Abstract
Whole exome/genome sequencing has been fundamental in the identification of somatic mutations in the spliceosome machinery in myelodysplastic syndromes (MDSs) and other hematologic disorders. SF3B1, splicing factor 3b subunit 1 is mutated in 60%-80% of refractory anemia with ring sideroblasts (RARS) and RARS associated with thrombocytosis (RARS-T), 2 distinct subtypes of MDS and MDS/myeloproliferative neoplasms (MDSs/MPNs). An idiosyncratic feature of RARS/RARS-T is the presence of abnormal sideroblasts characterized by iron overload in the mitochondria, called RS. Based on the high frequency of mutations of SF3B1 in RARS/RARS-T, we investigated the consequences of SF3B1 alterations. Ultrastructurally, SF3B1 mutants showed altered iron distribution characterized by coarse iron deposits compared with wild-type RARS patients by transmission electron microscopy. SF3B1 knockdown experiments in K562 cells resulted in down-regulation of U2-type intron-splicing by RT-PCR. RNA-sequencing analysis of SF3B1 mutants showed differentially used genes relevant in MDS pathogenesis, such as ASXL1, CBL, EZH, and RUNX families. A SF3B pharmacologic inhibitor, meayamycin, induced the formation of RS in healthy BM cells. Further, BM aspirates of Sf3b1 heterozygous knockout mice showed RS by Prussian blue. In conclusion, we report the first experimental evidence of the association between SF3B1 and RS phenotype. Our data suggest that SF3B1 haploinsufficiency leads to RS formation.
Introduction
Whole exome sequencing technology has been instrumental in the identification of somatic mutations involving genes important in RNA splicing.1–4 The first and most frequently identified mutation in hematologic malignancies involves the SF3B1 gene and was found in 60%-80% of refractory anemia with ring sideroblasts (RARS) and RARS associated with thrombocytosis (RARS-T) patients. Both RARS and RARS-T are subtypes of myelodysplastic syndromes (MDSs) and MDS/myeloproliferative neoplasm (MDS/MPN) overlap diseases. SF3B1 is a crucial component of the spliceosomal U2snRNP complex, which participates in normal RNA splicing.5 Intron splicing that culminates in the production of the mature mRNA product is catalyzed by spliceosomes. The major spliceosomes consist of U1, U2, U4/U6, and U5snRNPs and carry out splicing of major class or U2-type introns. Minor spliceosomes, which consist of U11, U12snRNPs, U4atac/U6atac and U5 snRNPs, carry out the splicing of minor class introns or U12-type introns. RNA splicing is crucial in maintaining normal genetic diversity.6 Because the fidelity of the final protein product depends highly on an intact and accurate RNA splicing machinery, somatic genetic errors in key components of the splicing pathway can potentially lead to the formation of dysfunctional proteins that can ultimately predispose to diseases. Indeed, emerging discoveries of genetic polymorphisms or mutations altering regulatory sequences or producing splice variants have been associated with certain types of hereditary diseases and neoplastic disorders.7
The presence of 15% or more ring sideroblasts (RSs) of nucleated erythroid precursors in the BM is a cardinal feature of RARS and RARS-T, even though RSs are also seen in 25% of patients with other subtypes of MDS, but rarely in other hematologic disorders.8–10 RS occurs because of abnormal localization of crystalline iron deposits in the mitochondria of erythoid precursors. Even though there is evidence of dysfunction in mitochondrial metabolism, the pathogenesis of RS remains unclear.11,12 The discovery of SF3B1 mutations in RARS and RARS-T has opened a new area of investigation in these diseases, and this article focuses on experimental evidence linking dysfunction in SF3B1 and formation of RS in RARS/RARS-T.
Methods
Patient samples
BM and peripheral blood cells were obtained with written informed consent in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the Cleveland Clinic. Diagnosis was assigned according to 2008 World Health Organization classification criteria. We studied a total of 456 patients. Overall, the MDS group includes: RARS, n = 19; refractory anemia with excess blasts 1/2, n = 34; refractory cytopenia with unilineage dysplasia/refractory cytopenia with multilineage dysplasia, n = 23; and MDS-U/5q, n = 17. The MDS/MPN group includes: RARS-T, n = 26; chronic myelomonocytic leukemia 1/2, n = 59; and MDS/MPN-U, n = 20. The acute myeloid leukemia (AML) group includes: secondary AML, n = 51; and primary AML, n = 43. Others includes: MPN, n = 69; mast cell disease, n = 31; and paroxysmal nocturnal hemoglobinuria/aplastic anemia/T-cell large granular lymphocytic leukemia, n = 64. The median age at sample collection was 52 years (range, 13-77 years).
Mutational detection
Genomic sequencing was performed on coding regions for SF3B1 (exons 13-16) SF3B14 (all exons), SF3B4 (all exons), and DYRK1A (all exons). Primer sequences and conditions used can be supplied on request. Bidirectional sequencing was performed by standard techniques using an ABI 3730xl DNA analyzer (Applied Biosystems). Somatic nature of novel mutations was confirmed using germline DNA from CD3+ cells. All mutations were scored as pathogenic on the basis of the observation that they were not detected in normal samples and were not found in published SNP databases (dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP) and/or they were not reported as SNPs in previous publications.
Transmission electron microscopy
One million BM mononuclear cells were first subjected to fixation in 3.75% glutaraldehyde, 0.1M sodium cacodylate, 6% sucrose (pH 7.2-7.4) for a period of 4 hours to overnight. Specimens were post-fixed in 1% osmium tetroxide and 0.1M cacodylate (pH 7.2-7.4) for 1 hour. After each fixation step, specimens were washed twice in cacodylate buffered sucrose, 10 minutes each. After the final wash, specimens were dehydrated in ascending graded alcohols, followed by immersion in propylene oxide, then passed through ascending graded mixtures of propylene oxide and EMBed 812 epoxy resin, and finally embedded in pure epoxy resin. Specimen blocks were polymerized overnight at 60°C. Specimens were initially evaluated by light microscopy of 1-μm plastic sections stained in a combination of toluidine blue and basic fuchsin. Thin sections from selected tissue blocks were cut at 60-80 nm and supported on 200 mesh copper grids. Sections were stained in uranyl acetate and lead citrate and examined using a Philips CM-12 electron microscope. Electron micrographs were taken using a Gatan Orius CCD digital camera.
Lentiviral infection and quantitative RT-PCR
Virus containing supernatant of 293T cells was used to infect 5 × 105 K562 cells. Five different shRNAs were constructed by Sigma-Aldrich. To assess the effect of shRNAs, cells were harvested 2 days after puromycin selection and SF3B1 mRNA level was measured by RT-PCR. Total RNA was isolated using the High Pure RNA Isolation kit (Roche Diagnostics). For quantitative RT-PCR, 0.5 μL of the complementary DNA was added to SYBR@advantage qPCR mix (Clontech Laboratories) and amplified in a Light Cycler 480 instrument (Roche Diagnostics). The U2- and U12-dependent introns were selected from the online database (http://genome.crg.es/datasets). The RT-PCR primers spanning the exon-intron junctions of several genes were designed using PrimerQuest (www.idtdna.com). And their specificity was assessed in silico (http://genome.ucsc.edu) and by standard PCR. A total of 40-50 PCR cycles were performed in a 2-step cycling procedure with an initial denaturation at 94°C for 3 minutes, subsequently at 94°C for 15 seconds, and 60°C for 30 seconds. Data were analyzed based on relative δ Ct values using GAPDH as control gene. Results are shown as mean ± SD of duplicate experiments.
Human colony formation assays
A total of 100 000 BM cells derived from 4 healthy persons were plated in duplicate in 1 mL methylcellulose with cytokines (StemCell Technologies) in 35-mm culture plates for 2 weeks at 37°C with 5% CO2. Meayamycin was added at the beginning of the culture at a range of concentrations (1, 2, 5, 10, and 50nM). Colony-forming cells were harvested, washed 5 times in PBS, and spotted on cytospin slides before Prussian blue staining.
Preparation of meayamycin
Meayamycin was synthesized according to the literature13 and purified on a C18 reverse-phase preparative HPLC. The fractions containing meayamycin were concentrated under vacuum to afford 16.3 mg of solid meayamycin. Using this solid, a 10mM stock solution of meayamycin in DMSO was prepared. A 10μM solution of meayamycin in DMSO was prepared through serial dilutions and stored at a −20°C.
Mice
BM aspirates from Sf3b1 heterozygous14 and wild-type (WT) C57BL/6 mice were kindly provided by Dr Koseki (RIKEN) and were subjected to Prussian blue staining using standard pathology procedures for the detection of RS.
RNA-Seq analysis
Mapping.
RNA sequencing (RNA-Seq) was performed by Otogenetics Corporation (Tucker); 100-bp paired-end RNA-Seq reads were mapped to the hg19 RefSeq human transcriptome and spliceosome by DNAnexus (http://dnanexus.com) using a Bayesian method,15 in which a read was mapped when its posterior probability of mapping exceeded 0.9. These filtered posterior probabilities were summed to generate fractional read counts per gene and per exon, with probabilities from splice-junction spanning reads counted for each relevant exon. We used rounded gene and exon read counts as inputs for our differential expression analyses.
Differential gene expression analysis.
We used the R package DESeq16 version 1.6.1 (http://www.bioconductor.org/packages/release/bioc/html/DESeq.html) and R version 2.14.1, to perform differential gene expression analysis of mutant versus normal samples. DESeq uses a negative binomial (NB) distribution to model the gene read counts and shrinkage estimators to estimate the per-gene NB dispersion (square of the coefficient of biologic variation) parameters. Specifically, we estimated the per-gene dispersion parameters using a shrinkage approach with the default maximum sharing mode and default parametric mean-dispersion relationship, where raw dispersion values were calculated using the samples in the mutant group. Before testing, we dropped all genes with read counts below the 40th percentile to improve testing power while maintaining type I error rates. We used the nbinomTest function, which does conditional testing assuming gene counts follow an NB distribution with fitted dispersions, to estimate P values and adjusted P values obtained using the Benjamini-Hochberg method17 for each filtered gene under the null hypothesis of common expression intensity across groups. Genes with adjusted P < .05 were declared significant. Moderated logarithm base 2-fold changes (mutant/normal) were estimated using a variance stabilizing transformation as implemented in the getVarianceStabilizedData function of DESeq.
Differential exon usage analysis.
We used the R package DEXSeq Version 1.1.4 (http://www.bioconductor.org/packages/release/bioc/html/DEXSeq.html), to perform differential exon usage analysis of mutant versus normal samples. DEXSeq uses an NB distribution to model the exon read counts and shrinkage estimators to estimate the per-exon NB dispersion parameters. Specifically, we estimated the per-exon dispersion parameters using a shrinkage approach with the maximum sharing mode and parametric mean-dispersion relationship, where raw dispersion values were calculated using the samples in the mutant group. We defined a testable exon as one that had a total sum of at least 10 mapped reads across samples and was in a gene with no more than 70 exons. Before exon usage testing, we dropped any exons that were not testable or were in genes with less than 2 testable exons to improve testing power while maintaining type I error rates. We used the testGeneForDEU function, which compares deviances from generalized linear model fits (assuming NB likelihood) to a χ2 reference distribution, to estimate P values and adjusted P values obtained using the Benjamini-Hochberg method for each exon under the null hypothesis of common usage across groups. Exons with adjusted P < .05 were declared significant. Logarithm base 2-fold changes (mutant/normal) for each exon were estimated using the function estimatelog2FoldChanges.
Gene set and exon set ORA.
We used the R package goseq18 Version 1.6.0 (http://www.bioconductor.org/packages/release/bioc/html/goseq.html) to perform over- and under-representation analysis (ORA) of gene sets taking length bias into account. We conducted separate ORAs for the gene-level and exon-level differential expression (DE) findings. ORA analysis requires a definition of significance for each gene member of the gene set. We defined this similarly for ORA of the gene-level and exon-level DE findings: (1) a gene with an adjusted P value of < .5 from the gene-level DE analysis and (2) a gene with at least one exon with an adjusted P value from the exon-level DE analysis < .5, respectively. Goseq requires a probability weighting function, which is used to adjust the gene set results to reflect the lengths of genes in the gene sets, reducing the common bias in gene set analyses because of longer genes having higher read counts and subsequently a higher chance of differential detection. For ORA of the gene-level DE findings, we defined length as the sum of the lengths of the observed exons within each gene, where the exon lengths were obtained from DNAnexus. For ORA of the exon-level DE findings, we defined length as the median exon length of detected exons within the specified gene, where exon lengths were obtained from DNAnexus. We used the MSigDB database19 Version 3.0 (http://www.broadinstitute.org/gsea/msigdb/index.jsp), gene set collections c2 through c5 in our ORA. For each MSigDB collection of gene sets, we ran the goseq function with the Wallenius approximation to estimate P values for over-representation and under-representation of significant genes (where significant is defined earlier in this paragraph) for each gene set within the collection. Adjusted P values were calculated on the gene-set P values per collection using the Benjamini-Hochberg method to control for the number of gene sets tested within a collection. These adjustments were made separately for the P values from the over-represented tests and for the P values from the under-represented tests. Any gene sets with adjusted P values < .05 from either the over- or under-represented tests were declared significant.
Statistical analyses
Overall survival was measured from the day of initial sampling to death from any cause (patients lost to follow-up were censored) or last follow-up and was summarized using the Kaplan-Meier method. Results were analyzed for data collected as of December 2011. All P values were 2-sided, and P values ≤ .05 indicated statistical significance. Statistical analyses were performed using JMP8 (SAS Inc). RNA-Seq data statistical analysis is discussed extensively in “RNA-Seq analysis.”
Results
SF3B1 mutations are frequently found in patients with MDS and RS
We first examined SF3B1 mutational status in our cohort of MDS and other hematologic malignancies (n = 456) using Sanger sequencing. Consistent with earlier studies,1–3 we found 50 cases with somatic mutations in SF3B1; 68% of RARS and 81% of patients with RARS-T harbored mutations, whereas SF3B1 mutations were infrequently found in higher-risk MDS and other hematologic conditions (supplemental Figure 1A-D, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The K700E was the most frequent mutation detected in SF3B1 among patients with RARS and RARS-T (34%). Further, SF3B1 did not predict for progression as underlined by the low prevalence of mutations in patients with secondary AML (5.9%) and de novo AML (4.7%; supplemental Figure 1E). Analysis of overall survival comparing mutant versus WT patients showed longer survival in patients with SF3B1 mutations (supplemental Figure 3). Interestingly, we next found 2 non-MDS cases in which SF3B1 mutations were detected: one with paroxysmal nocturnal hemoglobinuria (K666Q)20 and one with post-polycythemic myelofibrosis (K700E).1 Both displayed RS (17% and rare, respectively). This finding led us to investigate more closely the association of SF3B1 mutations and RS. In conclusion, SF3B1 is mainly associated with RARS and RARS-T MDS and MDS/MPN subtypes. In a total of 45 patients, 11 typical RARS and RARS-T patients with more than 15% RS were WTs for SF3B1. Thus, we sought to investigate whether SF3B1 might accompany distinctive features.
