monoclonal anti-CDK4 antibody 1:200 (clone DCS-31; Sigma-Aldrich; St Louis, MO). Horseradish peroxidase (HRP) conjugated rabbit polyclonal anti-GAPDH 1:200 (Abcam; Cambridge, MA). The next morning membranes were washed 3 times with PBS containing 0.1% (v/v) Tween 20. When necessary, membranes were incubated for 1 hour with HRP conjugated goat anti-mouse (Chemicon; Bedford, MA) secondary antibody diluted 1:5000 in blocking buffer. Bound antibody was detected using the Visualizer Spray & Glow system chemiluminescence assay (Upstate; Bedford, MA).
ELISA assays
ELISAs were performed using Human RB ELISA and Human RB ELISA pT821 assays (both from Invitrogen) according to the manufacturer’s protocol. Briefly, appropriately treated cells were grown in T-175 plates (Nunc) and cultured for 3 days in complete culture media with the previously stated supplements. Following intracellular protein extraction, assays were performed using a 96 well pre-coated plate. Samples were incubated in antibody-coated wells, washed and then incubated with a secondary antibody. Following a second round of washes the wells were incubated with a colorimetric solution for 1 hour. The reaction was then stopped using the appropriate stop solution. Quantification was performed by measuring absorbance of each well at 450 nm and then subtracting out baseline absorbance at 544 nm, using an automated plate reader (Fluostar Optima, BMG Labtech). A standard curve was generated using recombinant human RB or using Jurkat cell extracts supplied by the manufacturer.
Immunofluorescent Microscopy
For immunofluorescence microscopy, cells grown on chamber slides were washed in PBS and fixed in phosphate-buffered 4% (v/v) paraformaldehyde for 15 min. Mouse anti-human CDKN2A (Clone DCS-50: Sigma-Aldrich), mouse anti-human CDK2B (Clone 15P06; Abcam) were used as primary antibodies, and goat anti-mouse IgG conjugated to AlexaFluor594 (Molecular Probes; Carlsbad, Ca) was used as a secondary antibody. Images were acquired on an upright epifluorescent spinning disk confocal microscope (Hamamatsu EM-CCD C9100; Hamamatsu City, Japan) using StereoInvestigator software (MBF Bioscience, Williston, VT).
Apoptosis analysis
hMSCs grown on 4 mm2 chamber slides were washed in PBS and fixed in phosphate-buffered 4% (v/v) paraformaldehyde for 15 min. Cells were then permeabilized using PBS containing 0.02% (v/v) Triton X-100 (Sigma). Apoptosis was assayed using the DeadEnd™ Colorimetric TUNEL System (Promega; Madison, WI) according to the manufacturer’s protocol. A positive control (cells treated with 100ul of DNASE1) and a negative control (cells not treated with Triton X-100) were included. Following the staining protocol, cells were visualized using a on an upright epifluorescent microscope (Hamamatsu EM-CCD C9100; Hamamatsu City, Japan).
Statistical Analysis
All statistical analysis was performed using Student’s t-test, using a two-sample method, assuming unequal variances and with p-values generated for two tails. Sample sizes and p-values are noted in figure legends and specific p-values are also included in the text. For PCR using SuperArray technology, statistical analysis was performed using pre-designed templates available on the SuperArray website.
Supplementary Material
Acknowledgments
Some of the materials employed in this work were provided by the Tulane Center for Gene Therapy through a grant from NCRR of the NIH, Grant # P40RR017447 and grants NIH AR 47796 and AR 48323, the Oberkotter Foundation, the HCA the Health Care Company and the Louisiana Gene Therapy Research Consortium to DJP and Grant from Louisiana cancer research to RP. We would like to thank Rossi lab (Duarte, CA) for the gift of construct targeting Drosha (Aagaard et al., 2007) containing the tetracycline activator and the tetracycline-response element with an shRNA targeting Drosha, as well as a constitutively expressed GFP construct.
