Abstract
To understand how sucrose (Suc) is transported from source leaves to developing tap roots of carrot (Daucus carota L.), we cloned two cDNAs (DcSUT1 and DcSUT2) for proteins with homologies to plant Suc/H+ symporters. The deduced polypeptide sequences are 52% identical and have 12 predicted membrane-spanning domains each. Transport activities were confirmed by expression of the clones in yeast cells. Both transporters had optimal activity below pH 5.0 and Michaelis constant values of 0.5 mm. Suc uptake was inhibited by protonophores, suggesting that Suc transport is linked to the proton electrochemical potential across the plasma membrane. DcSUT1 and DcSUT2 had markedly different expression patterns. Transcripts of DcSUT1 were found only in the green parts of plants, with highest levels in the lamina of source leaves, indicating that DcSUT1 is required for the loading of Suc into the phloem. In leaf lamina expression was diurnally regulated, suggesting that Suc export from the leaves is higher during the day than during the night. The mRNA of DcSUT2 was found mainly in sink organs, and no diurnal expression pattern was detected in the storage root. Here, expression was not restricted to the phloem but was much higher in storage parenchyma tissues of phloem and xylem. The close relationship of DcSUT2 with a Suc/H+ symporter from fava bean, which facilitates Suc uptake into the cotyledons of developing seeds, indicates that this carrot Suc transporter may be involved in loading Suc into storage parenchyma cells.
Although higher plants are photoautotrophic, some tissues are dependent on the import of photoassimilates. In most plants, these heterotrophic tissues (sink tissues) are supplied with carbon in the form of Suc (Avigad, 1982). Long-distance transport of the disaccharide from source leaves to sink tissues takes place in the sieve tube/companion cell complex (phloem) and may require apoplastic loading and unloading steps (Ho, 1988). Passage of Suc through membranes is facilitated by energy-dependent, cargo-specific transport proteins (Giaquinta, 1977; Bush, 1993; Sauer et al., 1994; Bush et al., 1996; Rentsch et al., 1998).
A few years ago, the first plant sugar transporters were cloned by using engineered yeast cells, which can only live on Suc when they express a functional Suc transport protein. With this method, cDNA clones for Suc/H+ symporters were isolated from spinach (Riesmeier et al., 1992) and potato (Riesmeier et al., 1993). Subsequently, these clones were used as probes for the heterologous cloning of Suc transporters from other plant species such as Arabidopsis (Sauer and Stolz, 1994), Plantago major (Gahrtz et al., 1994), and castor bean (Weig and Komor, 1996). Most of these clones were predominantly expressed in the vascular system (Truernit and Sauer, 1995) and seem to be involved in loading Suc into the phloem (Riesmeier et al., 1994; Stadler and Sauer, 1996). Only two cDNA clones were isolated that were preferentially expressed in sink organs: a flower-specific transporter of P. major (Gahrtz et al., 1996), and a seed-specific transporter of fava beans (Weber et al., 1997). Characterization of these clones in yeast cells showed that the two transport proteins had different biochemical characteristics: the transporter from P. major was fairly insensitive to pH changes (neutral Suc transporter), whereas the Suc transporter from fava beans had an acidic pH optimum.
The transporters involved in the import of Suc into storage cells of roots and tubers are not known. Therefore, we set out to identify a tap-root-specific Suc transporter in carrot (Daucus carota), a plant that we use as a model for studying Suc metabolism and partitioning (Sturm, 1996). Our cloning approach was based on conserved peptide domains of plant Suc/H+ symporters. Here we describe two cDNA clones and demonstrate their functions in cells of Saccharomyces cerevisiae. DcSUT1 encodes a Suc transporter for the loading of Suc into the phloem of source leaves, whereas DcSUT2 is highly expressed in phloem and xylem parenchyma tissues of developing tap roots.
MATERIALS AND METHODS
Plant Material
Carrot plants (Daucus carota L. cv Nantaise) were grown in a garden near Basel. After harvest, plant organs were directly frozen in liquid nitrogen and stored at −80°C.
Isolation of cDNA Clones for Suc/H+ Symporters
Total RNA was isolated from leaves and roots of carrot plants with developing tap roots. For this purpose, the method described by Prescott and Martin (1987) was modified by adding 20 mg of Polyclar AT (Serva, Heidelberg, Germany) per gram of tissue before grinding in liquid nitrogen. Poly(A+) RNA was purified according to the batch protocol of the Oligotex kit (Qiagen, Chatsworth, CA). First-strand cDNA was obtained by reverse transcription of poly(A+) RNA with an oligo(dT25) primer (Amersham). A DNA fragment corresponding to a 330-bp-long domain of plant Suc/H+ symporters was amplified by PCR with the degenerated primers 5′-CA(AG)TT(CT)GG(GT)TGGGC(CT)(CT)T(AGT)CA-3′ and 5′-GC(AC)AC(AG)TC(AG)AG(AG)ATCCA(GT)AA-3′. Amplification was achieved in a DNA thermal cycler (Perkin-Elmer Cetus) with the following conditions: 30 cycles of denaturation at 94°C for 1 min, annealing at 48°C for 2 min, and elongation at 72°C for 2 min. Finally, the reaction mixture was kept at 72°C for 10 min. Reverse-transcription PCR products were analyzed by agarose-gel electrophoresis. Amplified DNA bands of the appropriate length were cloned into the pCR2.1 vector (Invitrogen, San Diego, CA). The plasmids generated were transformed into the Escherichia coli strain InVαF′ (Invitrogen).
