Abstract
BACKGROUND:
Organisms expressing Klebsiella pneumoniae carbapenemase (KPC) are found in several regions worldwide but are rarely detected in Canada. The first outbreak of KPC-expressing strains of Enterobacteriaceae clinical isolates in a university-affiliated hospital intensive care unit (ICU) in Canada is described.
METHODS:
Enterobacteriaceae isolates that were flagged by the Vitek 2 (bioMérieux, France) system as possible carbapenemase producers were subjected to the modified Hodge test. Modified Hodge test-positive organisms were analyzed by pulsed-field gel electrophoresis, tested for KPC and other beta-lactamase genes by polymerase chain reaction analysis and underwent subsequent nucleic acid sequencing. Antimicrobial susceptibility profiles were determined by Vitek 2 and Etest (bioMérieux, France). A chart review was conducted to establish epidemiological links.
RESULTS:
During the study period, 10 unique Enterobacteriaceae isolates expressing KPC were detected from nine ICU patients. Five patients had infections (three pneumonias, one surgical site infection, one urinary tract infection). Isolates included Escherichia coli (5), Klebsiella oxytoca (2), Serratia marcescens (2) and Citrobacter freundii (1). Polymerase chain reaction analysis and sequencing confirmed the presence of KPC-3 in all isolates; four also carried TEM, two CTX-M and one CMY-2. The imipenem minimum inhibitory concentrations as determined by Etest ranged from 0.75 μg/mL to ≥32 μg/mL. Pulsed field gel electrophoresis clonal patterns and patient location in the ICU revealed presumptive horizontal transmission events.
CONCLUSIONS:
In the present study, Enterobacteriaceae isolates with KPC are emerging and can result in serious infections. The KPC gene can spread via plasmids to different genera of the Enterobacteriaceae family. The dissemination of KPC in Enterobacteriaceae and the consequences for treatment and infection control measures warrant a high degree of vigilance among clinicians and microbiologists.
Keywords: Beta-lactamases KPC, Carbapenemase, Disease outbreaks
Abstract
HISTORIQUE :
On trouve des organismes producteurs de carbapénèmases de type Klebsiella pneumoniae (KPC) dans diverses régions du monde, mais rarement au Canada. Les auteurs décrivent la première flambée de souches exprimant le KPC dans des isolats cliniques d’entérobactéries prélevés à l’unité de soins intensifs (USI) d’un hôpital universitaire du Canada.
MÉTHODOLOGIE :
Les chercheurs ont soumis au test de Hodge modifié les isolats d’entérobactéries que le système Vitek 2 (bioMérieux, France) signalait comme de possibles producteurs de carbapénèmases. Les chercheurs ont analysé les organismes positifs au test de Hodge par électrophorèse sur gel en champ pulsé, ont vérifié la présence de KPC et d’autres gènes de bêta-lactamase par analyse de la réaction en chaîne de la polymérase et ont ensuite effectué un séquençage de l’acide nucléique. Ils ont déterminé les profils de susceptibilité antimicrobienne par le système Vitek 2 et le test E (bioMérieux, France), puis procédé à une analyse des dossiers pour établir des liens épidémiologiques.
RÉSULTATS :
Pendant la période de l’étude, les chercheurs ont décelé dix isolats uniques d’entérobactéries productrices de KPC chez neuf patients de l’USI. Cinq patients avaient une infection (trois pneumonies, une infection au foyer d’une opération, une infection urinaire). Les isolats incluaient l’Escherichia coli (5), le Klebsiella oxytoca (2), le Serratia marcescens (2) et le Citrobacter freundii (1). L’analyse et le séquençage de la réaction en chaîne de la polymérase ont confirmé la présence de KPC-3 dans tous les isolats. Quatre contenaient un TEM, deux, un CTX-M, et un, un CMY-2. Les concentrations inhibitrices minimales de l’imipénem déterminées par test E variaient entre 0,75 μg/mL et au moins 32 μg/mL. Les schémas clonaux de l’électrophorèse en champ pulsé et la présence du patient à l’USI révélaient des événements présomptifs de transmission horizontale.
CONCLUSIONS :
Dans la présente étude, les isolats d’entérobactéries productrices de KPC sont émergents et peuvent donner lieu à de graves infections. Le gène de KPC peut se propager par les plasmides à divers genres de la famille des entérobactéries. La dissémination du KPC dans les entérobactéries et les conséquences pour les traitements et les mesures de contrôle des infections justifient un degré élevé de vigilance chez les cliniciens et les microbiologistes.
Klebsiella pneumoniae carbapenemase (KPC) belongs to the Ambler class A β-lactamase group and is capable of hydrolyzing all β-lactam antibiotics. KPC is endemic in the eastern United States and is emerging internationally since its characterization in 2001 (1,2). KPC producers may be missed using routine phenotypic tests, which can lead to inappropriate antibiotic prescriptions and delays in infection control interventions. The the blaKPC gene, which is flanked by transposon Tn4401 and located on conjugative plasmids, is most frequently found in Klebsiella pneumoniae and is also transferable among genera in the Enterobacteriaceae family and even Pseudomonas species (3,4). KPC-producing K pneumoniae has been reported in Canada as isolated cases but remains rare (5,6). To date, other Enterobacteriaceae-expressing KPC have not been reported in Canada. In the present study, we describe the first Canadian outbreak of KPC-3-producing Enterobacteriaceae (KPC-Ent) isolates from patients in an intensive care unit (ICU).