Ultrastructural analysis of BM of SF3B1 mutant and WT RARS patients
Given the high frequency of SF3B1 mutations in patients with RARS,1–3,21 we then investigated the association of spliceosomal protein mutations with altered mitochondrial iron distribution manifested as RS. We analyzed ultrastructural differences of BM erythroid precursors between SF3B1 mutant and WT patients with the intent of evaluating whether SF3B1 mutations might alter iron distribution. Transmission electron microscopy was performed on BM cells derived from an RARS patient carrying an SF3B1 mutation (K700E) and an RARS patient WT for SF3B1. Abundant foci of iron deposits were observed around the nucleus of BM cells in the mutant case compared with scanty deposits in the WT patient. Although Prussian blue staining is traditionally used to identify RS in BM samples, it does not distinguish between SF3B1 mutant and WT cases (Figure 1 top right panel). The SF3B1 mutations are associated with more dense iron particles. It is possible that the topographic configuration of the iron might predict the presence of SF3B1 mutations.
Figure 1.
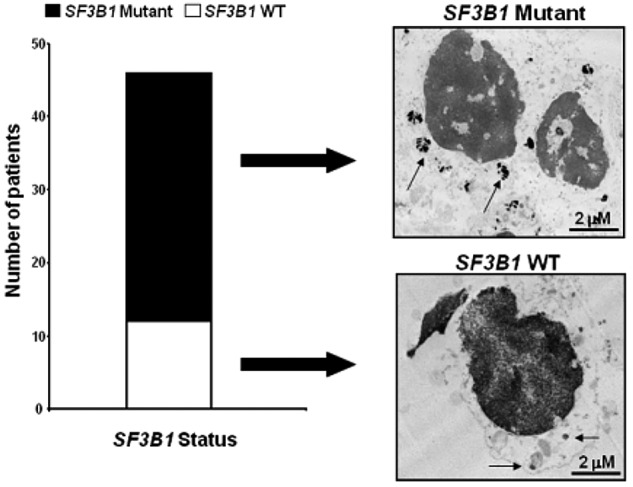
Mutations in SF3B1 are frequent in patients with RARS and RARS-T and lead to an alteration in the BM ultrastructure. Left panel: We performed Sanger sequencing on exons 13, 14, 15, and 16 of SF3B1 in 456 patients, finding that the frequency of SF3B1 mutations is higher in patients with RARS (68%) and RARS-T (81%) compared with the other groups. White bars and black bars represent WT and mutant RARS and RARS-T patients, respectively. Right panel: Transmission electron microscopy of BM cells from a representative WT and mutant RARS patient. Arrows indicate the presence of abundant perinuclear iron deposits in the mutant compared with the WT patient. Specimens' cuts are 2-μm-thick sections.
In vitro effects of SF3B1 knockdown
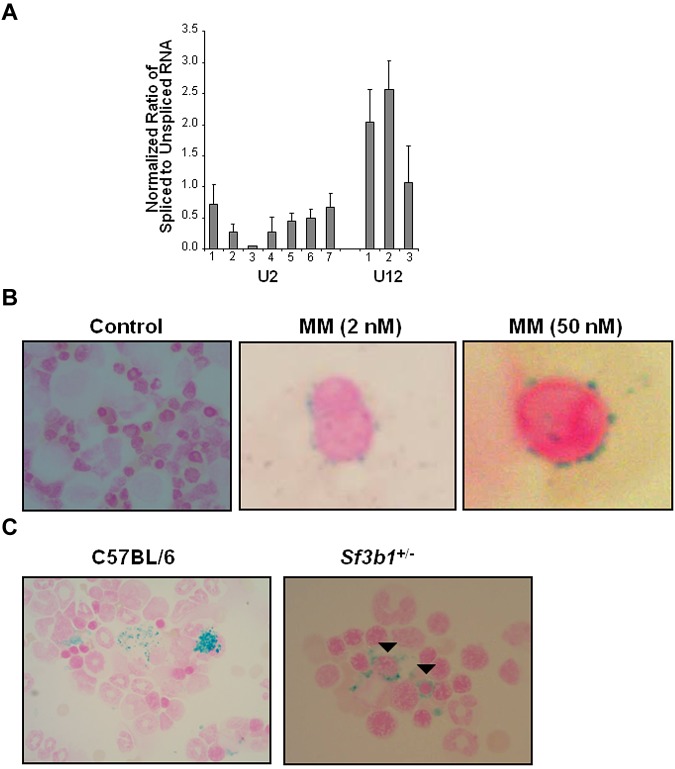
We then investigated the role of SF3B1 in pre-mRNA splicing of endogenous human genes. First we knocked down the level of SF3B1 mRNA in cells using RNAi. We applied shRNA lentiviral infection in K562 cells using 5 different shRNAs specific for the human SF3B1 gene. We performed quantitative RT-PCR to measure the level of knockdown. SF3B1 mRNA knockdown of 50% was consistently achieved on one shRNA clone. Using this selected shRNA, we quantified the ratio of spliced and unspliced RNA in 7 and 3 exemplary U2- and U12-introns. All examined U2-type introns were less efficiently spliced, whereas U12-introns were not significantly affected (Figure 2A; Table 1). We then analyzed the phenotype of K562 cells with decreased expression of SF3B1 mRNA. K562 cells were first cocultured with hemin, an inducer of hemoglobinization and erythroid differentiation, and then infected with different shRNAs specific for SF3B1. A 50% reduction of SF3B1 mRNA level was again achieved. Transfectant cells were sent for morphologic analysis and Prussian blue staining. Increased mitotic figures were noted, but RSs were not observed. Transmission electron microscopy of the transfectant cells also showed no iron deposits compared with controls (ie, K562 cells infected with a vector only; data not shown).
Figure 2.
In vitro and in vivo alterations of SF3B1 gene lead to an RARS phenotype. (A) K562 cells were transfected with shRNA constructs. After 48 hours of puromycin selection, RT-PCR assay was performed using random oligos for endogenous U2 (1-7) and U12 (1-3) introns (Table 1). The ratio of spliced to unspliced pre-mRNA for U2- and U12-dependent introns is shown. Average reaches 0.42 and 1.89 for U2-introns and U12-introns, respectively. Data are mean ± SD, calculated from 2 independent experiments. K562 transfected with vector only was used as control, and the ratio of spliced to unspliced pre-mRNA was set to 1. (B) Human colony-forming unit cell assay was performed on BM cells derived from 4 healthy persons. Cells were treated with different doses of meayamycin (2, 10, and 50nM). Representative pictures at 2 and 50nM are shown. Colonies were harvested after 2 weeks, spotted on cytospin slides, and subjected to Prussian blue staining. (C) Prussian blue staining of BM aspirates shows numerous RSs in Sf3b1+/− compared with C57BL/6 mice. Closed arrowheads indicate perinuclear RS. BM cytospin slides were kindly provided by Dr H. Koseki from Japan. Slides were analyzed using an Olympus system microscope (model BX41; objective lens, ×100; camera, SPOT Idea Model No 28.2). Images were acquired using a SPOT Imaging software by Diagnostic Instruments Inc (www.Diaginc.com).
Table 1.
U2- and U12-dependent genes and introns
| No. | Gene name | Introns |
|---|---|---|
| U2 introns | ||
| 1 | CTNNBL1 | 2 |
| 2 | CTNNBL1 | 3 |
| 3 | EFNA5 | 2 |
| 4 | ITPR1 | 3 |
| 5 | ITPR1 | 4 |
| 6 | UTR | 43 |
| 7 | THOC2 | 36 |
| U12 introns | ||
| 1 | ATXN | 10 |
| 2 | CTNNBL1 | 4 |
| 3 | THOC2 | 37 |
U2- and U12-intron numbers are indicated for each gene. Primers were designed to quantify the levels of spliced and unspliced introns in K562 transfected with shRNA constructs.
CTNNBL1 indicates catenin, β-like 1; EFNA5, ephrin-A5; ITPR1, inositol 1,4,5-trisphosphate receptor 1; UTR, untranslated region; THOC2, THO complex 2; and ATXN, ataxin-1.
In vitro treatment of healthy BM cells with meayamycin
To further explore the relationship between SF3B1 mutations and RS, we used meayamycin, a chemical analog of FR901464.13,22 FR901464 is a potent natural product that inhibits pre-mRNA splicing via SF3b factor binding.23,24 We performed human colony-forming unit cell assay using BM mononuclear cells from 4 healthy subjects without and with different concentrations of meayamycin (2, 10, and 50nM). The colonies were spotted on cytospin slides and stained with Prussian blue. As shown by the representative pictures, meayamycin consistently induced RS in all 4 healthy subjects compared with controls (Figure 2B). The percentages of RS per each meayamycin concentration are shown in supplemental Figure 2.
Characteristics of a heterozygous mouse model for SF3B1
To next examine the role of SF3B1 in RS formation, we analyzed the phenotype of a heterozygous mouse model for Sf3b1 (Sf3b1+/−) insufficiency in vivo.14 Sf3b1+/− mice showed only minimal differences in blood counts, which includes a mild lymphopenia compared with WT C57BL/6 mice (21.9 × 102/μL ± 2.9 vs 42.3 × 102/μL ± 11.6; n = 3). Indeed, hemoglobin levels (8.5 g/dL ± 0.2 vs 7.9 g/dL ± 0.2) and platelets counts (60.2 × 104/μL ± 13.4 vs 59.8 × 104/μL ± 5.4) were comparable between heterozygous and WT mice. C57BL/6 mice served as controls because Sf3b1+/− mice were backcrossed to this mouse strain. BM aspirates were provided by Dr Koseki (RIKEN) and stained for Prussian blue. We found numerous RS in Sf3b1+/− compared with C57BL/6 mice as shown in Figure 2C (right panel).
RNA-Seq analysis of SF3B1 mutant RARS patients
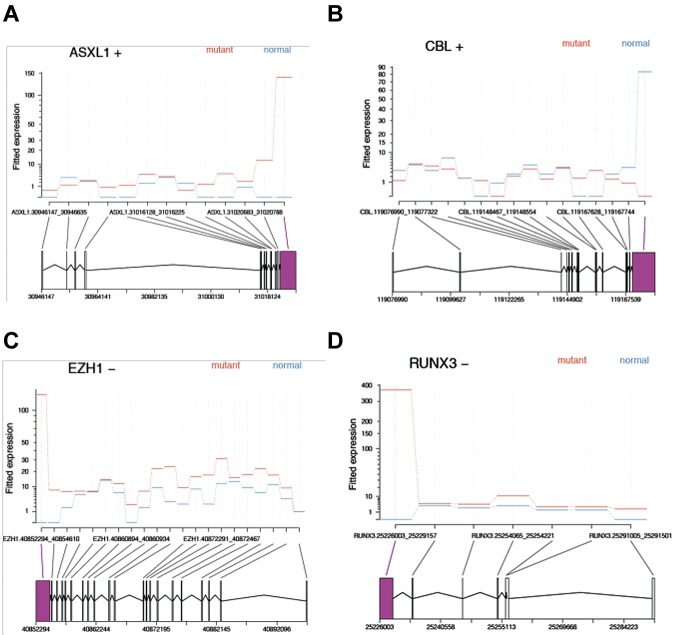
To further understand the functional consequences of SF3B1 mutations, we used RNA-Seq to investigate the differential exon usage and gene expression levels in BM cells of 2 SF3B1 mutant RARS patients. Using a false discovery rate level of 0.05, we tested 81 564 exons in 9069 genes and found that 423 exons derived from 350 genes were differentially used in mutants, compared with a representative healthy donor. A total of 67% of these exons had higher usage in mutant versus normal, and 56 genes were significantly affected in more than 1 exon (Table 2). Of note, 4 genes relevant to MDS pathophysiology, namely, ASXL1, CBL, EZH1, and RUNX3, were found to be alternatively spliced in at least one exon (Figure 3). Gene level analysis conducted on 12 870 reference genes showed that none of these 4 genes had differences in expression level. However, we identified 130 differentially expressed genes, of which 94% had lower expression in SF3B1 mutant patients (supplemental Table 1). Gene set analysis was also performed on 11 773 genes showing many gene sets related to myeloma or MDS (supplemental Table 2). In addition, a cytokine gene set, cytokine production, was the 16th most significant among the c5 collection of gene sets. This observation supports prior findings that cytokines play an important role in MDS pathogenesis.
Table 2.