For the isolation of full-length cDNAs, we generated cDNA libraries from poly(A+) RNA isolated from roots and leaves of carrot plants with developing tap roots (Amersham). The double-stranded cDNA was ligated to EcoRI adaptors. Excess adapters and cDNA smaller than 1 kb were removed by a spin column. cDNA was ligated into λ MOSSlox vector arms and packed into phage particles (Amersham). The libraries were screened with the corresponding 32P-labeled leaf- or root-specific PCR fragments.
Analysis of DNA and Protein Sequences
Both strands of the PCR fragments and the cDNA clones were automatically sequenced by the dideoxynucleotide chain-termination method (Messing, 1983). Computer-assisted analysis of DNA and protein sequences was performed using the Wisconsin package (version 9.1, Genetics Computer Group, Madison, WI).
Analysis of RNA and DNA
For RNA gel-blot analysis, total RNA (10 μg/lane) was separated on a 1.2% agarose gel containing 6% formaldehyde (Sambrook et al., 1989). For DNA gel-blot analysis, restricted DNA was separated on a 0.8% agarose gel (10 μg/lane). RNA and DNA were transferred to nylon membranes (Hybond N, Amersham). The gene-specific PCR fragments (approximately 330 bp) of DcSUT1 and DcSUT2 were labeled with 32P by random priming (Sambrook et al., 1989). As a probe for a constitutively expressed gene, a 1700-bp-long fragment (EcoRI/EcoRI) of the 18S rRNA gene from tomato was used (Schmidt-Puchta et al., 1989). The blots were prehybridized at 65°C for 5 h in 6× SSC, 5× Denhardt's solution, 100 mg/mL denatured calf thymus DNA, and 0.5% SDS (Sambrook et al., 1989). Hybridization was carried out in the same buffer overnight at 65°C. Blots were washed at 65°C with 1× SSC and 0.1% SDS, and 0.1× SSC and 0.1% SDS for 30 min each.
Expression of DcSUT1 and DcSUT2 in Saccharomyces cerevisiae
For the expression of DcSUT1 and DcSUT2 cDNAs in yeast cells, DNA fragments corresponding to the open reading frames of the carrot cDNA clones were isolated by PCR. PCR primers for the DcSUT1a sense construct were designed by introducing a PstI site upstream of the translation initiation codon ATG and a BamHI site beyond the stop codon, a BamHI site and a PstI site for the antisense constructs for DcSUT1a, an EcoRI site and a NotI site for the sense construct for DcSUT2, and a SmaI site and an EcoRI site for the antisense construct for DcSUT2. The following primers were used for the sense construct for DcSUT1a, 5′-CAAGCTGCAGATAGCAGCGATAACAG-3′ and 5′-GTCTTGGATCCTAACCAAGTCTGACAG-3′; for the antisense construct for DcSUT1a, 5′-CAAGGGATCCATAGCAGCCATAACAG-3′ and 5′-GTCTTCTGCAGTAACCAAGTCTGACAG-3′; for the sense construct for DcSUT2, 5′-CACAGAATTCACCATAAACAGATAAATC-3′ and 5′-CTAAAATGCGGCCGCTAACCACTTCACAG-3′; and for the antisense construct for DcSUT2, 5′-CACACCCGGGACCATAAACAGATAAATC-3′ and 5′-CTAAATGAATTCTAACCACTTCACAG-3′. Amplification was achieved with the following conditions: 30 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 2 min, and elongation at 72°C for 2 min. Finally, the reaction mixture was kept at 72°C for 10 min. The PCR fragments obtained were cleaved and ligated into the corresponding restriction sites of the E. coli/S. cerevisiae shuttle vector 112 A1NE (Riesmeier et al., 1992).
The S. saccharomyces stain SUSY7 (Riesmeier et al., 1992), which lacks the Trp gene for selection of recombinant cells, was transformed with the cDNAs in sense or antisense orientation according to the method of Gietz et al. (1992). As an additional control, the empty vector 112 A1NE was used. Cells were grown in 300 mL of yeast extract, peptone, and dextrose to an A600 of 0.5, washed in sterile water, and resuspended in 1.5 mL of freshly prepared lithium acetate buffer (1 volume of 10× Tris-EDTA, 1 volume of 1 m lithium acetate, and 8 volumes of sterile water). Ethanol-precipitated plasmid DNA (0.6–0.8 μg) was mixed with 200 μg of carrier DNA in a total volume of 20 μL of water. Yeast cells (200 μL) were added to each DNA sample, followed by 1.2 mL of 50% PEG 600 in water. Samples were incubated on a shaker for 30 min at 30°C, and then for an additional 15 min at 42°C. Finally, the cells were harvested by centrifugation and resuspended in 1 mL of 1× Tris-EDTA buffer. Different volumes of cells were plated on 2% agar in minimal medium (2% Glc, 0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, and 0.13% Trp minus dropout powder). After 4 d of incubation at 30°C, large colonies were picked and replated on solidified minimal medium with 2% Suc (pH 5.5). Fast-growing colonies from sense constructs were picked after incubation for 4 d at 30°C.
[14C]Suc Uptake Studies
Yeast cells were grown in 20 to 30 mL of minimal medium up to an A600 of 0.9 to 1.0, harvested by centrifugation, washed with 25 mm sodium phosphate buffer, pH 5.5, and resuspended in 1 mL of the same buffer. Suc uptake was determined as described by Cirillo (1989). For each assay, 1 mg of cells in 200 μL of buffer was used. The assays were started by the addition of 0.5 μCi of [14C]Suc (final concentration, 0.2 mm). Cells were incubated for 0, 2, 4, 6, and 8 min at 30°C in a shaking water bath. Sugar uptake was stopped by transfer of the cells into 10 mL of ice-cold water over a Whatman glass microfiber filter (0.7 μm) in a filtration apparatus. Cells were rapidly washed three times with 5 mL of ice-cold water. Finally, filters were transferred to liquid-scintillation vials and counted. In some experiments, d-Glc (10 mm) was added 5 min before the addition of labeled Suc.