Footnotes
All authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References list
- Aagaard L, Amarzguioui M, Sun G, Santos LC, Ehsani A, Prydz H, Rossi JJ. A facile lentiviral vector system for expression of doxycycline-inducible shRNAs: knockdown of the pre-miRNA processing enzyme Drosha. Mol Ther. 2007;15:938–945. doi: 10.1038/sj.mt.6300118. [DOI] [PubMed] [Google Scholar]
- Al-Khaldi A, Eliopoulos N, Martineau D, Lejeune L, Lachapelle K, Galipeau J. Postnatal bone marrow stromal cells elicit a potent VEGF-dependent neoangiogenic response in vivo. Gene Ther. 2003;10:621–629. doi: 10.1038/sj.gt.3301934. [DOI] [PubMed] [Google Scholar]
- Awad HA, Zhang X, Reynolds DG, Guldberg RE, O’Keefe RJ, Schwarz EM. Recent advances in gene delivery for structural bone allografts. Tissue Eng. 2007;13:1973–1985. doi: 10.1089/ten.2006.0107. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De MR. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, Alder H, Volinia S, Rassenti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- Chamberlain G, Wright K, Rot A, Ashton B, Middleton J. Murine mesenchymal stem cells exhibit a restricted repertoire of functional chemokine receptors: comparison with human. PLoS One. 2008;3:e2934. doi: 10.1371/journal.pone.0002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005a;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopp M, Li Y. Treatment of neural injury with marrow stromal cells. Lancet Neurol. 2002;1:92–100. doi: 10.1016/s1474-4422(02)00040-6. [DOI] [PubMed] [Google Scholar]
- Chung EY, Dews M, Cozma D, Yu D, Wentzel EA, Chang TC, Schelter JM, Cleary MA, Mendell JT, Thomas-Tikhonenko A. c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol Ther. 2008;7 doi: 10.4161/cbt.7.11.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M, Montanaro L, Chilla A, Tosti E, Vici M, Barbieri S, Govoni M, Mazzini G, Trere D. Key role of the achievement of an appropriate ribosomal RNA complement for G1-S phase transition in H4-II-E-C3 rat hepatoma cells. J Cell Physiol. 2005;202:483–491. doi: 10.1002/jcp.20144. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le BK, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716– 23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- Gnecchi M, He H, Liang OD, Melo LG, Morello F, Mu H, Noiseux N, Zhang L, Pratt RE, Ingwall JS, Dzau VJ. Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat Med. 2005;11:367–368. doi: 10.1038/nm0405-367. [DOI] [PubMed] [Google Scholar]
- Goldstein RH, Reagan MR, Anderson K, Kaplan DL, Rosenblatt M. Human bone marrow-derived MSCs can home to orthotopic breast cancer tumors and promote bone metastasis. Cancer Res. 2010;70:10044–10050. doi: 10.1158/0008-5472.CAN-10-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman M, Kutner R, gha-Mohammadi S, Reiser J, Sehgal I. A doxycycline-inducible urokinase receptor (uPAR) upregulates uPAR activities including resistance to anoikis in human prostate cancer cell lines. Mol Cancer. 2007;6:34. doi: 10.1186/1476-4598-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Hung SC, Pochampally RR, Hsu SC, Sanchez C, Chen SC, Spees J, Prockop DJ. Short-term exposure of multipotent stromal cells to low oxygen increases their expression of CX3CR1 and CXCR4 and their engraftment in vivo. PLoS One. 2007;2:e416. doi: 10.1371/journal.pone.0000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird T, Stabile E, Burnett MS, Epstein SE. Bone-marrow-derived cells for enhancing collateral development: mechanisms, animal data, and initial clinical experiences. Circ Res. 2004;95:354–363. doi: 10.1161/01.RES.0000137878.26174.66. [DOI] [PubMed] [Google Scholar]
- Kocher AA, Schuster MD, Szabolcs MJ, Takuma S, Burkhoff D, Wang J, Homma S, Edwards NM, Itescu S. Neovascularization of ischemic myocardium by human bone-marrow-derived angioblasts prevents cardiomyocyte apoptosis, reduces remodeling and improves cardiac function. Nat Med. 2001;7:430–436. doi: 10.1038/86498. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673– 677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kurokawa M, Kokubu F, Matsukura S, Kawaguchi M, Ieki K, Suzuki S, Odaka M, Watanabe S, Takeuchi H, Akabane T, Asano K, Iwase M, Honma I, Adachi M. Effects of corticosteroid on the expression of thymus and activation-regulated chemokine in a murine model of allergic asthma. Int Arch Allergy Immunol. 2005;137(Suppl 1):60–68. doi: 10.1159/000085434. [DOI] [PubMed] [Google Scholar]