Exons comparison between SF3B1 mutant and normal groups
| Gene | Chromosome | Strand | Exon location | No. of exon test | No. of exons | Exon no. | Adjusted P | Base M | Base N | Log2fold (mutant/healthy donor) | P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SIRPB1 | 20 | − | 1577987-1578928 | 11 | 11 | 6 | 0.00 | 125 | 0 | 33.46 | 0.00 |
| SIRPB1 | 20 | − | 1584456-1584788 | 11 | 11 | 4 | 1.63 × 10−12 | 74 | 0 | 32.79 | 7.77 × 10−16 |
| SIRPB1 | 20 | − | 1585388-1585705 | 11 | 11 | 3 | 2.52 × 10−07 | 47 | 0 | 32.00 | 2.81 × 10−10 |
| SIRPB1 | 20 | − | 1579468-1579582 | 11 | 11 | 5 | 1.44 × 10−05 | 34 | 0 | 31.72 | 2.33 × 10−08 |
| SIRPB1 | 20 | − | 1592003-1592359 | 11 | 11 | 2 | 2.58 × 10−04 | 95 | 0 | 32.88 | 5.54 × 10−07 |
| AKAP13 | 15 | + | 86287859-86292586 | 36 | 38 | 38 | 0.00 | 258 | 0 | 30.91 | 0.00 |
| HBG2 | 11 | − | 5275867-5276011 | 3 | 3 | 1 | 0.00 | 15 658 | 0 | 30.03 | 0.00 |
| HBG2 | 11 | − | 5274421-5274635 | 3 | 3 | 3 | 1.64 × 10−05 | 29 067 | 344 | −0.14 | 2.73 × 10−08 |
| PHF3 | 6 | + | 64421482-64424405 | 15 | 15 | 15 | 0.00 | 141 | 0 | 29.99 | 0.00 |
| CD37 | 19 | + | 49838677-49838832 | 9 | 9 | 1 | 0.00 | 111 | 0 | 29.65 | 0.00 |
| NIPAL3 | 1 | + | 24795476-24799472 | 12 | 12 | 12 | 0.00 | 173 | 0 | 29.40 | 0.00 |
| VPS36 | 13 | − | 52986739-52990058 | 13 | 14 | 14 | 0.00 | 222 | 0 | 28.94 | 0.00 |
| HELZ | 17 | − | 65066555-65074702 | 29 | 33 | 33 | 7.76 × 10−13 | 153 | 0 | 29.12 | 3.33 × 10−16 |
| CTTN | 11 | + | 70281132-70282689 | 16 | 19 | 19 | 1.51 × 10−12 | 419 | 0 | 31.66 | 6.66 × 10−16 |
| RSAD2 | 2 | + | 7035909-7038363 | 6 | 6 | 6 | 2.26 × 10−12 | 308 | 0 | 27.95 | 1.11 × 10−15 |
| RPS17 | 15 | − | 83207631-83207736 | 7 | 9 | 2 | 3.23 × 10−12 | 220 | 0 | 33.49 | 1.67 × 10−15 |
| POU2F2 | 19 | − | 42592673-42595743 | 10 | 13 | 13 | 6.74 × 10−12 | 280 | 0 | 29.46 | 3.55 × 10−15 |
| PARP14 | 3 | + | 122447155-122449686 | 17 | 17 | 17 | 2.06 × 10−11 | 128 | 0 | 29.10 | 1.11 × 10−14 |
| RAPGEF6 | 5 | − | 130759616-130762984 | 29 | 30 | 30 | 2.72 × 10−11 | 127 | 0 | 30.14 | 1.50 × 10−14 |
| CARD8 | 19 | − | 48711344-48715232 | 10 | 10 | 10 | 5.15 × 10−11 | 115 | 0 | 29.40 | 3.03 × 10−14 |
| TSC1 | 9 | − | 135766735-135772141 | 15 | 24 | 24 | 6.01 × 10−11 | 164 | 0 | 30.44 | 3.69 × 10−14 |
| ATM | 11 | + | 108236052-108239826 | 54 | 64 | 64 | 1.13 × 10−10 | 200 | 0 | 31.49 | 7.21 × 10−14 |
| RABL3 | 3 | − | 120405530-120408735 | 7 | 8 | 8 | 1.58 × 10−10 | 102 | 0 | 30.15 | 1.07 × 10−13 |
| CCDC25 | 8 | − | 27590835-27593762 | 7 | 9 | 9 | 1.58 × 10−10 | 108 | 0 | 29.79 | 1.07 × 10−13 |
| MEGF9 | 9 | − | 123363091-123367919 | 5 | 6 | 6 | 2.41 × 10−10 | 262 | 0 | 31.12 | 1.68 × 10−13 |
| IL32 | 16 | + | 3119282-3119667 | 4 | 11 | 11 | 3.05 × 10−10 | 345 | 0 | 31.09 | 2.21 × 10−13 |
| IL32 | 16 | + | 3118991-3119667 | 4 | 11 | 10 | 1.53 × 10−02 | 665 | 85 | −0.12 | 6.40 × 10−05 |
| DDX54 | 12 | − | 113594984-113596914 | 21 | 21 | 21 | 3.19 × 10−10 | 90 | 0 | 28.84 | 2.34 × 10−13 |
| TTC14 | 3 | + | 180327418-180328917 | 13 | 13 | 13 | 4.73 × 10−10 | 95 | 0 | 28.73 | 3.54 × 10−13 |
| ADPGK | 15 | − | 73043710-73045236 | 4 | 9 | 8 | 6.16 × 10−10 | 102 | 0 | 32.16 | 4.69 × 10−13 |
| ADPGK | 15 | − | 73043710-73045233 | 4 | 9 | 9 | 2.35 × 10−05 | 101 | 98 | −0.11 | 4.10 × 10−08 |
| EZH1 | 17 | − | 40852294-40854610 | 19 | 21 | 21 | 2.04 × 10−09 | 131 | 0 | 28.95 | 1.63 × 10−12 |
| MADD | 11 | + | 47350597-47351582 | 35 | 38 | 38 | 5.39 × 10−09 | 90 | 0 | 29.52 | 4.43 × 10−12 |
| RABL2B | 22 | − | 51205920-51207302 | 13 | 13 | 13 | 5.49 × 10−09 | 80 | 0 | 29.36 | 4.58 × 10−12 |
| RABL2B | 22 | − | 51207883-51207977 | 13 | 13 | 10 | 2.93 × 10−02 | 0 | 16 | −28.17 | 1.39 × 10−04 |
| ZNF638 | 2 | + | 71649944-71651168 | 24 | 28 | 22 | 6.95 × 10−09 | 84 | 0 | 30.73 | 5.88 × 10−12 |
| SON | 21 | + | 34948671-34949812 | 13 | 13 | 13 | 8.44 × 10−09 | 76 | 0 | 32.49 | 7.25 × 10−12 |
| IDE | 10 | − | 94211441-94214296 | 24 | 25 | 25 | 9.99 × 10−09 | 71 | 0 | 29.31 | 8.69 × 10−12 |
| MOBKL1A | 4 | + | 71847697-71853890 | 6 | 6 | 6 | 2.45 × 10−08 | 268 | 0 | 30.54 | 2.22 × 10−11 |
| ZNF592 | 15 | + | 85345094-85349661 | 10 | 11 | 11 | 2.45 × 10−08 | 81 | 0 | 29.21 | 2.20 × 10−11 |
| RPRD1B | 20 | + | 36718128-36720764 | 7 | 7 | 7 | 2.62 × 10−08 | 112 | 0 | 29.42 | 2.41 × 10−11 |
| ARHGEF7 | 13 | + | 111955338-111958080 | 11 | 23 | 23 | 2.99 × 10−08 | 120 | 0 | 32.24 | 2.78 × 10−11 |
| KDM5D | Y | − | 21867303-21868231 | 27 | 28 | 28 | 5.42 × 10−08 | 92 | 0 | 28.78 | 5.25 × 10−11 |
| ZNF295 | 21 | − | 43406941-43414217 | 3 | 5 | 4 | 1.51 × 10−07 | 28 | 0 | 30.45 | 1.55 × 10−10 |
| ZNF295 | 21 | − | 43406941-43411998 | 3 | 5 | 5 | 6.31 × 10−06 | 0 | 80 | −27.93 | 9.20 × 10−09 |
| ARHGAP27 | 17 | − | 43471269-43472999 | 11 | 17 | 17 | 1.51 × 10−07 | 154 | 0 | 29.46 | 1.56 × 10−10 |
| CEP350 | 1 | + | 180080132-180084014 | 22 | 36 | 36 | 1.62 × 10−07 | 102 | 0 | 32.63 | 1.69 × 10−10 |
| R3HDM1 | 2 | + | 136481507-136482836 | 19 | 25 | 25 | 1.85 × 10−07 | 66 | 0 | 29.65 | 1.97 × 10−10 |
| TRMT5 | 14 | − | 61438169-61441912 | 4 | 5 | 5 | 2.25 × 10−07 | 180 | 0 | 30.11 | 2.43 × 10−10 |
| TRMT5 | 14 | − | 61445949-61446604 | 4 | 5 | 2 | 5.94 × 10−07 | 25 | 23 | −0.36 | 6.99 × 10−10 |
| CA5B | X | + | 15800608-15805747 | 7 | 8 | 8 | 2.31 × 10−07 | 114 | 0 | 30.37 | 2.52 × 10−10 |
| KIAA0319L | 1 | − | 35899092-35900682 | 20 | 21 | 21 | 2.52 × 10−07 | 73 | 0 | 27.72 | 2.81 × 10−10 |
| CD3E | 11 | + | 118186201-118186889 | 8 | 8 | 8 | 4.06 × 10−07 | 238 | 0 | 27.14 | 4.57 × 10−10 |
| GTF2E1 | 3 | + | 120499890-120501915 | 4 | 5 | 5 | 4.96 × 10−07 | 138 | 0 | 32.16 | 5.72 × 10−10 |
| GTF2E1 | 3 | + | 120469370-120469847 | 4 | 5 | 2 | 1.39 × 10−06 | 11 | 20 | 0.11 | 1.83 × 10−09 |
| SLC9A8 | 20 | + | 48504366-48508772 | 9 | 16 | 16 | 5.36 × 10−07 | 65 | 0 | 30.44 | 6.25 × 10−10 |
| ST6GALNA | 9 | − | 130647601-130649067 | 5 | 7 | 7 | 7.62 × 10−07 | 136 | 0 | 28.77 | 9.06 × 10−10 |
| FCHSD1 | 5 | − | 141018870-141021130 | 15 | 20 | 20 | 8.83 × 10−07 | 94 | 0 | 27.20 | 1.07 × 10−09 |
| RUNX3 | 1 | − | 25226003-25229157 | 4 | 7 | 7 | 1.01 × 10−06 | 375 | 0 | 30.61 | 1.26 × 10−09 |
| CHD9 | 16 | + | 53357932-53361413 | 15 | 38 | 38 | 1.16 × 10−06 | 71 | 0 | 32.47 | 1.47 × 10−09 |
| ATG10 | 5 | + | 81272012-81272146 | 8 | 9 | 2 | 1.39 × 10−06 | 437 | 1440 | −0.66 | 1.79 × 10−09 |
| ATG10 | 5 | + | 81267844-81268125 | 8 | 9 | 1 | 3.04 × 10−05 | 201 | 155 | 1.44 | 5.40 × 10−08 |
| NAA16 | 13 | + | 41949541-41951166 | 18 | 22 | 22 | 1.39 × 10−06 | 55 | 0 | 29.50 | 1.83 × 10−09 |
| LPIN1 | 2 | + | 11964758-11967531 | 14 | 20 | 20 | 1.98 × 10−06 | 80 | 0 | 29.54 | 2.67 × 10−09 |
| 2-Mar | 19 | + | 8503272-8503897 | 5 | 6 | 6 | 2.42 × 10−06 | 126 | 0 | 30.44 | 3.29 × 10−09 |
| AFG3L1 | 16 | + | 90061101-90063028 | 11 | 15 | 13 | 2.43 × 10−06 | 75 | 0 | 28.76 | 3.34 × 10−09 |
| LDLRAP1 | 1 | + | 25893339-25895375 | 5 | 9 | 9 | 3.44 × 10−06 | 127 | 0 | 31.25 | 4.79 × 10−09 |
| FRYL | 4 | − | 48499380-48501697 | 36 | 63 | 63 | 4.11 × 10−06 | 99 | 0 | 31.64 | 5.80 × 10−09 |
| ITK | 5 | + | 156679617-156682109 | 17 | 17 | 17 | 4.46 × 10−06 | 237 | 0 | 31.53 | 6.35 × 10−09 |
| MCTP2 | 15 | + | 95022195-95027180 | 13 | 24 | 24 | 4.64 × 10−06 | 55 | 0 | 31.32 | 6.71 × 10−09 |
| HDDC3 | 15 | − | 91474157-91474786 | 4 | 4 | 4 | 6.58 × 10−06 | 80 | 0 | 28.62 | 9.68 × 10−09 |
| RAP1GAP2 | 17 | + | 2936686-2941034 | 9 | 25 | 25 | 6.98 × 10−06 | 164 | 0 | 31.13 | 1.04 × 10−08 |
| ARFRP1 | 20 | − | 62329996-62332054 | 9 | 9 | 8 | 8.08 × 10−06 | 85 | 0 | 27.86 | 1.22 × 10−08 |
| CHST15 | 10 | − | 125767186-125769855 | 7 | 7 | 7 | 8.68 × 10−06 | 81 | 0 | 29.59 | 1.32 × 10−08 |
| SLC5A6 | 2 | − | 27422457-27423447 | 19 | 20 | 19 | 1.02 × 10−05 | 77 | 0 | 30.99 | 1.58 × 10−08 |
| SLC5A6 | 2 | − | 27422973-27423447 | 19 | 20 | 18 | 1.46 × 10−03 | 54 | 0 | 30.43 | 4.39 × 10−06 |
| BTN3A3 | 6 | + | 26451903-26453642 | 8 | 11 | 11 | 1.02 × 10−05 | 140 | 0 | 29.53 | 1.58 × 10−08 |
| GCFC1 | 21 | − | 34106213-34107378 | 13 | 18 | 18 | 1.39 × 10−05 | 62 | 0 | 29.83 | 2.19 × 10−08 |
| MKNK2 | 19 | − | 2037470-2037828 | 8 | 13 | 13 | 1.39 × 10−05 | 55 | 0 | 28.44 | 2.19 × 10−08 |
| TRIM58 | 1 | + | 248039202-248043436 | 4 | 6 | 6 | 1.44 × 10−05 | 880 | 0 | 31.19 | 2.34 × 10−08 |
| TRIM58 | 1 | + | 248023919-248024014 | 4 | 6 | 2 | 1.24 × 10−02 | 10 | 3 | −0.88 | 4.92 × 10−05 |
| OXSR1 | 3 | + | 38294308-38296978 | 5 | 18 | 18 | 1.64 × 10−05 | 72 | 0 | 32.32 | 2.70 × 10−08 |
| POLR2A | 17 | + | 7416340-7417929 | 27 | 29 | 29 | 1.64 × 10−05 | 67 | 0 | 28.77 | 2.72 × 10−08 |