To determine the pH dependence of Suc uptake, cells were treated as described above but were washed and resuspended in 25 mm sodium phosphate buffer of pH 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, or 8.0. To disrupt the proton gradient across membranes, CCCP was added 30 s before the addition of Suc at a final concentration of 5, 50, or 500 μm. To measure the Km values of DcSUT1 and DcSUT2, Suc concentrations of 0.125, 0.25, 0.5, 1.0, 2.0, 4.0, 6.0, 8.0, and 10.0 mm were used.
RESULTS
Isolation of cDNA Clones for Suc/H+ Symporters from Carrot
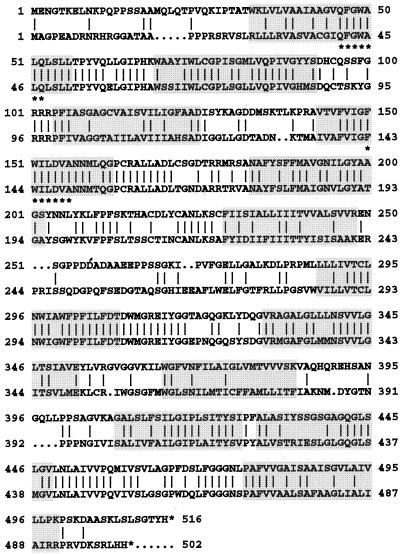
A comparison of the amino acid sequences deduced from the sequences of cDNA clones for plant Suc/H+ symporters (the sequences used are listed in the legend to Fig. 3) identified two highly conserved heptapeptides, QFGWALQ and FWILDVA, located in the first and fourth membrane-spanning domains, respectively (marked by asterisks in Fig. 1). Degenerated primers generated from these peptide sequences and first-strand cDNA made from mRNA from leaves or roots of carrot plants with developing tap roots were used to amplify DNA fragments of about 330 bp, which were subcloned and sequenced. Two types of sequences were obtained with clear homologies to the sequences of plant Suc/H+ symporters and that share about 58% sequence identity at the DNA level. Most of the fragments amplified from leaf cDNA were of one type (leaf-specific PCR fragment), whereas the majority of fragments generated from root cDNA were of the other type (root-specific fragment). Subsequently, the two PCR fragments were used to isolate full-length cDNA clones from a leaf- and a root-specific cDNA library.
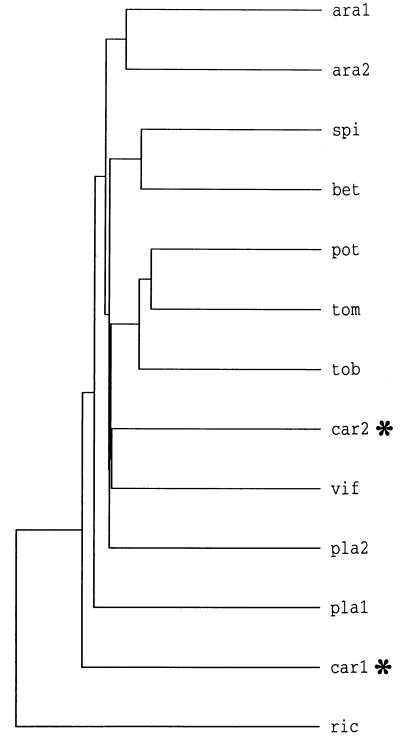
Figure 3.
Comparison of amino acid sequences of Suc/H+ symporters from dicotyledonous plants. The dendrogram was generated by comparison of the sequences by the PileUp program of the Genetics Computer Group sequence analysis software package. ara1, Arabidopsis (accession no. X75365); ara2, Arabidopsis (accession no. X75382); spi, spinach (accession no. X67125); bet, Beta vulgaris (accession no. X83850); pot, potato (accession no. X69165); tom, tomato (accession no. X82275); tob, tobacco (accession no. X82276); car2, DcSUT2 (accession no. Y16768); vif, fava bean (accession no. Z93774); pla2, P. major (accession no. X75764); pla1, P. major (accession no. X84379); car1, DcSUT1 (accession no. Y16766); and ric, castor bean (accession no. Z31561). The two carrot polypeptides are marked by asterisks. ara1 and pla1 have neutral pH optima, whereas all other transporters have maximal activities below pH 5.0.
Figure 1.
Comparison of the amino acid sequences derived from the cDNA clones DcSUT1 (bottom line) and DcSUT2 (top line). The amino acid sequences are in the one-letter code and have been aligned by introducing gaps (… ) to maximize identity. Vertical bars indicate identical amino acid residues, and the highlighted regions represent putative membrane-spanning domains. The two peptide sequences marked with asterisks were used for the generation of degenerated primers for the isolation of the cDNA clones.
Screening of the leaf library resulted in the isolation of two clones of different lengths (DcSUT1a, 1861 bp; DcSUT1b, 2132 bp). No differences were found when their 1503-bp-long open reading frames were compared. However, the two clones showed sequence deviations in the 5′ upstream noncoding sequences and were completely different in their 3′ noncoding domains. The two open reading frames code for a polypeptide of 54,066 D with a pI of 8.33. Only one type of cDNA clone was isolated from the root library (DcSUT2, 1858 bp). Its open reading frame of 1545 bp codes for a protein of 54,424 D with a pI of 8.85. The two polypeptide sequences are 52% identical (62.0% similar) and have 12 predicted membrane-spanning domains each (Fig. 1).