- Lama VN, Smith L, Badri L, Flint A, Andrei AC, Murray S, Wang Z, Liao H, Toews GB, Krebsbach PH, Peters-Golden M, Pinsky DJ, Martinez FJ, Thannickal VJ. Evidence for tissue-resident mesenchymal stem cells in human adult lung from studies of transplanted allografts. J Clin Invest. 2007;117:989–996. doi: 10.1172/JCI29713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson BL, Ylostalo J, Prockop DJ. Human multipotent stromal cells undergo sharp transition from division to development in culture. Stem Cells. 2008;26:193–201. doi: 10.1634/stemcells.2007-0524. [DOI] [PubMed] [Google Scholar]
- le SC, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, Farace MG, Agami R. Regulation of the p27 (Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RH, Hsu SC, Munoz J, Jung JS, Lee NR, Pochampally R, Prockop DJ. A subset of human rapidly self-renewing marrow stromal cells preferentially engraft in mice. Blood. 2006;107:2153–2161. doi: 10.1182/blood-2005-07-2701. [DOI] [PubMed] [Google Scholar]
- Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Smibert P, Yalcin A, Tyler DM, Schafer U, Tuschl T, Lai EC. A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways. Mol Cell Biol. 2009;29:861–870. doi: 10.1128/MCB.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Bar-Eli M, Sood AK. The dicey role of Dicer: implications for RNAi therapy. Cancer Res. 2010;70:2571–2574. doi: 10.1158/0008-5472.CAN-09-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Mazzini G, Barbieri S, Vici M, Nardi-Pantoli A, Govoni M, Donati G, Trere D, Derenzini M. Different effects of ribosome biogenesis inhibition on cell proliferation in retinoblastoma protein- and p53-deficient and proficient human osteosarcoma cell lines. Cell Prolif. 2007;40:532–549. doi: 10.1111/j.1365-2184.2007.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosna F, Sensebe L, Krampera M. Human bone marrow and adipose tissue mesenchymal stem cells: a user’s guide. Stem Cells Dev. 2010;19:1449–1470. doi: 10.1089/scd.2010.0140. [DOI] [PubMed] [Google Scholar]
- Moss T. At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261– 269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon AJ, Goodrich LR, Scimeca MS, Witte TH, Schnabel LV, Watts AE, Robbins PD. Gene therapy in musculoskeletal repair. Ann N Y Acad Sci. 2007;1117:310–327. doi: 10.1196/annals.1402.065. [DOI] [PubMed] [Google Scholar]
- Ohtaki H, Ylostalo JH, Foraker JE, Robinson AP, Reger RL, Shioda S, Prockop DJ. Stem/progenitor cells from bone marrow decrease neuronal death in global ischemia by modulation of inflammatory/immune responses. Proc Natl Acad Sci U S A. 2008;105:14638–14643. doi: 10.1073/pnas.0803670105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Lai EC. The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs. Cell Cycle. 2008;7:2840–2845. doi: 10.4161/cc.7.18.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskowitz A, McFerrin H, Gutschow M, Carter ML, Pochampally R. Serum-deprived human multipotent mesenchymal stromal cells (MSCs) are highly angiogenic. Stem Cell Res. 2011 doi: 10.1016/j.scr.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskowitz AZ, Lu J, Penfornis P, Ylostalo J, McBride J, Flemington EK, Prockop DJ, Pochampally R. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci U S A. 2008;105:18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen M, Friedenstein AJ. Stromal stem cells: marrow-derived osteogenic precursors. Ciba Found Symp. 1988;136:42–60. doi: 10.1002/9780470513637.ch4. [DOI] [PubMed] [Google Scholar]
- Ozawa K, Sato K, Oh I, Ozaki K, Uchibori R, Obara Y, Kikuchi Y, Ito T, Okada T, Urabe M, Mizukami H, Kume A. Cell and gene therapy using mesenchymal stem cells (MSCs) J Autoimmun. 2008;30:121–127. doi: 10.1016/j.jaut.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce CM, Fusco A. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- Parekkadan B, Milwid JM. Mesenchymal stem cells as therapeutics. Annu Rev Biomed Eng. 2010;12:87–117. doi: 10.1146/annurev-bioeng-070909-105309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cells. 2007;25:2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- Pochampally RR, Horwitz EM, DiGirolamo CM, Stokes DS, Prockop DJ. Correction of a mineralization defect by overexpression of a wild-type cDNA for COL1A1 in marrow stromal cells (MSCs) from a patient with osteogenesis imperfecta: a strategy for rescuing mutations that produce dominant-negative protein defects. Gene Ther. 2005;12:1119–1125. doi: 10.1038/sj.gt.3302514. [DOI] [PubMed] [Google Scholar]