| RNF219 | 13 | − | 79188421-79191257 | 6 | 6 | 6 | 1.91 × 10−05 | 63 | 0 | 27.84 | 3.23 × 10−08 |
| GNPTAB | 12 | − | 102139278-102141019 | 13 | 20 | 20 | 2.00 × 10−05 | 85 | 0 | 29.10 | 3.40 × 10−08 |
| CXorf15 | X | + | 16859551-16862640 | 7 | 10 | 10 | 2.29 × 10−05 | 64 | 0 | 28.14 | 3.97 × 10−08 |
| RHCE | 1 | − | 25747130-25747363 | 10 | 10 | 1 | 2.46 × 10−05 | 75 | 0 | 27.80 | 4.31 × 10−08 |
| FHDC1 | 4 | + | 153895827-153900848 | 11 | 11 | 11 | 4.27 × 10−05 | 317 | 0 | 31.91 | 7.79 × 10−08 |
| CCNT1 | 12 | − | 49086752-49088219 | 4 | 8 | 8 | 4.49 × 10−05 | 45 | 0 | 30.46 | 8.25 × 10−08 |
| CAPRIN2 | 12 | − | 30862486-30863404 | 15 | 19 | 19 | 4.70 × 10−05 | 73 | 0 | 28.41 | 8.71 × 10−08 |
| RXRB | 6 | − | 33161366-33162606 | 7 | 10 | 10 | 5.98 × 10−05 | 64 | 0 | 27.55 | 1.12 × 10−07 |
| GIGYF2 | 2 | + | 233721503-233725285 | 23 | 33 | 33 | 5.99 × 10−05 | 43 | 0 | 28.04 | 1.13 × 10−07 |
| DNAJB6 | 7 | + | 157208710-157210132 | 12 | 12 | 12 | 6.27 × 10−05 | 165 | 90 | 1.87 | 1.19 × 10−07 |
| FAM63B | 15 | + | 59146681-59149732 | 8 | 10 | 9 | 6.56 × 10−05 | 76 | 0 | 30.03 | 1.25 × 10−07 |
| USP37 | 2 | − | 219314974-219319740 | 10 | 26 | 26 | 7.33 × 10−05 | 53 | 0 | 30.01 | 1.41 × 10−07 |
| EME1 | 17 | + | 48458124-48458820 | 6 | 10 | 10 | 8.83 × 10−05 | 43 | 0 | 30.84 | 1.71 × 10−07 |
| SQLE | 8 | + | 126010720-126011936 | 11 | 11 | 1 | 9.10 × 10−05 | 32 | 0 | 27.62 | 1.77 × 10−07 |
| GATAD2A | 19 | + | 19616153-19619740 | 5 | 11 | 11 | 1.55 × 10−04 | 104 | 0 | 28.45 | 3.07 × 10−07 |
| RFC1 | 4 | − | 39289076-39290464 | 25 | 25 | 25 | 1.73 × 10−04 | 42 | 0 | 28.28 | 3.46 × 10−07 |
| RORA | 15 | − | 60780485-60789818 | 10 | 12 | 12 | 2.42 × 10−04 | 140 | 0 | 28.47 | 5.02 × 10−07 |
| SBNO1 | 12 | − | 123773657-123780597 | 28 | 32 | 32 | 2.43 × 10−04 | 335 | 128 | 1.24 | 5.06 × 10−07 |
| PSMB9 | 6 | + | 32821938-32822014 | 8 | 8 | 1 | 2.46 × 10−04 | 48 | 0 | 27.86 | 5.15 × 10−07 |
| ADRBK2 | 22 | + | 26118256-26125256 | 4 | 16 | 16 | 2.52 × 10−04 | 186 | 0 | 33.06 | 5.35 × 10−07 |
| PRDM1 | 6 | + | 106552700-106553808 | 6 | 8 | 5 | 2.57 × 10−04 | 41 | 18 | 0.04 | 5.49 × 10−07 |
| PRDM1 | 6 | + | 106554786-106557809 | 6 | 8 | 7 | 1.50 × 10−02 | 110 | 0 | 29.90 | 6.27 × 10−05 |
| PRDM1 | 6 | + | 106554786-106557814 | 6 | 8 | 8 | 1.50 × 10−02 | 110 | 0 | 29.90 | 6.27 × 10−05 |
| PPID | 4 | − | 159630280-159630976 | 10 | 10 | 10 | 2.63 × 10−04 | 76 | 0 | 27.35 | 5.68 × 10−07 |
| PITPNA | 17 | − | 1421285-1423832 | 5 | 10 | 10 | 2.82 × 10−04 | 55 | 0 | 30.22 | 6.20 × 10−07 |
| LHFPL2 | 5 | − | 77781040-77784976 | 2 | 5 | 5 | 2.82 × 10−04 | 86 | 0 | 29.15 | 6.19 × 10−07 |
| LHFPL2 | 5 | − | 77805607-77806221 | 2 | 5 | 4 | 2.82 × 10−04 | 6 | 12 | −0.29 | 6.19 × 10−07 |
| CYTSB | 17 | + | 20217289-20218065 | 15 | 16 | 16 | 3.02 × 10−04 | 56 | 0 | 28.96 | 6.70 × 10−07 |
| SNTB1 | 8 | − | 121547986-121551209 | 5 | 7 | 7 | 3.07 × 10−04 | 70 | 0 | 27.79 | 6.88 × 10−07 |
| TSGA14 | 7 | − | 130036377-130038880 | 7 | 11 | 11 | 3.30 × 10−04 | 49 | 0 | 28.47 | 7.49 × 10−07 |
| CCND3 | 6 | − | 42016239-42016610 | 6 | 6 | 1 | 3.33 × 10−04 | 43 | 0 | 27.68 | 7.60 × 10−07 |
| ENO2 | 12 | + | 7031894-7032859 | 9 | 12 | 12 | 3.39 × 10−04 | 83 | 0 | 29.41 | 7.82 × 10−07 |
| FLVCR2 | 14 | + | 76112744-76114512 | 7 | 10 | 10 | 3.42 × 10−04 | 67 | 0 | 27.42 | 7.98 × 10−07 |
| PTPRC | 1 | + | 198671516-198671659 | 34 | 34 | 7 | 3.42 × 10−04 | 165 | 29 | 2.08 | 8.02 × 10−07 |
| SMAD4 | 18 | + | 48604626-48611409 | 8 | 12 | 12 | 3.77 × 10−04 | 53 | 0 | 31.76 | 8.91 × 10−07 |
| FCRL1 | 1 | − | 157764196-157765960 | 4 | 13 | 13 | 4.18 × 10−04 | 66 | 0 | 29.44 | 1.00 × 10−06 |
| C3orf63 | 3 | − | 56654161-56657871 | 18 | 26 | 26 | 4.22 × 10−04 | 124 | 0 | 28.62 | 1.02 × 10−06 |
| ZNF45 | 19 | − | 44416777-44419352 | 5 | 10 | 10 | 4.62 × 10−04 | 55 | 0 | 30.37 | 1.12 × 10−06 |
| CREBZF | 11 | − | 85374859-85376182 | 7 | 7 | 1 | 4.96 × 10−04 | 67 | 0 | 31.33 | 1.22 × 10−06 |
| SLC37A1 | 21 | + | 44000455-44001549 | 8 | 20 | 20 | 5.27 × 10−04 | 35 | 0 | 32.09 | 1.32 × 10−06 |
| FAM122B | X | − | 133903597-133905384 | 6 | 12 | 12 | 5.54 × 10−04 | 94 | 0 | 29.22 | 1.39 × 10−06 |
| TIMM17B | X | − | 48755350-48755426 | 8 | 8 | 1 | 5.79 × 10−04 | 44 | 0 | 27.31 | 1.46 × 10−06 |
| CHD3 | 17 | + | 7814782-7816075 | 9 | 37 | 37 | 5.99 × 10−04 | 51 | 0 | 32.62 | 1.53 × 10−06 |
| RNF160 | 21 | − | 30300466-30302832 | 20 | 30 | 30 | 5.99 × 10−04 | 42 | 0 | 28.92 | 1.53 × 10−06 |
| SORT1 | 1 | − | 109852192-109856679 | 3 | 20 | 20 | 6.31 × 10−04 | 108 | 0 | 32.48 | 1.62 × 10−06 |
| POM121C | 7 | − | 75046066-75048176 | 2 | 9 | 9 | 6.33 × 10−04 | 59 | 0 | 31.72 | 1.64 × 10−06 |
| POM121C | 7 | − | 75050792-75052486 | 2 | 9 | 7 | 6.33 × 10−04 | 9 | 14 | −0.29 | 1.64 × 10−06 |
| ZBTB40 | 1 | + | 22852695-22857650 | 10 | 18 | 18 | 7.60 × 10−04 | 90 | 0 | 29.12 | 2.00 × 10−06 |
| APPL2 | 12 | − | 105567075-105568226 | 7 | 21 | 21 | 9.03 × 10−04 | 32 | 0 | 29.36 | 2.39 × 10−06 |
| MCART1 | 9 | − | 37877572-37880745 | 3 | 6 | 6 | 9.16 × 10−04 | 29 | 0 | 30.07 | 2.44 × 10−06 |
| GOPC | 6 | − | 117881435-117884547 | 7 | 8 | 8 | 9.16 × 10−04 | 65 | 0 | 27.39 | 2.45 × 10−06 |
| PDGFD | 11 | − | 103777915-103780547 | 5 | 8 | 8 | 9.54 × 10−04 | 60 | 0 | 29.12 | 2.60 × 10−06 |
| CD247 | 1 | − | 167399878-167400983 | 6 | 7 | 7 | 9.89 × 10−04 | 361 | 0 | 27.72 | 2.71 × 10−06 |
| CA1 | 8 | − | 86262040-86262114 | 12 | 12 | 3 | 1.07 × 10−03 | 58 | 0 | 26.32 | 2.95 × 10−06 |
| SYVN1 | 11 | − | 64894753-64895960 | 12 | 17 | 17 | 1.11 × 10−03 | 42 | 0 | 27.32 | 3.09 × 10−06 |
| SP2 | 17 | + | 46005090-46006321 | 6 | 6 | 6 | 1.11 × 10−03 | 54 | 0 | 26.89 | 3.13 × 10−06 |
| ST8SIA4 | 5 | − | 100142640-100147833 | 7 | 7 | 7 | 1.11 × 10−03 | 129 | 71 | 1.56 | 3.13 × 10−06 |
| ESRRA | 11 | + | 64083179-64084210 | 4 | 7 | 7 | 1.12 × 10−03 | 69 | 0 | 30.36 | 3.19 × 10−06 |
| DDX26B | X | + | 134715453-134716459 | 17 | 17 | 17 | 1.16 × 10−03 | 45 | 0 | 26.74 | 3.32 × 10−06 |
| RNPEPL1 | 2 | + | 241517016-241518141 | 4 | 11 | 11 | 1.18 × 10−03 | 64 | 0 | 30.68 | 3.37 × 10−06 |
| ZKSCAN5 | 7 | + | 99128731-99131444 | 5 | 7 | 7 | 1.26 × 10−03 | 35 | 0 | 29.99 | 3.64 × 10−06 |
| POM121 | 7 | + | 72416676-72418837 | 2 | 8 | 8 | 1.33 × 10−03 | 57 | 0 | 30.76 | 3.87 × 10−06 |
| POM121 | 7 | + | 72412376-72414061 | 2 | 8 | 6 | 1.33 × 10−03 | 10 | 14 | −0.22 | 3.87 × 10−06 |
| TJAP1 | 6 | + | 43472499-43474294 | 10 | 14 | 14 | 1.38 × 10−03 | 52 | 0 | 28.80 | 4.07 × 10−06 |
| TBCCD1 | 3 | − | 186263858-186264744 | 8 | 9 | 9 | 1.38 × 10−03 | 47 | 0 | 27.32 | 4.07 × 10−06 |
| SHQ1 | 3 | − | 72798430-72799987 | 10 | 11 | 11 | 1.42 × 10−03 | 36 | 0 | 27.01 | 4.23 × 10−06 |
| CUL4A | 13 | + | 113917801-113919391 | 10 | 18 | 18 | 1.44 × 10−03 | 83 | 0 | 30.07 | 4.31 × 10−06 |
| NLRP12 | 19 | − | 54310749-54311959 | 10 | 11 | 4 | 1.50 × 10−03 | 32 | 0 | 27.88 | 4.56 × 10−06 |
| ATF7IP | 12 | + | 14650588-14651696 | 13 | 15 | 15 | 1.50 × 10−03 | 39 | 0 | 27.30 | 4.58 × 10−06 |
| ZNHIT6 | 1 | − | 86115107-86119781 | 7 | 11 | 11 | 1.50 × 10−03 | 47 | 0 | 26.99 | 4.55 × 10−06 |
| MSH3 | 5 | + | 80171570-80172633 | 23 | 24 | 24 | 1.59 × 10−03 | 41 | 0 | 28.75 | 4.85 × 10−06 |
| KIAA2026 | 9 | − | 5919010-5923309 | 3 | 6 | 6 | 1.63 × 10−03 | 67 | 0 | 29.99 | 5.01 × 10−06 |
| SH2D1B | 1 | − | 162365056-162367105 | 4 | 4 | 4 | 1.75 × 10−03 | 313 | 0 | 26.58 | 5.39 × 10−06 |
| SH2D1B | 1 | − | 162381673-162381928 | 4 | 4 | 1 | 4.00 × 10−02 | 47 | 6 | −1.06 | 1.99 × 10−04 |
| TGFBR3 | 1 | − | 92145902-92149414 | 16 | 17 | 17 | 2.06 × 10−03 | 99 | 19 | 0.03 | 6.44 × 10−06 |
| ALDOA | 16 | + | 30066105-30066248 | 16 | 16 | 2 | 2.21 × 10−03 | 26 | 0 | 27.87 | 6.95 × 10−06 |
| SEPSECS | 4 | − | 25121628-25125847 | 5 | 10 | 10 | 2.43 × 10−03 | 35 | 0 | 30.43 | 7.69 × 10−06 |
| PIK3C2A | 11 | − | 17108126-17111467 | 3 | 32 | 32 | 2.51 × 10−03 | 51 | 0 | 30.24 | 7.98 × 10−06 |
| PIK3C2A | 11 | − | 17190224-17191354 | 3 | 32 | 1 | 1.94 × 10−02 | 18 | 15 | 0.17 | 8.75 × 10−05 |
| SAPS3 | 11 | + | 68382100-68382799 | 27 | 28 | 28 | 2.55 × 10−03 | 54 | 0 | 29.21 | 8.17 × 10−06 |
| C11orf49 | 11 | + | 47183015-47183796 | 12 | 12 | 11 | 2.58 × 10−03 | 40 | 0 | 30.77 | 8.28 × 10−06 |
| TCF3 | 19 | − | 1609263-1611368 | 9 | 19 | 19 | 2.80 × 10−03 | 46 | 0 | 29.10 | 9.02 × 10−06 |
| RB1CC1 | 8 | − | 53535019-53536419 | 18 | 25 | 25 | 3.02 × 10−03 | 79 | 0 | 29.68 | 9.77 × 10−06 |
| VPRBP | 3 | − | 51433298-51434555 | 21 | 26 | 26 | 3.13 × 10−03 | 35 | 0 | 28.00 | 1.02 × 10−05 |
| C17orf68 | 17 | − | 8128140-8131637 | 22 | 23 | 23 | 3.23 × 10−03 | 46 | 0 | 27.31 | 1.06 × 10−05 |