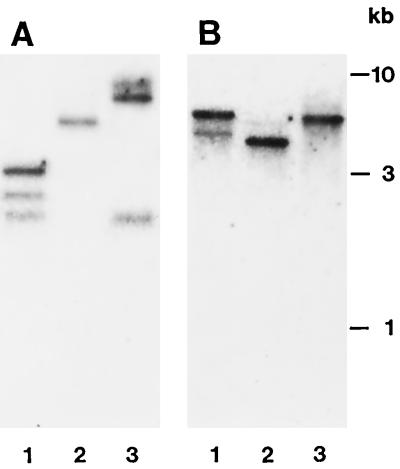
The identification of two closely related but different leaf-specific cDNA clones and only one root-specific clone fits very well with the results of DNA gel-blot analysis of the respective sequences in the carrot genome. The number of hybridization signals obtained with the leaf-specific PCR fragment as a probe (Fig. 2A) suggests the presence of at least two genes per carrot genome, whereas hybridization of a similar DNA gel blot with the root-specific fragment indicates the presence of only one gene.
Figure 2.
DNA gel-blot analysis of Suc/H+ symporter sequences in the carrot genome. Genomic DNA (10 μg/lane) was digested with HindII (lanes 1), HindIII (lanes 2), or PstI (lanes 3). The blot was hybridized with a 32P-labeled PCR fragment of DcSUT1 (A) or DcSUT2 (B), neither of which contained cleavage sites for the restriction enzymes used.
The amino acid sequences deduced from the leaf- and root-specific cDNA clones and the published amino acid sequences of Suc/H+ symporters from dicotyledonous plants were used to generate the phylogenetic tree shown in Figure 3. The 13 sequences appear to be divided into several subclasses with a very close relationship between DcSUT2 (car2) and the Suc/H+ symporter from fava bean (vif). In contrast, the sequence of DcSUT1 (car1) appears to be fairly different from the sequences of plant Suc/H+ symporters.
Characterization of DcSUT1 and DcSUT2 in S. cerevisiae
To determine whether the carrot cDNA clones code for Suc/H+ symporters, they were expressed in the S. cerevisiae strain SUSY7 (Riesmeier et al., 1992), which lacks the invertase gene (SUC2) and contains a functional plant gene for Suc synthase for its expression in the cytoplasm. These yeast cells can only live on Suc as the sole carbon source when they also express a functional Suc transporter gene.
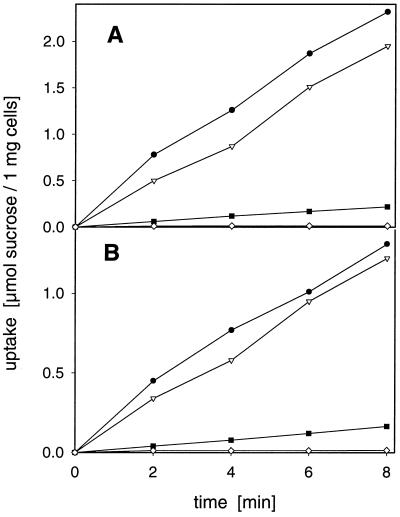
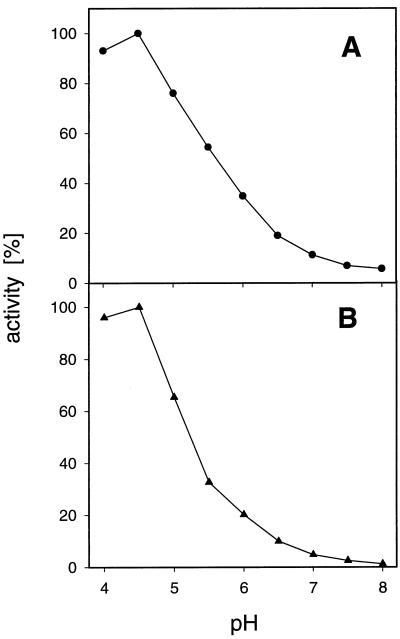
DNA fragments containing the open reading frames of DcSUT1 or DcSUT2 were cloned into the E. coli/S. cerevisiae shuttle vector 112 A1NE (Riesmeier et al., 1992), containing the ADH promoter in front of a multiple cloning site and the ADH terminator. Both carrot cDNA clones allowed the growth of SUSY7 cells on Suc-containing agar plates. Uptake studies with radiolabeled Suc showed up to a 200-fold accumulation of the disaccharide compared with yeast cells containing only the vector or the clones in the antisense orientation (Fig. 4). Suc uptake catalyzed by both carrot transporters was optimal below pH 5.0 and was less than 10% of optimal activity at pH 7.0 (Fig. 5). Low concentrations of the proton uncoupler CCCP markedly inhibited Suc uptake (Fig. 4), indicating that Suc transport is linked to the proton electrochemical potential across the plasma membrane. The Km of DcSUT1 for Suc at pH 5.5 was 0.5 mm and that of DcSUT2 was 0.7 mm (data not shown). Both transporter activities were stimulated by Glc, which is in agreement with the results obtained by Riesmeier et al. (1992) and Sauer and Stolz (1994).
Figure 4.