- Pochampally RR, Smith JR, Ylostalo J, Prockop DJ. Serum deprivation of human marrow stromal cells (hMSCs) selects for a subpopulation of early progenitor cells with enhanced expression of OCT-4 and other embryonic genes. Blood. 2004;103:1647–1652. doi: 10.1182/blood-2003-06-1967. [DOI] [PubMed] [Google Scholar]
- Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- Prockop DJ, Olson SD. Clinical trials with adult stem/progenitor cells for tissue repair: let’s not overlook some essential precautions. Blood. 2007;109:3147–3151. doi: 10.1182/blood-2006-03-013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti B, Funari A, Michienzi S, Di CS, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, Bianco P. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Penfornis P, Oskowitz AZ, Boonjindasup AG, Cai DZ, Dhule S, Rowan BG, Kelekar A, Krause DS, Pochampally RR. Activation of Autophagy in Mesenchymal Stem Cells Provides Tumor Stromal Support. Carcinogenesis. 2011 doi: 10.1093/carcin/bgr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Abe R, Fujita Y, Ando S, Inokuma D, Shimizu H. Mesenchymal stem cells are recruited into wounded skin and contribute to wound repair by transdifferentiation into multiple skin cell type. J Immunol. 2008;180:2581–2587. doi: 10.4049/jimmunol.180.4.2581. [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Subramaniyam S, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Schneider A, rias-Pulido H, Murty VV. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer. 2008;7:58. doi: 10.1186/1476-4598-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya I, Larson BL, Smith JR, Pochampally R, Cui JG, Prockop DJ. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- Sessarego N, Parodi A, Podesta M, Benvenuto F, Mogni M, Raviolo V, Lituania M, Kunkl A, Ferlazzo G, Bricarelli FD, Uccelli A, Frassoni F. Multipotent mesenchymal stromal cells from amniotic fluid: solid perspectives for clinical application. Haematologica. 2008;93:339–346. doi: 10.3324/haematol.11869. [DOI] [PubMed] [Google Scholar]
- Shapiro JS, Varble A, Pham AM, Tenoever BR. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16:2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Shoji M, Oskowitz A, Malone CD, Prockop DJ, Pochampally R. Human Mesenchymal Stromal Cells (MSCs) Reduce Neointimal Hyperplasia in a Mouse Model of Flow-Restriction by Transient Suppression of Anti-Inflammatory Cytokines. J Atheroscler Thromb. 2011 doi: 10.5551/jat.6213. [DOI] [PubMed] [Google Scholar]
- Spees JL, Whitney MJ, Sullivan DE, Lasky JA, Laboy M, Ylostalo J, Prockop DJ. Bone marrow progenitor cells contribute to repair and remodeling of the lung and heart in a rat model of progressive pulmonary hypertension. Faseb J. 22:1226–1236. doi: 10.1096/fj.07-8076com. [DOI] [PubMed] [Google Scholar]
- Srikantan S, Marasa BS, Becker KG, Gorospe M, Abdelmohsen K. Paradoxical microRNAs: Individual gene repressors, global translation enhancers. Cell Cycle. 2011;10:751–759. doi: 10.4161/cc.10.5.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugito N, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Kurehara H, Ando T, Mori R, Takashima N, Ogawa R, Fujii Y. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Russo L, Pallante P, De MI, Ferraro A, Leone V, Borbone E, Petrocca F, Alder H, Croce CM, Fusco A. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney MJ, Lee A, Ylostalo J, Zeitouni S, Tucker A, Gregory CA. Leukemia inhibitory factor secretion is a predictor and indicator of early progenitor status in adult bone marrow stromal cells. Tissue Eng Part A. 2009;15:33–44. doi: 10.1089/ten.tea.2007.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang J, Scott PG, Tredget EE. Bone marrow-derived stem cells in wound healing: a review. Wound Repair Regen. 2007;15(Suppl 1):S18–26. doi: 10.1111/j.1524-475X.2007.00221.x. [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Ylostalo J, Bazhanov N, Prockop DJ. Reversible commitment to differentiation by human multipotent stromal cells in single-cell-derived colonies. Exp Hematol. 2008;36:1390–1402. doi: 10.1016/j.exphem.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Anderson TA. MicroRNA: a new player in stem cells. J Cell Physiol. 2006;209:266–269. doi: 10.1002/jcp.20713. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