| ZSCAN18 | 19 | − | 58601232-58601753 | 3 | 8 | 2 | 3.26 × 10−03 | 15 | 6 | 0.32 | 1.07 × 10−05 |
| SERINC5 | 5 | − | 79434552-79439633 | 7 | 13 | 12 | 3.41 × 10−03 | 79 | 0 | 30.17 | 1.13 × 10−05 |
| LINS1 | 15 | − | 101113856-101114446 | 6 | 9 | 7 | 3.55 × 10−03 | 44 | 0 | 28.77 | 1.18 × 10−05 |
| ATP10D | 4 | + | 47593059-47595503 | 11 | 22 | 22 | 3.57 × 10−03 | 34 | 0 | 30.41 | 1.19 × 10−05 |
| CYHR1 | 8 | − | 145689200-145689995 | 6 | 9 | 6 | 3.67 × 10−03 | 43 | 0 | 32.53 | 1.23 × 10−05 |
| MINA | 3 | − | 97660661-97664186 | 10 | 12 | 12 | 4.03 × 10−03 | 70 | 0 | 27.71 | 1.37 × 10−05 |
| ELP3 | 8 | + | 28047166-28048667 | 13 | 15 | 15 | 4.03 × 10−03 | 34 | 0 | 26.78 | 1.37 × 10−05 |
| AHNAK | 11 | − | 62283375-62301546 | 7 | 7 | 5 | 4.04 × 10−03 | 1458 | 606 | 0.31 | 1.38 × 10−05 |
| POLH | 6 | + | 43581397-43588259 | 8 | 11 | 11 | 4.15 × 10−03 | 60 | 0 | 29.11 | 1.42 × 10−05 |
| SCO1 | 17 | − | 10583651-10584570 | 6 | 6 | 6 | 4.28 × 10−03 | 44 | 0 | 27.43 | 1.47 × 10−05 |
| PAPOLG | 2 | + | 61024197-61026096 | 14 | 22 | 22 | 4.37 × 10−03 | 41 | 0 | 28.47 | 1.51 × 10−05 |
| TTYH2 | 17 | + | 72256268-72258155 | 8 | 15 | 15 | 4.57 × 10−03 | 53 | 0 | 29.97 | 1.59 × 10−05 |
| KIAA0355 | 19 | + | 34843552-34846470 | 11 | 14 | 14 | 4.59 × 10−03 | 73 | 0 | 28.43 | 1.60 × 10−05 |
| SH2D1A | X | + | 123505201-123507008 | 5 | 5 | 5 | 5.12 × 10−03 | 88 | 0 | 26.73 | 1.79 × 10−05 |
| GRK6 | 5 | + | 176868738-176869848 | 12 | 16 | 16 | 5.20 × 10−03 | 36 | 0 | 29.55 | 1.83 × 10−05 |
| KIAA0907 | 1 | − | 155882836-155884111 | 9 | 14 | 14 | 5.27 × 10−03 | 51 | 0 | 26.56 | 1.86 × 10−05 |
| DDX19B | 16 | + | 70363734-70363971 | 12 | 12 | 9 | 5.72 × 10−03 | 0 | 27 | −28.00 | 2.05 × 10−05 |
| DDX19B | 16 | + | 70367424-70367729 | 12 | 12 | 12 | 8.84 × 10−03 | 22 | 0 | 29.10 | 3.38 × 10−05 |
| MANEA | 6 | + | 96053624-96057326 | 2 | 5 | 5 | 6.18 × 10−03 | 46 | 0 | 29.45 | 2.24 × 10−05 |
| MANEA | 6 | + | 96034278-96034859 | 2 | 5 | 2 | 6.18 × 10−03 | 6 | 11 | 0.03 | 2.24 × 10−05 |
| COPB1 | 11 | − | 14521155-14521404 | 24 | 24 | 3 | 6.18 × 10−03 | 23 | 0 | 27.40 | 2.22 × 10−05 |
| ZNF37B | 10 | − | 43008963-43016575 | 2 | 8 | 8 | 6.49 × 10−03 | 106 | 0 | 29.56 | 2.37 × 10−05 |
| ZNF37B | 10 | − | 43047021-43047262 | 2 | 8 | 4 | 6.49 × 10−03 | 6 | 7 | 0.04 | 2.37 × 10−05 |
| FCAR | 19 | + | 55399374-55400437 | 8 | 8 | 7 | 6.49 × 10−03 | 66 | 540 | 1.16 | 2.36 × 10−05 |
| ZNF498 | 7 | + | 99226814-99230030 | 4 | 8 | 8 | 7.64 × 10−03 | 53 | 0 | 27.07 | 2.83 × 10−05 |
| TEP1 | 14 | − | 20833827-20836718 | 5 | 46 | 46 | 7.91 × 10−03 | 57 | 0 | 31.96 | 2.96 × 10−05 |
| ZDHHC7 | 16 | − | 85008067-85010125 | 3 | 9 | 9 | 7.91 × 10−03 | 69 | 0 | 30.62 | 2.96 × 10−05 |
| MOGS | 2 | − | 74688185-74690139 | 3 | 5 | 5 | 8.02 × 10−03 | 63 | 0 | 26.72 | 3.01 × 10−05 |
| BCL7B | 7 | − | 72950686-72951720 | 5 | 6 | 6 | 8.24 × 10−03 | 63 | 0 | 28.92 | 3.10 × 10−05 |
| FAM13A | 4 | − | 89647106-89649810 | 14 | 24 | 24 | 8.39 × 10−03 | 96 | 0 | 30.28 | 3.17 × 10−05 |
| N4BP2L2 | 13 | − | 33016525-33018263 | 10 | 14 | 11 | 8.39 × 10−03 | 30 | 0 | 27.06 | 3.18 × 10−05 |
| NFAT5 | 16 | + | 69725652-69728142 | 3 | 10 | 8 | 9.53 × 10−03 | 26 | 0 | 31.02 | 3.66 × 10−05 |
| CTNS | 17 | + | 3563530-3566396 | 7 | 14 | 13 | 9.57 × 10−03 | 67 | 0 | 30.69 | 3.68 × 10−05 |
| KIAA1467 | 12 | + | 13233559-13236381 | 7 | 12 | 12 | 1.03 × 10−02 | 72 | 0 | 31.36 | 4.00 × 10−05 |
| ZNF333 | 19 | + | 14829040-14831770 | 5 | 12 | 12 | 1.12 × 10−02 | 34 | 0 | 30.32 | 4.42 × 10−05 |
| PRKCQ | 10 | − | 6469105-6470324 | 12 | 18 | 18 | 1.23 × 10−02 | 51 | 0 | 27.05 | 4.88 × 10−05 |
| SLC26A2 | 5 | + | 149359856-149366959 | 2 | 3 | 3 | 1.24 × 10−02 | 31 | 0 | 29.85 | 4.97 × 10−05 |
| SLC26A2 | 5 | + | 149357191-149357914 | 2 | 3 | 2 | 1.24 × 10−02 | 5 | 19 | 0.22 | 4.97 × 10−05 |
| TMCO7 | 16 | + | 69117388-69119083 | 16 | 18 | 18 | 1.24 × 10−02 | 37 | 0 | 26.44 | 4.94 × 10−05 |
| PDZD11 | X | − | 69506211-69506966 | 7 | 7 | 7 | 1.24 × 10−02 | 39 | 0 | 26.32 | 4.97 × 10−05 |
| FCGR3B | 1 | − | 161595935-161596192 | 5 | 5 | 4 | 1.24 × 10−02 | 74 | 196 | 1.06 | 4.98 × 10−05 |
| LPAR2 | 19 | − | 19734468-19735378 | 2 | 3 | 3 | 1.36 × 10−02 | 57 | 0 | 27.47 | 5.54 × 10−05 |
| LPAR2 | 19 | − | 19737352-19738093 | 2 | 3 | 2 | 1.36 × 10−02 | 32 | 14 | 0.04 | 5.54 × 10−05 |
| NAPEPLD | 7 | − | 102740023-102744001 | 3 | 7 | 6 | 1.38 × 10−02 | 14 | 0 | 29.38 | 5.60 × 10−05 |
| NAPEPLD | 7 | − | 102740023-102743569 | 3 | 7 | 7 | 2.30 × 10−02 | 0 | 12 | −28.98 | 1.05 × 10−04 |
| PIK3CA | 3 | + | 178951882-178952495 | 10 | 21 | 21 | 1.40 × 10−02 | 33 | 0 | 31.17 | 5.72 × 10−05 |
| STAG3L2 | 7 | − | 74306509-74306731 | 7 | 8 | 1 | 1.41 × 10−02 | 36 | 0 | 26.66 | 5.77 × 10−05 |
| DNAJC16 | 1 | + | 15894273-15898226 | 6 | 15 | 15 | 1.44 × 10−02 | 79 | 0 | 30.26 | 5.94 × 10−05 |
| ZNF227 | 19 | + | 44738855-44741420 | 4 | 6 | 6 | 1.49 × 10−02 | 44 | 0 | 27.42 | 6.17 × 10−05 |
| NPC1 | 18 | − | 21111464-21112248 | 25 | 25 | 25 | 1.53 × 10−02 | 36 | 0 | 27.05 | 6.43 × 10−05 |
| SHPRH | 6 | − | 146205947-146207923 | 9 | 29 | 29 | 1.54 × 10−02 | 25 | 0 | 29.37 | 6.51 × 10−05 |
| YPEL4 | 11 | − | 57412561-57413543 | 4 | 5 | 5 | 1.54 × 10−02 | 72 | 0 | 28.59 | 6.50 × 10−05 |
| MPHOSPH | 13 | + | 20245974-20247598 | 14 | 14 | 14 | 1.54 × 10−02 | 69 | 0 | 27.28 | 6.49 × 10−05 |
| GOLM1 | 9 | − | 88641060-88642808 | 5 | 11 | 11 | 1.66 × 10−02 | 54 | 0 | 28.28 | 7.03 × 10−05 |
| VPS13B | 8 | + | 100887646-100889807 | 35 | 56 | 56 | 1.66 × 10−02 | 27 | 0 | 29.68 | 7.06 × 10−05 |
| NLRX1 | 11 | + | 119050402-119050997 | 5 | 10 | 6 | 1.68 × 10−02 | 10 | 9 | 0.05 | 7.17 × 10−05 |
| C18orf8 | 18 | + | 21110351-21110576 | 16 | 20 | 19 | 1.74 × 10−02 | 32 | 0 | 27.64 | 7.45 × 10−05 |
| ZBTB20 | 3 | − | 114056947-114058273 | 5 | 12 | 12 | 1.74 × 10−02 | 41 | 0 | 27.40 | 7.51 × 10−05 |
| ZFYVE19 | 15 | + | 41099274-41100066 | 9 | 11 | 1 | 1.76 × 10−02 | 27 | 0 | 27.83 | 7.61 × 10−05 |
| RBM12 | 20 | − | 34236888-34243266 | 2 | 2 | 2 | 1.79 × 10−02 | 97 | 0 | 28.98 | 7.76 × 10−05 |
| RBM12 | 20 | − | 34252682-34252838 | 2 | 2 | 1 | 1.79 × 10−02 | 34 | 32 | −0.02 | 7.76 × 10−05 |
| MEPCE | 7 | + | 100031125-100031740 | 4 | 4 | 4 | 1.83 × 10−02 | 52 | 0 | 27.62 | 8.03 × 10−05 |
| ELMOD3 | 2 | + | 85617261-85618873 | 11 | 17 | 16 | 1.85 × 10−02 | 48 | 0 | 30.27 | 8.15 × 10−05 |
| NPLOC4 | 17 | − | 79523915-79526442 | 2 | 15 | 15 | 1.88 × 10−02 | 202 | 0 | 32.35 | 8.38 × 10−05 |
| NPLOC4 | 17 | − | 79555970-79556130 | 2 | 15 | 10 | 1.88 × 10−02 | 4 | 3 | 0.30 | 8.38 × 10−05 |
| GTF3C4 | 9 | + | 135564268-135565468 | 2 | 4 | 4 | 1.89 × 10−02 | 13 | 0 | 28.70 | 8.45 × 10−05 |
| GTF3C4 | 9 | + | 135553364-135555190 | 2 | 4 | 1 | 1.89 × 10−02 | 0 | 10 | −26.38 | 8.45 × 10−05 |
| IGHMBP2 | 11 | + | 68707002-68708069 | 15 | 15 | 15 | 1.94 × 10−02 | 35 | 0 | 28.12 | 8.71 × 10−05 |
| TRIM37 | 17 | − | 57075562-57076820 | 14 | 26 | 25 | 2.28 × 10−02 | 36 | 0 | 29.83 | 1.04 × 10−04 |
| ENG | 9 | − | 130577292-130578332 | 14 | 16 | 15 | 2.38 × 10−02 | 30 | 0 | 30.61 | 1.09 × 10−04 |
| C14orf45 | 14 | + | 74516405-74516898 | 9 | 12 | 8 | 2.44 × 10−02 | 30 | 0 | 27.12 | 1.13 × 10−04 |
| CCDC91 | 12 | + | 28701996-28703098 | 11 | 12 | 12 | 2.54 × 10−02 | 30 | 0 | 28.77 | 1.18 × 10−04 |
| ZSCAN21 | 7 | + | 99661411-99662661 | 2 | 4 | 4 | 2.92 × 10−02 | 41 | 0 | 28.95 | 1.38 × 10−04 |
| ZSCAN21 | 7 | + | 99654534-99655028 | 2 | 4 | 2 | 2.92 × 10−02 | 8 | 9 | 0.10 | 1.38 × 10−04 |
| RRN3P2 | 16 | + | 29127448-29128036 | 8 | 14 | 14 | 3.14 × 10−02 | 31 | 0 | 28.51 | 1.52 × 10−04 |
| DIP2A | 21 | + | 47987283-47989926 | 7 | 40 | 40 | 3.14 × 10−02 | 116 | 0 | 33.06 | 1.52 × 10−04 |
| ZNF611 | 19 | − | 53206067-53210117 | 4 | 8 | 8 | 3.25 × 10−02 | 140 | 0 | 28.67 | 1.58 × 10−04 |
| RASGRP3 | 2 | + | 33787796-33789797 | 15 | 20 | 20 | 3.31 × 10−02 | 36 | 0 | 30.57 | 1.61 × 10−04 |
| CHD7 | 8 | + | 61777575-61779463 | 3 | 32 | 32 | 3.37 × 10−02 | 26 | 0 | 32.90 | 1.65 × 10−04 |
| FLCN | 17 | − | 17124486-17124942 | 15 | 15 | 9 | 3.59 × 10−02 | 45 | 0 | 30.05 | 1.77 × 10−04 |
| C10orf119 | 10 | − | 121588972-121591112 | 7 | 11 | 11 | 3.78 × 10−02 | 35 | 0 | 26.46 | 1.87 × 10−04 |
| FIGNL1 | 7 | − | 50511834-50514995 | 3 | 5 | 5 | 3.80 × 10−02 | 42 | 0 | 29.09 | 1.88 × 10−04 |
| NOL9 | 1 | − | 6581409-6586063 | 6 | 10 | 10 | 4.14 × 10−02 | 169 | 0 | 28.77 | 2.06 × 10−04 |
| GLUL | 1 | − | 182360310-182360539 | 9 | 9 | 2 | 4.40 × 10−02 | 79 | 121 | 1.52 | 2.20 × 10−04 |