Uptake of Suc in transgenic S. cerevisiae cells. Uptake of [14C]Suc (0.2 mm final concentration) in yeast cells transformed with DcSUT1 (A) or DcSUT2 (B) in the presence (•) or absence (▾) of 10 mm d-Glc, or the presence of 5 μm of the protonophore CCCP (▪). ⋄, Suc uptake into control cells transformed with DcSUT1 or DcSUT2 in the antisense orientation.
Figure 5.
pH dependence of Suc transport in transgenic S. cerevisiae cells expressing either DcSUT1 (•, A) or DcSUT2 (▴, B). Measurements were performed at 0.2 mm Suc and 10 mm Glc in 25 mm sodium phosphate buffer between pH 4.0 and 8.0.
Tissue-Specific Expression of DcSUT1 and DcSUT2 mRNA
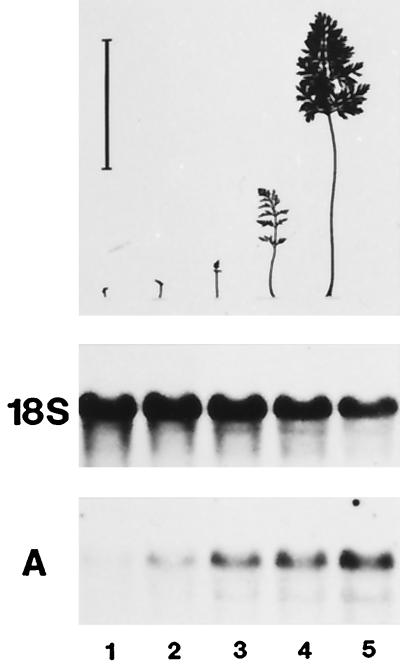
The development of leaves from leaf primordia to the mature organ is characterized by a gradual transition from net importers of photoassimilates (sink leaves) to net exporters (source leaves) (Turgeon, 1989). To determine when DcSUT1 and DcSUT2 are expressed during carrot leaf development, leaf lamina from approximately 12-week-old carrot plants were harvested and separated according to their size into five classes (Fig. 6, top). Total RNA was isolated and analyzed on RNA gel blots with the leaf- or root-specific PCR fragment (Fig. 6A; only the blot probed with the fragment of DcSUT1 is shown). Because the probes are fragments of the open reading frames, a distinction between transcripts of the two highly related leaf clones DcSUT1a and DcSUT1b was not possible. Instead, the common expression pattern of DcSUT1 was detected. The highest transcript levels for DcSUT1 were found in the lamina size classes 4 and 5, which are thought to be source leaves. DcSUT2 mRNA levels were very low but detectable in all five tissue samples (data not shown).
Figure 6.
Steady-state mRNA levels of DcSUT1 in lamina of different size classes of carrot leaves. Leaf lamina from 12-week-old carrot plants were separated into five size classes as shown in the schematic representation (upper panel). The bar above the smallest leaf (class 1) indicates a length of 10 cm. A northern blot with total RNA (10 μg/lane) was hybridized with the 32P-labeled probe for DcSUT1 (A). As a probe for a constitutively expressed gene, a fragment of the 18S rRNA gene from tomato was used (middle).
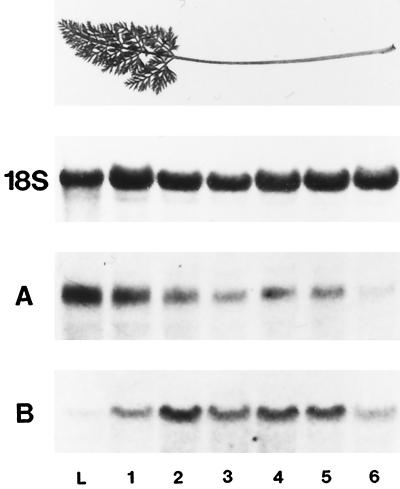
Source leaves, which had not yet reached their final size (between size classes 4 and 5), were dissected into lamina and six consecutive stem segments (Fig. 7, top). Total RNA from these tissues was analyzed on RNA gel blots. The highest levels of DcSUT1 transcripts were found in the leaf lamina (Fig. 7A), indicating a function of the Suc/H+ symporter in loading of Suc into the phloem. Lower transcript levels were found in the stem, with a decreasing gradient toward the stem base. In contrast, only very low mRNA levels of DcSUT2 were found in the leaf lamina, indicating that the encoded polypeptide is functionally different from DcSUT1. High transcript levels were found in the central stem segments, and lower levels at the two stem ends (Fig. 7B).
Figure 7.
Steady-state mRNA levels of DcSUT1 and DcSUT2 in leaf lamina and sections of the stem. Top, Leaves of about 12-week-old plants that had not yet reached their mature size were dissected into lamina and six consecutive stem sections. A, A northern blot with total RNA (10 μg/lane) from leaf lamina (L) and stem sections (sections 1–6 from top to bottom; section 1 was a highly branched stem segment connecting the numerous leaf lamina) was hybridized with a 32P-labeled probe for DcSUT1. B, A blot identical to that in A was hybridized with a 32P-labeled probe for DcSUT2. As a probe for a constitutively expressed gene, a fragment of the 18S rRNA gene from tomato was used.