| TRAF5 | 1 | + | 211545470-211548284 | 3 | 11 | 11 | 4.40 × 10−02 | 68 | 0 | 30.21 | 2.21 × 10−04 |
| DHRS7B | 17 | + | 21094261-21094835 | 6 | 7 | 7 | 4.51 × 10−02 | 35 | 0 | 26.40 | 2.27 × 10−04 |
| NLRC3 | 16 | − | 3589038-3591915 | 2 | 19 | 19 | 4.51 × 10−02 | 106 | 0 | 31.12 | 2.28 × 10−04 |
| NLRC3 | 16 | − | 3613010-3614759 | 2 | 19 | 4 | 4.51 × 10−02 | 31 | 6 | 0.34 | 2.28 × 10−04 |
| ABCD1 | X | + | 152990323-152991621 | 7 | 10 | 1 | 4.53 × 10−02 | 19 | 0 | 27.49 | 2.30 × 10−04 |
| GRIPAP1 | X | − | 48847338-48847514 | 23 | 27 | 8 | 4.56 × 10−02 | 24 | 0 | 25.88 | 2.32 × 10−04 |
| CHAC2 | 2 | + | 54001279-54002287 | 3 | 3 | 3 | 4.63 × 10−02 | 46 | 0 | 27.32 | 2.37 × 10−04 |
| SLC33A1 | 3 | − | 155571012-155572167 | 6 | 6 | 1 | 4.78 × 10−02 | 48 | 0 | 26.51 | 2.45 × 10−04 |
| ASXL1 | 20 | + | 31022235-31027121 | 4 | 13 | 13 | 4.88 × 10−02 | 141 | 0 | 32.33 | 2.51 × 10−04 |
| CASC3 | 17 | + | 38296507-38297032 | 13 | 14 | 1 | 4.94 × 10−02 | 28 | 0 | 26.09 | 2.55 × 10−04 |
| SEC22A | 3 | + | 122990369-122992980 | 5 | 7 | 7 | 4.98 × 10−02 | 23 | 0 | 27.20 | 2.58 × 10−04 |
| RPL21P28 | 13 | + | 27830573-27830698 | 2 | 2 | 2 | 0.00 | 1736 | 0 | 2.38 | 0.00 |
| RPL21P28 | 13 | + | 27825693-27825722 | 2 | 2 | 1 | 0.00 | 231 | 202 | −5.86 | 0.00 |
| HNRNPA1L | 13 | + | 53201573-53201713 | 4 | 7 | 3 | 0.00 | 6 | 4 | 1.00 | 0.00 |
| HNRNPA1L | 13 | + | 53196124-53196339 | 4 | 7 | 2 | 0.00 | 6 | 9 | 0.10 | 0.00 |
| HNRNPA1L | 13 | + | 53191605-53191817 | 4 | 7 | 1 | 0.00 | 3 | 10 | −1.17 | 0.00 |
| HNRNPA1L | 13 | + | 53216541-53217919 | 4 | 7 | 7 | 0.00 | 0 | 1743 | −34.56 | 0.00 |
| TXLNA | 1 | + | 32653555-32653725 | 7 | 11 | 5 | 0.00 | 15 | 5 | 0.76 | 0.00 |
| TXLNA | 1 | + | 32658258-32658332 | 7 | 11 | 8 | 0.00 | 4 | 2 | 0.63 | 0.00 |
| TXLNA | 1 | + | 32657912-32658031 | 7 | 11 | 7 | 0.00 | 9 | 3 | 0.62 | 0.00 |
| TXLNA | 1 | + | 32655656-32655850 | 7 | 11 | 6 | 0.00 | 18 | 9 | 0.35 | 0.00 |
| TXLNA | 1 | + | 32646843-32647178 | 7 | 11 | 3 | 0.00 | 7 | 7 | −0.64 | 0.00 |
| TXLNA | 1 | + | 32660503-32663885 | 7 | 11 | 11 | 0.00 | 0 | 345 | −33.27 | 0.00 |
| MIER1 | 1 | + | 67452086-67454302 | 20 | 21 | 21 | 0.00 | 0 | 447 | −31.97 | 0.00 |
| MIER1 | 1 | + | 67450255-67450335 | 20 | 21 | 19 | 2.43 × 10−10 | 21 | 24 | 0.59 | 1.73 × 10−13 |
| MIER1 | 1 | + | 67450255-67450275 | 20 | 21 | 18 | 5.31 × 10−08 | 12 | 15 | 0.51 | 5.08 × 10−11 |
| MIER1 | 1 | + | 67447462-67447601 | 20 | 21 | 17 | 1.44 × 10−05 | 25 | 36 | 0.34 | 2.30 × 10−08 |
| MIER1 | 1 | + | 67411833-67411978 | 20 | 21 | 8 | 1.82 × 10−02 | 29 | 43 | 0.23 | 7.98 × 10−05 |
| CSF3R | 1 | − | 36931644-36932509 | 20 | 20 | 18 | 0.00 | 0 | 1003 | −32.06 | 0.00 |
| CSF3R | 1 | − | 36932831-36932912 | 20 | 20 | 16 | 1.63 × 10−12 | 44 | 206 | 0.35 | 7.77 × 10−16 |
| CSF3R | 1 | − | 36931644-36932428 | 20 | 20 | 19 | 5.15 × 10−11 | 174 | 999 | 0.05 | 3.01 × 10−14 |
| CSF3R | 1 | − | 36933423-36933563 | 20 | 20 | 14 | 4.53 × 10−06 | 61 | 318 | 0.20 | 6.50 × 10−09 |
| CSF3R | 1 | − | 36933159-36933252 | 20 | 20 | 15 | 5.23 × 10−04 | 48 | 244 | 0.22 | 1.30 × 10−06 |
| SLC25A36 | 3 | + | 140695105-140698785 | 8 | 8 | 8 | 0.00 | 0 | 241 | −30.23 | 0.00 |
| SLC25A36 | 3 | + | 140695102-140698785 | 8 | 8 | 7 | 4.68 × 10−04 | 155 | 242 | 0.16 | 1.14 × 10−06 |
| CDV3 | 3 | + | 133306003-133309116 | 4 | 6 | 5 | 0.00 | 372 | 283 | 0.15 | 0.00 |
| CDV3 | 3 | + | 133306740-133309116 | 4 | 6 | 6 | 0.00 | 0 | 275 | −33.17 | 0.00 |
| NUFIP2 | 17 | − | 27582855-27591609 | 3 | 4 | 4 | 0.00 | 0 | 196 | −30.52 | 0.00 |
| NUFIP2 | 17 | − | 27613010-27614734 | 3 | 4 | 2 | 2.14 × 10−09 | 68 | 94 | 0.11 | 1.73 × 10−12 |
| H3F3A | 1 | + | 226252030-226252180 | 3 | 3 | 2 | 0.00 | 822 | 2293 | 0.09 | 0.00 |
| H3F3A | 1 | + | 226250421-226250512 | 3 | 3 | 1 | 0.00 | 255 | 835 | −0.15 | 0.00 |
| H3F3A | 1 | + | 226259052-226259702 | 3 | 3 | 3 | 0.00 | 0 | 4290 | −33.08 | 0.00 |
| POU5F1 | 6 | − | 31133704-31134615 | 6 | 6 | 2 | 0.00 | 0 | 299 | −30.99 | 0.00 |
| POU5F1 | 6 | − | 31134311-31134615 | 6 | 6 | 1 | 3.44 × 10−06 | 335 | 273 | 0.09 | 4.80 × 10−09 |
| QKI | 6 | + | 163984452-163999627 | 8 | 9 | 7 | 0.00 | 0 | 273 | −31.04 | 0.00 |
| QKI | 6 | + | 163991726-163999627 | 8 | 9 | 9 | 7.22 × 10−10 | 171 | 212 | 0.06 | 5.58 × 10−13 |
| SUMO1 | 2 | − | 203070803-203071526 | 7 | 7 | 7 | 0.00 | 0 | 328 | −30.28 | 0.00 |
| SUMO1 | 2 | − | 203070903-203072044 | 7 | 7 | 6 | 4.59 × 10−02 | 506 | 811 | 0.01 | 2.34 × 10−04 |
| C1orf9 | 1 | + | 172578956-172580971 | 22 | 25 | 25 | 0.00 | 0 | 200 | −29.70 | 0.00 |
| LGALS3 | 14 | + | 55612030-55612147 | 9 | 9 | 9 | 0.00 | 0 | 139 | −30.94 | 0.00 |
| NCOA1 | 2 | + | 24991093-24993568 | 14 | 23 | 23 | 0.00 | 0 | 97 | −33.15 | 0.00 |
| WDR45 | X | − | 48957143-48957492 | 13 | 14 | 2 | 1.63 × 10−12 | 0 | 57 | −32.27 | 7.77 × 10−16 |
| CREM | 10 | + | 35500584-35501885 | 9 | 15 | 15 | 2.43 × 10−12 | 0 | 177 | −31.41 | 1.22 × 10−15 |
| RAD17 | 5 | + | 68665124-68665291 | 20 | 21 | 1 | 4.15 × 10−11 | 0 | 78 | −30.64 | 2.34 × 10−14 |
| ATXN7 | 3 | + | 63985126-63989134 | 7 | 12 | 12 | 5.93 × 10−11 | 0 | 77 | −31.51 | 3.56 × 10−14 |
| GON4L | 1 | − | 155734577-155736500 | 23 | 33 | 22 | 9.48 × 10−11 | 0 | 84 | −32.55 | 5.93 × 10−14 |
| PIP4K2C | 12 | + | 57995709-57997210 | 13 | 14 | 14 | 1.29 × 10−10 | 0 | 61 | −31.00 | 8.36 × 10−14 |
| ATP13A3 | 3 | − | 194123405-194126845 | 18 | 30 | 30 | 2.25 × 10−10 | 0 | 313 | −30.91 | 1.54 × 10−13 |
| ZNF641 | 12 | − | 48733795-48737510 | 5 | 8 | 8 | 1.70 × 10−09 | 0 | 49 | −32.63 | 1.34 × 10−12 |
| ZNF641 | 12 | − | 48741690-48741898 | 5 | 8 | 3 | 2.78 × 10−02 | 14 | 0 | 27.56 | 1.30 × 10−04 |
| AMPD3 | 11 | + | 10527255-10529126 | 15 | 16 | 16 | 1.51 × 10−08 | 0 | 80 | −32.15 | 1.33 × 10−11 |
| TCEAL4 | X | + | 102841582-102842653 | 5 | 6 | 6 | 3.54 × 10−08 | 0 | 107 | −29.05 | 3.34 × 10−11 |
| PRDM2 | 1 | + | 14104913-14109326 | 2 | 9 | 8 | 6.57 × 10−08 | 195 | 137 | 0.12 | 6.52 × 10−11 |
| PRDM2 | 1 | + | 14113010-14114573 | 2 | 9 | 9 | 6.57 × 10−08 | 0 | 59 | −31.18 | 6.52 × 10−11 |
| LRCH4 | 7 | − | 100171636-10017292 | 18 | 18 | 18 | 1.36 × 10−07 | 0 | 60 | −29.52 | 1.37 × 10−10 |
| MAP4 | 3 | − | 47892180-47894529 | 17 | 20 | 19 | 1.66 × 10−07 | 0 | 53 | −30.48 | 1.75 × 10−10 |
| MAP4 | 3 | − | 47892180-47894428 | 17 | 20 | 20 | 1.92 × 10−06 | 82 | 0 | 29.48 | 2.55 × 10−09 |
| MXD3 | 5 | − | 176732501-176734196 | 6 | 7 | 7 | 4.26 × 10−07 | 0 | 82 | −28.74 | 4.85 × 10−10 |
| SLC4A1 | 17 | − | 42325759-42327906 | 20 | 20 | 20 | 8.44 × 10−07 | 3390 | 1884 | −0.83 | 1.01 × 10−09 |
| LIPA | 10 | − | 91011464-91011660 | 12 | 12 | 2 | 8.84 × 10−07 | 0 | 78 | −28.02 | 1.09 × 10−09 |
| STRADA | 17 | − | 61780194-61781111 | 13 | 15 | 15 | 8.84 × 10−07 | 0 | 58 | −28.26 | 1.09 × 10−09 |
| LSM5 | 7 | − | 32529637-32530475 | 6 | 7 | 3 | 1.03 × 10−06 | 0 | 72 | −29.98 | 1.31 × 10−09 |
| ERI2 | 16 | − | 20807662-20810389 | 5 | 11 | 9 | 1.96 × 10−06 | 0 | 75 | −32.21 | 2.62 × 10−09 |
| SC5DL | 11 | + | 121177766-121184117 | 5 | 5 | 5 | 7.40 × 10−06 | 0 | 133 | −28.25 | 1.11 × 10−08 |
| PKN2 | 1 | + | 89298928-89301937 | 18 | 22 | 22 | 1.03 × 10−05 | 0 | 55 | −29.92 | 1.60 × 10−08 |
| CEACAM1 | 19 | − | 43011459-43013380 | 7 | 9 | 9 | 1.59 × 10−05 | 0 | 146 | −28.49 | 2.59 × 10−08 |
| NF2 | 22 | + | 30090741-30094583 | 3 | 18 | 18 | 1.88 × 10−05 | 0 | 162 | −33.73 | 3.16 × 10−08 |
| NF2 | 22 | + | 30070825-30070930 | 3 | 18 | 12 | 3.50 × 10−02 | 7 | 2 | 2.02 | 1.72 × 10−04 |
| PRKCB | 16 | + | 24231282-24231930 | 15 | 18 | 18 | 2.10 × 10−05 | 0 | 39 | −34.39 | 3.60 × 10−08 |
| CBL | 11 | + | 119170205-119178858 | 6 | 16 | 16 | 2.75 × 10−05 | 0 | 84 | −30.64 | 4.86 × 10−08 |
| TGFBR2 | 3 | + | 30732912-30735631 | 4 | 7 | 7 | 3.34 × 10−05 | 0 | 72 | −30.44 | 6.02 × 10−08 |
| TGFBR2 | 3 | + | 30713130-30713929 | 4 | 7 | 4 | 4.94 × 10−02 | 14 | 10 | 0.21 | 2.55 × 10−04 |
| AKAP2 | 9 | + | 112898407-112900819 | 2 | 3 | 2 | 3.34 × 10−05 | 44 | 80 | −0.05 | 6.06 × 10−08 |
| AKAP2 | 9 | + | 112930679-112934789 | 2 | 3 | 3 | 3.34 × 10−05 | 0 | 71 | −28.15 | 6.06 × 10−08 |
| UHRF1BP1 | 6 | + | 34840090-34845289 | 9 | 21 | 21 | 4.90 × 10−05 | 0 | 49 | −31.33 | 9.13 × 10−08 |
| SMCR7 | 17 | + | 18167098-18169093 | 4 | 5 | 5 | 1.03 × 10−04 | 0 | 23 | −29.44 | 2.02 × 10−07 |
| SMCR7 | 17 | + | 18167024-18169093 | 4 | 5 | 4 | 7.14 × 10−03 | 22 | 0 | 27.99 | 2.62 × 10−05 |
| NDUFV1 | 11 | + | 67374323-67374520 | 11 | 11 | 1 | 1.60 × 10−04 | 0 | 4 | −28.18 | 3.19 × 10−07 |
| BCOR | X | − | 39931602-39934433 | 10 | 15 | 3 | 1.76 × 10−04 | 0 | 43 | −29.87 | 3.54 × 10−07 |
| FGD4 | 12 | + | 32793210-32798982 | 7 | 17 | 17 | 1.90 × 10−04 | 0 | 90 | −31.24 | 3.85 × 10−07 |
| SRCAP | 16 | + | 30748370-30751449 | 2 | 29 | 29 | 2.13 × 10−04 | 36 | 0 | 29.81 | 4.36 × 10−07 |
| SRCAP | 16 | + | 30734905-30736403 | 2 | 29 | 21 | 2.13 × 10−04 | 0 | 22 | −30.64 | 4.36 × 10−07 |
| OAS1 | 12 | + | 113357194-113357711 | 8 | 8 | 7 | 2.42 × 10−04 | 0 | 29 | −29.09 | 5.02 × 10−07 |
| ETS2 | 21 | + | 40194598-40196876 | 8 | 9 | 9 | 2.52 × 10−04 | 0 | 171 | −30.13 | 5.35 × 10−07 |
| C7orf28A | 7 | + | 5965263-5965601 | 5 | 7 | 7 | 2.98 × 10−04 | 0 | 46 | −30.72 | 6.59 × 10−07 |
| PIGN | 18 | − | 59711460-59713212 | 23 | 31 | 31 | 3.07 × 10−04 | 0 | 54 | −29.06 | 6.89 × 10−07 |
| SELP | 1 | − | 169558090-169558699 | 15 | 17 | 17 | 3.28 × 10−04 | 0 | 35 | −29.54 | 7.39 × 10−07 |
| XRCC4 | 5 | + | 82648950-82649577 | 9 | 10 | 10 | 3.38 × 10−04 | 0 | 40 | −28.80 | 7.76 × 10−07 |
| SIGLEC14 | 19 | − | 52149510-52149893 | 7 | 7 | 2 | 3.40 × 10−04 | 0 | 84 | −28.80 | 7.87 × 10−07 |
| CDK12 | 17 | + | 37686884-37690799 | 8 | 15 | 15 | 3.75 × 10−04 | 0 | 38 | −31.39 | 8.82 × 10−07 |
| ATF3 | 1 | + | 212792388-212794114 | 4 | 5 | 4 | 4.11 × 10−04 | 0 | 73 | −27.83 | 9.78 × 10−07 |
| MYH11 | 16 | − | 15818745-15818849 | 4 | 16 | 13 | 4.53 × 10−04 | 0 | 21 | −29.20 | 1.09 × 10−06 |
| ABR | 17 | − | 906760-909409 | 9 | 22 | 18 | 4.96 × 10−04 | 0 | 36 | −30.39 | 1.22 × 10−06 |
| OLFM4 | 13 | + | 53624104-53626186 | 5 | 5 | 5 | 5.23 × 10−04 | 0 | 405 | −29.11 | 1.30 × 10−06 |
| HIVEP1 | 6 | + | 12120123-12126103 | 2 | 7 | 3 | 6.98 × 10−04 | 13 | 0 | 27.93 | 1.83 × 10−06 |
| HIVEP1 | 6 | + | 12163516-12165231 | 2 | 7 | 7 | 6.98 × 10−04 | 0 | 26 | −29.98 | 1.83 × 10−06 |
| MIDN | 19 | + | 1256994-1259139 | 3 | 7 | 7 | 6.98 × 10−04 | 0 | 239 | −31.82 | 1.82 × 10−06 |
| CEP63 | 3 | + | 134204575-134204894 | 15 | 19 | 1 | 9.33 × 10−04 | 0 | 38 | −29.07 | 2.51 × 10−06 |
| RREB1 | 6 | + | 7248744-7252212 | 6 | 14 | 14 | 9.34 × 10−04 | 0 | 69 | −31.98 | 2.52 × 10−06 |
| NCOA6 | 20 | − | 33328167-33331145 | 4 | 10 | 6 | 9.41 × 10−04 | 0 | 27 | −30.79 | 2.55 × 10−06 |
| NISCH | 3 | + | 52525888-52527087 | 18 | 21 | 21 | 1.02 × 10−03 | 0 | 34 | −28.34 | 2.79 × 10−06 |
| RNF111 | 15 | + | 59323003-59323901 | 11 | 13 | 1 | 1.08 × 10−03 | 0 | 29 | −28.45 | 3.00 × 10−06 |
| YLPM1 | 14 | + | 75264283-75266400 | 15 | 21 | 5 | 1.11 × 10−03 | 0 | 33 | −28.11 | 3.13 × 10−06 |
| PML | 15 | + | 74327513-74328734 | 10 | 13 | 10 | 1.12 × 10−03 | 0 | 39 | −30.59 | 3.18 × 10−06 |
| SLC41A1 | 1 | − | 205758221-205760846 | 3 | 11 | 11 | 1.33 × 10−03 | 0 | 37 | −30.47 | 3.85 × 10−06 |
| RNF19B | 1 | − | 33402052-33402910 | 10 | 11 | 10 | 1.40 × 10−03 | 0 | 43 | −27.66 | 4.13 × 10−06 |
| SLC17A9 | 20 | + | 61598689-61599941 | 6 | 13 | 13 | 1.42 × 10−03 | 0 | 53 | −29.47 | 4.21 × 10−06 |
| RIN3 | 14 | + | 93154271-93155332 | 3 | 8 | 8 | 1.59 × 10−03 | 21 | 37 | −0.01 | 4.88 × 10−06 |
| RIN3 | 14 | + | 93117927-93119420 | 3 | 8 | 4 | 3.01 × 10−02 | 0 | 62 | −29.47 | 1.45 × 10−04 |
| RTN4 | 2 | − | 5525222-55254621 | 8 | 10 | 3 | 1.90 × 10−03 | 0 | 29 | −29.39 | 5.89 × 10−06 |
| DHX34 | 19 | + | 47856011-47856992 | 16 | 17 | 2 | 2.05 × 10−03 | 0 | 42 | −29.13 | 6.37 × 10−06 |
| TMEM68 | 8 | − | 56651320-56652777 | 4 | 6 | 6 | 2.24 × 10−03 | 0 | 36 | −29.39 | 7.05 × 10−06 |
| SSH2 | 17 | − | 2795296-27959949 | 3 | 11 | 11 | 2.54 × 10−03 | 0 | 101 | −29.84 | 8.10 × 10−06 |
| FCGR2C | 1 | + | 161569420-161570031 | 3 | 5 | 5 | 3.18 × 10−03 | 33 | 39 | −0.01 | 1.04 × 10−05 |
| SLCO4C1 | 5 | − | 101569694-101572722 | 6 | 13 | 13 | 3.48 × 10−03 | 0 | 66 | −29.04 | 1.15 × 10−05 |
| IL21R | 16 | + | 27459855-27462115 | 5 | 10 | 10 | 3.63 × 10−03 | 0 | 27 | −29.51 | 1.22 × 10−05 |
| MED6 | 14 | − | 71050958-71051660 | 7 | 8 | 8 | 3.80 × 10−03 | 0 | 39 | −27.26 | 1.28 × 10−05 |
| STEAP4 | 7 | − | 87905750-87908943 | 4 | 5 | 5 | 4.35 × 10−03 | 0 | 93 | −28.18 | 1.50 × 10−05 |
| PRNP | 20 | + | 4667157-4667382 | 4 | 4 | 3 | 4.40 × 10−03 | 0 | 40 | −27.50 | 1.53 × 10−05 |
| PAPD7 | 5 | + | 6754867-6757161 | 2 | 9 | 9 | 5.59 × 10−03 | 20 | 0 | 27.14 | 1.99 × 10−05 |
| PAPD7 | 5 | + | 6754764-6757161 | 2 | 9 | 8 | 5.59 × 10−03 | 0 | 17 | −28.94 | 1.99 × 10−05 |
| KIF24 | 9 | − | 34255733-34257979 | 3 | 12 | 10 | 5.60 × 10−03 | 0 | 14 | −29.00 | 2.00 × 10−05 |
| TMCC3 | 12 | − | 94960900-94965513 | 4 | 4 | 4 | 6.85−03 | 0 | 293 | −27.77 | 2.51 × 10−05 |
| TMCC3 | 12 | − | 94975398-94976314 | 4 | 4 | 2 | 2.26 × 10−02 | 8 | 100 | 0.08 | 1.03 × 10−04 |
| C10orf18 | 10 | + | 5804992-5806943 | 4 | 15 | 15 | 7.20 × 10−03 | 0 | 32 | −30.92 | 2.66 × 10−05 |
| SMYD5 | 2 | + | 73452924-73454355 | 5 | 13 | 13 | 7.73 × 10−03 | 0 | 24 | −29.40 | 2.87 × 10−05 |
| EP400 | 12 | + | 132561946-132565004 | 2 | 27 | 27 | 8.76 × 10−03 | 54 | 17 | 0.15 | 3.34 × 10−05 |
| EP400 | 12 | + | 132445130-132446499 | 2 | 27 | 1 | 8.76 × 10−03 | 0 | 18 | −32.52 | 3.34 × 10−05 |
| FRY | 13 | + | 32869339-32870775 | 9 | 57 | 57 | 9.80 × 10−03 | 0 | 40 | −31.47 | 3.78 × 10−05 |
| NPIPL3 | 16 | − | 21426278-21426406 | 4 | 5 | 3 | 9.91 × 10−03 | 7 | 3 | 0.19 | 3.84 × 10−05 |
| NPIPL3 | 16 | − | 21436621-21436658 | 4 | 5 | 1 | 2.92 × 10−02 | 4 | 3 | −0.42 | 1.38 × 10−04 |
| IL1R1 | 2 | + | 102792813-102796333 | 4 | 10 | 10 | 1.06 × 10−02 | 0 | 107 | −29.14 | 4.12 × 10−05 |
| SLC35E3 | 12 | + | 69158484-69159851 | 4 | 5 | 5 | 1.09 × 10−02 | 0 | 33 | −26.93 | 4.27 × 10−05 |
| ELF2 | 4 | − | 140058784-140059021 | 10 | 11 | 2 | 1.09 × 10−02 | 0 | 22 | −28.48 | 4.28 × 10−05 |
| LUZP1 | 1 | − | -23410516-23415546 | 3 | 5 | 5 | 1.19 × 10−02 | 0 | 36 | −26.87 | 4.70 × 10−05 |
| MLH3 | 14 | − | 75513079-75516421 | 4 | 12 | 2 | 1.41 × 10−02 | 0 | 19 | −31.99 | 5.81 × 10−05 |
| VCAM1 | 1 | + | 101203679-101204599 | 9 | 9 | 9 | 1.50 × 10−02 | 0 | 51 | −26.73 | 6.20 × 10−05 |
| ADPRH | 3 | + | 119306311-119308791 | 3 | 5 | 5 | 1.67 × 10−02 | 0 | 53 | −26.88 | 7.13 × 10−05 |
| ALCAM | 3 | + | 105085713-105086325 | 15 | 16 | 1 | 1.81 × 10−02 | 0 | 34 | −27.99 | 7.88 × 10−05 |
| CHKB | 22 | − | 51019849-51019982 | 8 | 8 | 4 | 1.81 × 10−02 | 0 | 20 | −28.43 | 7.89 × 10−05 |
| PPIP5K1 | 15 | − | 43825660-43827674 | 20 | 36 | 36 | 1.84 × 10−02 | 0 | 24 | −27.56 | 8.10 × 10−05 |
| CLEC5A | 7 | − | 141627157-141630018 | 6 | 7 | 7 | 1.87 × 10−02 | 0 | 252 | −28.64 | 8.27 × 10−05 |
| MICAL3 | 22 | − | 18342922-18347752 | 5 | 28 | 21 | 1.95 × 10−02 | 35 | 25 | −0.06 | 8.81 × 10−05 |
| LRP12 | 8 | − | 105501459-105503767 | 3 | 7 | 7 | 2.08 × 10−02 | 0 | 26 | −28.17 | 9.40 × 10−05 |
| SRGAP2 | 1 | + | 206634382-206637782 | 15 | 21 | 21 | 2.13 × 10−02 | 0 | 21 | −28.97 | 9.64 × 10−05 |
| GGCX | 2 | − | 85776193-85777249 | 12 | 15 | 15 | 2.32 × 10−02 | 0 | 25 | −27.44 | 1.06 × 10−04 |
| SLC8A1 | 2 | − | 40339287-40342769 | 2 | 7 | 7 | 2.49 × 10−02 | 26 | 29 | −0.34 | 1.16 × 10−04 |
| SLC8A1 | 2 | − | 40655613-40657444 | 2 | 7 | 1 | 2.49 × 10−02 | 0 | 41 | −28.24 | 1.16 × 10−04 |
| DHRS4 | 14 | + | 24437966-24438486 | 7 | 8 | 8 | 2.70 × 10−02 | 0 | 22 | −26.85 | 1.26 × 10−04 |
| VPS13A | 9 | + | 80030872-80032397 | 22 | 72 | 72 | 2.74 × 10−02 | 0 | 14 | −31.43 | 1.28 × 10−04 |
| HLA-DPB1 | 6 | + | 33054316-33054976 | 6 | 6 | 6 | 2.81 × 10−02 | 151 | 368 | −0.91 | 1.33 × 10−04 |
| SKA2 | 17 | − | 57232317-57232800 | 5 | 5 | 2 | 2.81 × 10−02 | 0 | 26 | −27.22 | 1.32 × 10−04 |
| METTL4 | 18 | − | 2537525-2539144 | 8 | 9 | 9 | 2.95 × 10−02 | 0 | 19 | −27.47 | 1.41 × 10−04 |
| CRAMP1L | 16 | + | 1705878-1706993 | 2 | 13 | 5 | 2.96 × 10−02 | 7 | 0 | 28.86 | 1.42 × 10−04 |
| CRAMP1L | 16 | + | 1723882-1727907 | 2 | 13 | 13 | 2.96 × 10−02 | 0 | 25 | −31.99 | 1.42 × 10−04 |
| IKBKG | X | + | 153792534-153793260 | 10 | 11 | 11 | 3.01 × 10−02 | 0 | 20 | −29.87 | 1.45 × 10−04 |
| LEPR | 1 | + | 65886248-65886423 | 20 | 22 | 1 | 3.34 × 10−02 | 0 | 16 | −28.60 | 1.63 × 10−04 |
| XPNPEP3 | 22 | + | 41322273-41323878 | 6 | 10 | 10 | 3.50 × 10−02 | 0 | 24 | −28.94 | 1.72 × 10−04 |
| CTSH | 15 | − | 79237293-79237420 | 13 | 14 | 1 | 4.08 × 10−02 | 0 | 25 | −27.42 | 2.03 × 10−04 |
| BEST1 | 11 | + | 61731576-61731939 | 3 | 9 | 9 | 4.40 × 10−02 | 0 | 75 | −27.40 | 2.21 × 10−04 |
| C11orf48 | 11 | − | 62439036-62439241 | 7 | 7 | 1 | 4.58 × 10−02 | 0 | 16 | −28.26 | 2.34 × 10−04 |
Differential exons comparison was done testing 81 564 exons from 9069 genes. The table illustrates the complete list of 423 exons from 350 genes found to be differentially used at the 0.05 False Discovery Rate (FDR) level (ie, adjusted P value less than .05) in the SF3B1 mutant relative to normal groups.