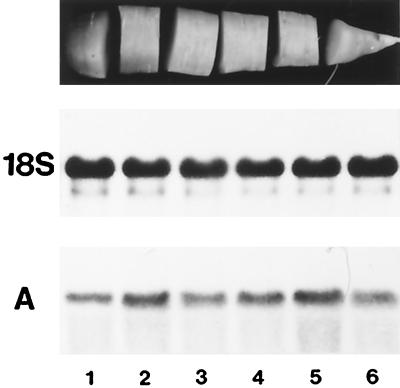
Developing tap roots were also divided into zones with different biological activities. The root elongated and thickened at the tip (Fig. 8, top, segment 6) (utilization sink for Suc) and storage of sugars occurred in the upper part (segments 1–5) (storage sink for Suc). These upper segments also increased in diameter (secondary root growth) but the increase was small compared with the rate of root elongation at the root tip. Transcripts for DcSUT1 were barely detectable (data not shown in this figure, but the lack of expression of DcSUT1 in roots can be seen in Fig. 11D). High and approximately equal levels of mRNA for DcSUT2 were found in all root sections (Fig. 8A).
Figure 8.
Steady-state mRNA levels of DcSUT2 in developing tap roots. Twelve-week-old carrot plants were harvested in the middle of a sunny, warm day. The tap roots were washed and divided into six equal, consecutive sections (top). Bottom, A northern blot with total RNA (10 μg/lane) from the six root sections (1–6) was hybridized with the 32P-labeled probe for DcSUT2 (A). As a probe for a constitutively expressed gene, a fragment of the 18S rRNA gene from tomato was used (middle).
Figure 11.
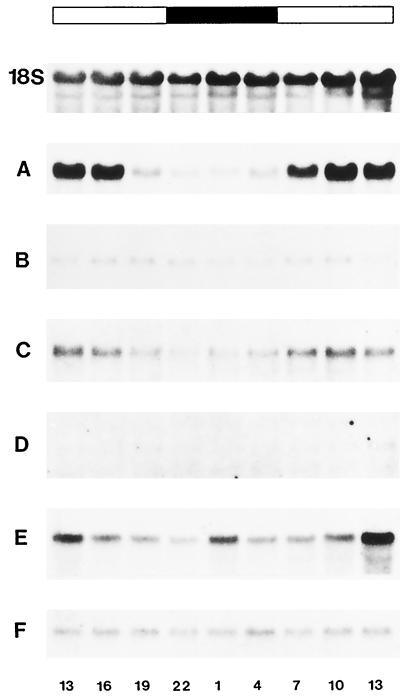
Light-dependent expression of DcSUT1 and DcSUT2 mRNA. Carrot plants (12 weeks old) were harvested at 3-h intervals between 1 pm of d 1 and 1 pm of d 2. Total RNA (10 μg/lane) from entire leaves (leaf lamina and stems) (A, C, and E) and the middle third of tap roots (excluding crown and tip) (B, D, and F) were hybridized on northern blots with a 32P-labeled probe for the carrot chlorophyll a/b-binding protein (A and B), DcSUT1 (C and D), and DcSUT2 (E and F). As a probe for a constitutively expressed gene, a fragment of the 18S rRNA gene from tomato was used (only the blot for leaf RNA is shown). The bar at the top shows the light conditions at the time of organ harvest (white bar, daylight; black bar, darkness). The numbers below the RNA gel blots indicate time of organ harvest (13 = 1 pm; 1 = 1 am).
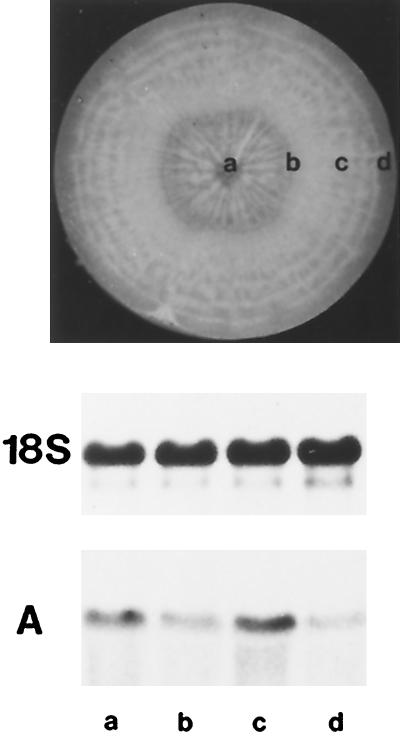
To determine whether DcSUT1 and DcSUT2 are tissue-specifically expressed in tap roots, 12-week-old roots were separated into xylem (a), cambium plus a few millimeters of phloem tissue (the phloem-containing tissue; Sturm et al., 1998) (b), phloem (c), and periderm (d) (Fig. 9, top). Transcripts for DcSUT1 were barely detectable, whereas mRNA for DcSUT2 was found in all four tissues, with the highest levels in phloem and xylem parenchyma tissues (Fig. 9A).
Figure 9.
Steady-state mRNA levels of DcSUT2 in developing tap roots. Top, The middle third sections of tap roots of 12-week-old carrot plants (excluding crown and tip) were dissected into xylem (a), cambium plus a few millimeters of adjacent phloem (b), the remaining phloem (c), and periderm (d). Bottom, A northern blot with total RNA (10 μg/lane) was hybridized with the 32P-labeled probe for DcSUT2 (A). As a probe for a constitutively expressed gene, a fragment of the 18S rRNA gene from tomato was used (middle).
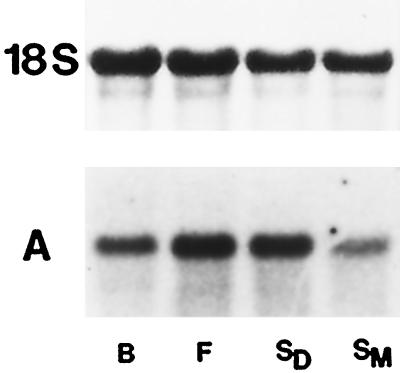
At different developmental stages of carrot reproductive organs only low levels of DcSUT1 transcripts were detected (data not shown). In contrast, DcSUT2 mRNA was found at all times during organ development, with the highest levels in flowers and developing seeds (Fig. 10A).