Figure 3.
RNA-Seq analysis shows differential exon usage of MDS-related genes. (A-D) A total of 81 564 exons were tested in 9069 genes for differential exon usage between normal and mutant groups. A total of 48 835 exons were excluded, being not testable or present in genes with less than 2 testable exons. A total of 423 exons from 350 genes were found to be differentially used at the 0.05 false discovery rate level (ie, adjusted P < .05; Table 2 shows the complete list). A total of 282 of these 423 exons (67%) had higher usage in the mutant group relative to a healthy person. A total of 56 genes contained more than one significant exon. Screenshots of fitted exon counts indicate that 4 genes, specifically ASXL1, CBL, EZH1, and RUNX3, contained at least 1 exon showing differential usage between SF3B1 mutants (red) and normals (blue). Positive (+) and negative (−) signs indicate the strand, giving the 3′ and 5′ ends of the gene. Area in magenta represents the differentially used exon.
Discussion
RS occurs because of abnormal localization of crystalline iron deposits around the nucleus of erythroid precursors. The presence of ≥ 15% RSs of nucleated erythroid precursors in the BM is one of the defined diagnostic criteria of RARS and RARS-T.25 These iron deposits are not confined to RARS and RARS-T, and are present in other diseases, especially congenital sideroblastic anemia. The biologic basis of RS in erythroid precursors is still unknown, even though accumulation of iron has been associated with alterations of the mitochondria and iron overloading. It has been speculated that the accumulated iron in sideroblastic anemia is in the ferric form. Physiologically, the enzymes involved in normal iron exchange within the mitochondria, such as ferrochelatase, only accept ferrous iron. Thus, the change in the biochemical status of iron can lead to an abnormal accumulation of iron and possibly iron overload.26,27
We demonstrate here the direct consequences of SF3B1 dysfunction on the disease phenotype. Electron microscopy is a sensitive and specific technique for the direct visualization of iron in biologic tissues. Furthermore, the concomitant application of energy dispersive X-ray analysis with electron microscopy permits analysis of iron composition and elemental status, which allows for the accurate detection of abnormal iron distribution and iron composition in various diseases.26 Indeed, RARS patients carrying SF3B1 mutations seemed to present with conspicuous iron particles as evidenced by analysis of the BM ultrastructure. It is possible that SF3B1 mutations might generate alterations in the chemical status of iron leading to a pathologic iron accumulation and other disease-related features. The further analysis of the transcriptome by RNA-Seq tool provided new information about MDS, and specifically RARS pathophysiology. This finding was supported by the RNA signature we defined by RNA-Seq, showing that the expression levels of genes associated with poor prognosis specifically ASXL1 and CBL were not changed. Second, we did not observe any differences in exon usage or gene expression in genes of the mitochondrial pathway purportedly linked in the pathogenesis of RARS, such as ALAS2, ABCB7, SLC25A38, and FTMT. This observation supports the distinct biologic difference between hereditary and acquired sideroblastic anemia, where mutations or defects in these mitochondrial genes are common.
In support of this, none of the known mitochondrial gene sets showed over- or under-representation of significant exon usage in total BM cells derived from SF3B1 mutant patients. Genes of the mitochondrial pathways, such as ALAS2 and ABCB7, have been previously described to be up-regulated and down-regulated, respectively, in RARS and RARS-T compared with healthy persons in CD34+ cells.12,28 Our RNA-Seq analysis was done on unfractionated BM cells because an adequate amount of RNA is required to perform the analysis. New emerging platforms of RNA-Seq are becoming available and may provide us the ability to investigate the RNA splicing patterns in CD34+ cells of RARS and RARS-T patients.
Third, we found that SF3B1 mutant patients showed an over-representation of differentially used exons that were the most 3′, that is, last 5′ to 3′ exon order among exons with any mapped reads (P = 2.10 × 10−7, 445 3′ significant exons of 810 significant exons, with 8668 3′ exons tested), with nonsignificant evidence for over-representation of the most 5′ exons. Finally, it also supports the idea that SF3B1 mutations may be founder mutations that can promote subsequent acquisition of additional genetic or pathway abnormalities, leading to the final disease phenotype. The analysis of RNA derived from clones of K562 cells expressing low level of SF3B1 by RT-PCR indicated that SF3B1 haploinsufficiency mainly affects U2-type introns. When we analyzed the phenotype of K562 with decreased SF3B1 expression by lentiviral infection, we could not recapitulate the RS phenotype. The absence of RS may be attributed to several factors. The K562 cell line is an immortalized myelogenous leukemia cell line derived from a patient with chronic myeloid leukemia in blast crisis. It is possible that the patency of additional genes or downstream pathways is necessary for the final formation of RS in patients with SF3B1 mutations and that these pathways may be altered in K562 cell lines. Thus, other cell lines may be more appropriate models. However, the phenotypic features of RS are confined to erythroid precursors that are not readily seen in other cell lines. It is also possible that more complete knockdown of SF3B1 may be necessary to achieve the RS phenotype. On the other hand, when we used meayamycin, a pharmacologic inhibitor of SF3b factors, we were able to induce the presence of RS in healthy BM cells, supporting the hypothesis that SF3B1 may have relevant function in RS formation. In support to this hypothesis is the finding of RS in the BM of Sf3b1+/− mice. Sf3b1+/− have not been associated with any hematologic phenotype but rather to skeletal abnormalities.14
The clinical impact of alterations in SF3B1 is most apparent in RARS/ RARS-T patients. However, the clinical heterogeneity of patients with these 2 disease subtypes is common. It is therefore conceivable that biologic and clinical differences between SF3B1 haploinsufficient patients and those with point mutations in SF3B1 may exist. Multiple molecular mutations involving other relevant pathways can occur in the same MDS patient. It is possible that these additional mutations, including those involving the polycomb trithorax complex, signaling pathways, transcription factors, and even other splicing factor genes, may contribute to the variability in the level of expression of SF3B1 within SF3B1 mutant cases and differences in the final disease phenotype and outcome. Similarly, it is also possible that the type of molecular mutations (ie, amino acid substitutions at codons 700, 662, 666, and others) may confer different effects on the final SF3B1 expression. The upstream and downstream genes that modulate SF3B1 expression in MDS and its effect on hematopoiesis are also not completely understood and may contribute to this difference.
We also present updated survival data on the clinical significance of SF3B1 mutations in MDS. So far, data regarding the survival impact of SF3B1 mutations in MDS are conflicting, with some groups demonstrating better survival2,21 and others showing no influence on overall survival.29–31In our hands,20 RARS and RARS-T patients carrying SF3B1 mutations portend a better clinical outcome similar to the findings of Papaemmanuil et al.2
Our current study, together with other recent discoveries, reinforces the idea that splicing factors, especially SF3B1, might be key regulators in the pathogenesis of RARS and RARS-T. We previously reported that SF3B1 is, so far, the only SF3b component to be frequently mutated in MDS and related disorders.1 No mutations were detected in other SF3b components and targets, such as SF3B14, SF3B4, and DYRK1A.1,32 In conclusion, SF3B1 is a commonly mutated gene in acquired sideroblastic anemia, such as RARS and RARS-T. We also show the first experimental evidence that haploinsufficiency or inhibition of SF3B1 leads to the formation of RS.
Supplementary Material
Acknowledgments
The authors thank Dr Velizar Shivarov of Laboratory of Hematopathology and Immunology, National Hematology Hospital, Sofia, Bulgaria, for reading the manuscript and providing valuable comments as well as Edy Hasrouni for preparing the figures.
This work was supported in full or in part by the Cleveland Clinic Seed Support, MDS Foundation (YIA grant), AA-MDS International Foundation (YIA grant, R.V.T.), the National Institutes of Health (grant R01 HL082983, J.P.M.; grant U54 RR019391, J.P.M. and M.A.S.; grant K24 HL077522, J.P.M.; grant R01 CA120792, K.K.; and grant RO1 GM093074, R.A.P.), and the Robert Duggan Cancer Research Foundation (charitable donation).
Footnotes
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: V.V. and R.V.T. conceived and designed the study, analyzed the data, and wrote the manuscript; H.J.R. analyzed clinical pathology data; J.M. performed transmission electron microscopy; J.B. analyzed RNA-Seq; V.V., J.S., F.T., H.M., H.S., A. Jankowska, and A. Jerez performed experiments; M.B. analyzed data; C.O. edited the manuscript; M.A.S., Y.S., A.S.A., and E.C. contributed to patient samples and edited the manuscript; R.A.P. edited the manuscript; H.K. and K.I. provided BM aspirates of Sf3b1 heterozygous mice; S.O. and K.K. synthesized meayamycin; and J.P.M. provided patient samples, contributed to the design of the project, provided substantial scientific advice, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ramon V. Tiu, Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, Cleveland Clinic, 9500 Euclid Ave, R40, Cleveland, OH 44195; e-mail: tiur@ccf.org.
References
- 1.Visconte V, Makishima H, Jankowska A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26(3):542–545. doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 4.Makishima H, Visconte V, Sakaguchi H, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119(14):3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Will CL, Schneider C, MacMillan AM, et al. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 2001;20(16):4536–4546. doi: 10.1093/emboj/20.16.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marz M, Kirsten T, Stadler PF. Evolution of spliceosomal snRNA genes in metazoan animals. J Mol Evol. 2008;67(6):594–607. doi: 10.1007/s00239-008-9149-6. [DOI] [PubMed] [Google Scholar]
- 7.Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220(2):152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mufti GJ, Bennett JM, Goasguen J, et al. Diagnosis and classification of myelodysplastic syndrome: International Working Group on Morphology of Myelodysplastic Syndrome (IWGM-MDS) consensus proposals for the definition and enumeration of myeloblasts and ring sideroblasts. Haematologica. 2008;93(11):1712–1717. doi: 10.3324/haematol.13405. [DOI] [PubMed] [Google Scholar]
- 9.Cazzola M, Invernizzi R. Ring sideroblasts and sideroblastic anemias. Haematologica. 2011;96(6):789–792. doi: 10.3324/haematol.2011.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall J, Foucar K. Diagnosing myelodysplastic/myeloproliferative neoplasms: laboratory testing strategies to exclude other disorders. Int J Lab Hematol. 2010;32(6):559–571. doi: 10.1111/j.1751-553X.2010.01251.x. [DOI] [PubMed] [Google Scholar]
- 11.Pellagatti A, Cazzola M, Giagounidis AA, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006;108(1):337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]
- 12.Boultwood J, Pellagatti A, Nikpour M, et al. The role of the iron transporter ABCB7 in refractory anemia with ring sideroblasts. PLoS One. 2008;3(4):e1970. doi: 10.1371/journal.pone.0001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albert BJ, Sivaramakrishnan A, Naka T, Czaicki NL, Koide K. Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J Am Chem Soc. 2007;129(9):2648–2659. doi: 10.1021/ja067870m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isono K, Mizutani-Koseki Y, Komori T, Schmidt-Zachmann MS, Koseki H. Mammalian polycomb-mediated repression of Hox genes requires the essential spliceosomal protein Sf3b1. Genes Dev. 2005;19(5):536–541. doi: 10.1101/gad.1284605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 16.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini YaH, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995:289–300. [Google Scholar]
- 18.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visconte V, Makishima H, Jankowska A, et al. Association of SF3B1 with ring sideroblasts in patients, in vivo, and in vitro models of spliceosomal dysfunction [abstract]. Blood (ASH Annual Meeting Abstracts) 2011;118:457. [Google Scholar]
- 21.Malcovati L, Papaemmanuil E, Bowen DT, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24):6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osman S, Albert BJ, Wang Y, Li M, Czaicki NL, Koide K. Structural requirements for the antiproliferative activity of pre-mRNA splicing inhibitor FR901464. Chem Eur J. 2011;17(3):895–904. doi: 10.1002/chem.201002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaida D, Motoyoshi H, Tashiro E, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 24.Albert BJ, McPherson PA, O'Brien K, et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol Cancer Ther. 2009;8(8):2308–2318. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazzola M, Malcovati L, Invernizzi R. Myelodysplastic/myeloproliferative neoplasms. Hematology Am Soc Hematol Educ Program. 2011;2011:264–272. doi: 10.1182/asheducation-2011.1.264. [DOI] [PubMed] [Google Scholar]
- 26.Grasso JA, Myers TJ, Hines JD, Sullivan AL. Energy-dispersive X-ray analysis of the mitochondria of sideroblastic anaemia. Br J Haematol. 1980;46(1):57–72. doi: 10.1111/j.1365-2141.1980.tb05935.x. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg PL, Young NS, Gattermann N. Myelodysplastic syndromes. Hematology Am Soc Hematol Edu Program. 2002;2002:136–161. doi: 10.1182/asheducation-2002.1.136. [DOI] [PubMed] [Google Scholar]
- 28.Malcovati L, Della Porta MG, Pietra D, et al. Molecular and clinical features of refractory anemia with ringed sideroblasts associated with marked thrombocytosis. Blood. 2009;114(17):3538–3545. doi: 10.1182/blood-2009-05-222331. [DOI] [PubMed] [Google Scholar]
- 29.Damm F, Thol F, Kosmider O, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26(5):1137–1140. doi: 10.1038/leu.2011.321. [DOI] [PubMed] [Google Scholar]
- 30.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 31.Patnaik MM, Lasho TL, Hodnefield JM, et al. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012;119(2):569–572. doi: 10.1182/blood-2011-09-377994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Graaf K, Czajkowska H, Rottmann S, et al. The protein kinase DYRK1A phosphorylates the splicing factor SF3b1/SAP155 at Thr434, a novel in vivo phosphorylation site. BMC Biochem. 2006;7:7. doi: 10.1186/1471-2091-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.