Figure 10.
Steady-state mRNA levels of DcSUT2 in reproductive organs of carrot plants. A northern blot with total RNA (10 μg/lane) from flower buds (lane B), flowers (lane F), developing seeds (lane SD), and mature seeds (lane SM) was hybridized with the 32P-labeled probe for DcSUT2 (A). As a probe for a constitutively expressed gene, a fragment of the 18S rRNA gene from tomato was used (top).
Light-Dependent Expression of DcSUT1 and DcSUT2 mRNA
To determine whether the expression of the carrot Suc/H+ symporters is coordinated with the diurnal changes in photosynthesis, steady-state mRNA levels for DcSUT1 and DcSUT2 were analyzed at 3-h intervals for 24 h (Fig. 11). As a positive control, RNA isolated from entire leaves (leaf lamina and stems) was probed with a PCR fragment of the cDNA for the chlorophyll a/b-binding (cab) protein (approximately 80% identical with the petunia gene; accession no. X04966). As expected, high transcript levels were found during the day and only low levels were found during the night (A). Low expression of the cab gene was also detected in roots but no diurnal regulation was seen (B). Like the cab gene in leaves, the levels of DcSUT1 transcripts were light dependent, with higher levels during the day and low levels during the night (C). In contrast, roots had only barely detectable levels of DcSUT1 mRNA and no diurnal concentration alterations (D). In leaves (in stems and not in lamina; see Fig. 7), DcSUT2 transcript levels were highest at midday and lower during the night (E). High, constant levels were found in roots throughout the 24-h time course (F).
DISCUSSION
The deposition of storage reserves in plant storage organs is intimately linked to photosynthesis and long-distance transport of photoassimilates such as Suc. In most plants, Suc transport from its place of biosynthesis in source leaves to the sink organs is accompanied by the passage of the disaccharide through several membranes. In general, it has been shown that this passage is facilitated by cargo-specific, energy-dependent transport proteins (Giaquinta, 1977; Bush, 1993; Sauer et al., 1994). In the present study, two Suc transporters were isolated from carrot, a plant that accumulates high levels of soluble sugars in parenchyma cells of a tap root. The two polypeptides have significant homology to previously described plant Suc carriers (Riesmeier et al., 1992, 1993; Sauer and Stolz, 1994). All belong to the large family of major facilitator proteins (Marger and Saier, 1993), with 12 membrane-spanning domains and a molecular mass of about 54 kD. Yeast cells expressing the carrot cDNA clones imported Suc efficiently and accumulated it at least 200-fold more than cells containing only the vector or the clones in the antisense orientation. The yeast-expressed transporters had optimal activities in the acidic pH range and Km values for Suc of about 0.5 mm. Protonophores such as CCCP inhibited Suc completely, strongly suggesting that the two polypeptides transport the disaccharide together with protons.
The amino acid sequences of DcSUT1 and DcSUT2 were only 52% identical and most sequence conservation was found in the transmembrane domains. In a phylogenetic tree generated from the deduced sequences of the two carrot proteins and other plant Suc/H+ symporters, transporters with similar biochemical characteristics were not grouped together (e.g. transporters with a neutral pH optimum). Because of this lack of correlation between amino acid sequences and transporter properties, it is not possible to make property predictions based on the position in the dendrogram.
One of the carrot transporters was predominantly expressed in source cells (DcSUT1) and the other in different sink tissues (DcSUT2), suggesting different biological functions. Analysis of steady-state transcript levels indicated that the expression of DcSUT1 is linked to photosynthesis and the provision of the plant with assimilated carbon. Like the Suc/H+ symporters of Arabidopsis (AtSUC2) (Sauer and Stolz, 1994; Truernit and Sauer, 1995) and potato (StSUT1) (Riesmeier et al., 1993), DcSUT1 may be involved in loading of Suc into the phloem of source leaves and retrieval of escaped Suc in the stem. Transport activity appears to be regulated in a diurnal fashion (Kühn et al., 1997) and, as a consequence, we speculate that transport rates may be high during the day and low during the night. This hypothesis fits very well with our recent finding of transient accumulation of starch in tap roots (Sturm et al., 1998). Our data suggest that during the day some of the Suc made in leaves is directly transported into vacuoles of storage parenchyma cells without intermediate cleavage by a cell wall invertase. Any excess is converted into transitory starch in tap root cells surrounding the phloem. During the night, the flow of Suc into storage cells is kept constant by the reconversion of starch into Suc.
High steady-state transcript levels of DcSUT2 were found only in sink organs such as the growing stems, developing tap roots, flowers, and developing seeds. In tap roots expression was clearly not restricted to the phloem. However, higher mRNA levels were found in phloem and xylem storage parenchyma tissues (Fig. 9). The amino acid sequence of DcSUT2 is closely related to that of a Suc/H+ symporter of fava bean (Weber et al., 1997), suggesting that these polypeptides may have a common function. VfSUT1 is highly expressed in transfer cells of maturing seeds, providing the developing cotyledons with a source of carbon and energy. Likewise, DcSUT2 may import Suc into storage parenchyma cells.
One of the questions still completely open is how Suc is unloaded from the phloem in carrot sink organs. Simple thermodynamics and knowledge of plant cell bioenergetics suggest that the proton-motive-force-driven symporters are never involved in efflux reactions. Thus, either an as-yet-unidentified transporter facilitates the exit of Suc from the phloem or the disaccharide may passively leak out into the apoplast because of the high concentration of Suc.
ACKNOWLEDGMENTS
We thank Wolf Frommer (University of Tübingen, Germany) for the yeast expression vector 112 A1NE, the yeast strain SUSY7, and helpful comments on the Suc uptake studies. We also acknowledge our colleagues Thomas Boller and Pat King for critical reading of the manuscript.
Abbreviation:
- CCCP
carbonylcyanide-m-chlorophenylhydrazone
Footnotes
LITERATURE CITED
- Avigad G. Sucrose and other disaccharides. In: Loewus FA, Tanner W, editors. Encyclopedia of Plant Physiology, Vol 13A. Berlin: Springer-Verlag; 1982. pp. 217–347. [Google Scholar]
- Bush DR, Chiou T-J, Chen L. Molecular analysis of plant sugar and amino acid transporters. J Exp Bot. 1996;47:1205–1210. doi: 10.1093/jxb/47.Special_Issue.1205. [DOI] [PubMed] [Google Scholar]
- Bush RH. Proton-coupled sugar and amino acid transporters in plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:513–542. [Google Scholar]
- Cirillo VP. Sugar transport in normal and mutant yeast cells. Methods Enzymol. 1989;174:617–622. doi: 10.1016/0076-6879(89)74040-4. [DOI] [PubMed] [Google Scholar]
- Gahrtz M, Schmelzer E, Stolz J, Sauer N. Expression of the PmSUC1 sucrose carrier gene from Plantago major L. is induced during seed development. Plant J. 1996;9:93–100. doi: 10.1046/j.1365-313x.1996.09010093.x. [DOI] [PubMed] [Google Scholar]
- Gahrtz M, Stolz J, Sauer N. A phloem-specific sucrose-H+ symporter from Plantago major L. supports a model of apoplastic phloem loading. Plant J. 1994;6:697–706. doi: 10.1046/j.1365-313x.1994.6050697.x. [DOI] [PubMed] [Google Scholar]
- Giaquinta R. Possible role of pH gradient and membrane ATPase in the loading of sucrose into the sieve tubes. Nature. 1977;267:369–370. [Google Scholar]
- Gietz D, Jean AS, Woods RA, Schiestl RH. Improved method for the high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LC. Metabolism and compartmentation of imported sugars in sink organs in relation to sink strength. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:355–378. [Google Scholar]
- Kühn C, Fanceschi VR, Schultz A, Lemoine R, Frommer WB. Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science. 1997;275:1298–1300. doi: 10.1126/science.275.5304.1298. [DOI] [PubMed] [Google Scholar]
- Marger MD, Saier MH., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Prescott A, Martin C. A rapid method for the quantitative assessment of levels of specific mRNAs in plants. Plant Mol Biol Rep. 1987;4:219–224. [Google Scholar]
- Rentsch D, Boorer KJ, Frommer WB. Structure and function of plasma membrane amino acid, oligopeptide and sucrose transporters from higher plants. J Membr Biol. 1998;162:177–190. doi: 10.1007/s002329900355. [DOI] [PubMed] [Google Scholar]
- Riesmeier JW, Hirner B, Frommer WB. Potato sucrose transporter expression in minor veins indicates a role in phloem loading. Plant Cell. 1993;5:1591–1598. doi: 10.1105/tpc.5.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992;11:4705–4713. doi: 10.1002/j.1460-2075.1992.tb05575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 1994;13:1–7. doi: 10.1002/j.1460-2075.1994.tb06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sauer N, Baier K, Gahrtz M, Stadler R, Stolz J, Truernit E. Sugar transporters across the plasma membrane of higher plants. Plant Mol Biol. 1994;26:1671–1679. doi: 10.1007/BF00016496. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stolz J. SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana: expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- Schmidt-Puchta W, Kütemeier G, Günther I, Haas B, Sänger HL. Cloning and sequence analysis of the 18S ribosomal RNA gene of tomato and the secondary structure model for the 18S rRNA of angiosperms. Mol Gen Genet. 1989;219:17–25. doi: 10.1007/BF00261152. [DOI] [PubMed] [Google Scholar]
- Stadler R, Sauer N. The Arabidopsis thaliana ATSUC2 gene is specifically expressed in companion cells. Bot Acta. 1996;109:299–306. [Google Scholar]
- Sturm A. Molecular characterization and functional analysis of sucrose-cleaving enzymes in carrot (Daucus carota L.) J Exp Bot. 1996;47:1187–1192. doi: 10.1093/jxb/47.Special_Issue.1187. [DOI] [PubMed] [Google Scholar]
- Sturm A, Lienhard S, Schatt S, Hardegger M (1998) Tissue-specific expression of two genes for sucrose synthase in carrot (Daucus carota L.). Plant Mol Biol (in press) [DOI] [PubMed]
- Truernit E, Sauer N. The promoter of the Arabidopsis thaliana SUS2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- Turgeon R. The sink-source transition in leaves. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:119–138. [Google Scholar]
- Weber H, Borisjuk L, Heim U, Sauer N, Wobus U. A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in fava bean seeds. Plant Cell. 1997;9:895–908. doi: 10.1105/tpc.9.6.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig A, Komor E. An active sucrose carrier (Scr1) that is predominantly expressed in the seedlings of Ricinus communis L. J Plant Physiol. 1996;147:685–690. [Google Scholar]