METHODS
The affected medical/surgical ICU has 25 single patient rooms and shared medical and nursing staff. Infection control screening strategies have been implemented for methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE) and Clostridium difficile, but active screening for carbapenemase-producing organisms did not exist at the time of the study. On December 8, 2009, an endotracheal tube aspirate from an ICU patient contained Escherichia coli resistant to all β-lactams but susceptible to amikacin and nitrofurantoin. The Vitek 2 (bioMérieux, France) advanced expert system provided the comment, “ESBL + carbapenemase (metallo- or KPC)”.
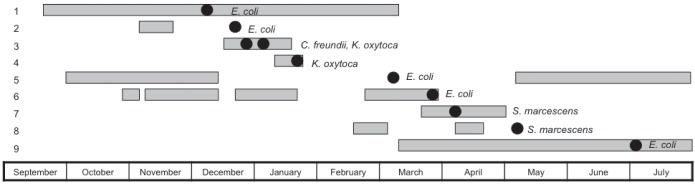
The modified Hodge test (MHT) was used to phenotypically determine carbapenemase production (7). A customized algorithm was added to Vitek 2 to alert technologists to perform the MHT if Enterobacteriaceae isolates had imipenem minimum inhibitory concentrations (MICs) ≥2 μg/mL (the AST085 card that incorporates imipenem as the only carbapenem was used) or were resistant to all β-lactams except imipenem. Using the laboratory information system, no additional isolates were found in the six months before this first isolate. Clinical data were recorded using standardized collection forms. To establish an epidemiological link, each patient’s length of stay in the ICU was plotted with an indicator marking when the first isolate was identified (Figure 1). The Centers for Disease Control and Prevention (CDC, Atlanta, Georgia, USA) surveillance definitions for classification of infections, were used (8). Species identification and antibiotic susceptibility were performed using Vitek 2. Additional MICs for colistin, tigecycline, meropenem and imipenem (9) were determined using Etest (bioMérieux, France). Isolates were further characterized by sequencing the blaKPC gene, and pulsed-field gel electrophoresis (PFGE). The blaKPC gene was detected using polymerase chain reaction primers, which were previously described and encompassed the entire coding region resulting in an amplicon of 893 bp (10). The amplified product was purified using a MinElute purification kit (Qiagen Inc, Germany) and underwent bidirectional sequencing (Applied Biosystems, USA). Internal primers were included for sequencing blaKPC (11). BLAST software from the National Center for Biotechnology Information (USA) was used for sequence identity. PFGE was performed after XbaI restriction enzyme digestion (Figure 2).
Figure 1).
Timeline depicting patient length of stay in the intensive care unit. Each horizontal gray bar represents length of stay, with bars on the same y-axis representing the same patient. Some patients were admitted to the intensive care unit more than once during the hospitalization. The black ovals indicate the date the Enterobacteriaceae isolate was obtained from the patient. Patient 3 has two isolates Citrobacter freundii and Klebsiella oxytoca. Patients 1 and 2 had Escherichia coli pulsovar A. Patients 3 and 4 had K oxytoca pulsovar D. Patient 5 had E coli pulsovar B. Patients 6 and 9 had E coli pulsovars D and D1 respectively. Patients 7 and 8 had Serratia marcescens pulsovars A
Figure 2).
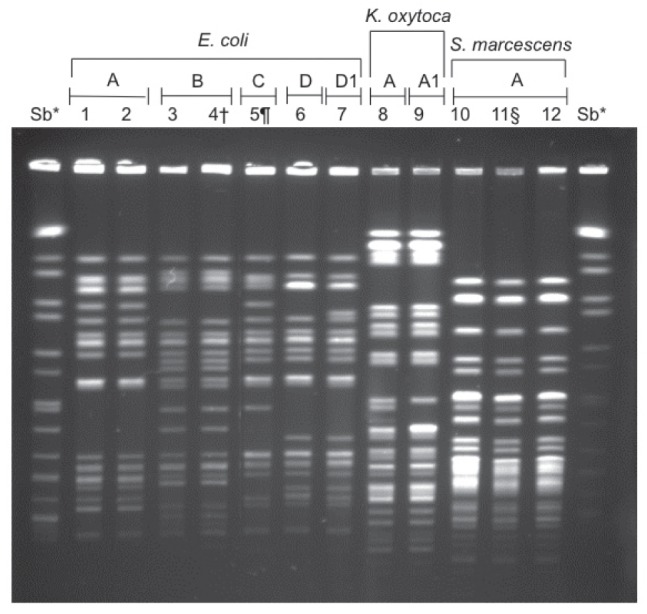
Macrorestriction analysis (XbaI) by pulsed-field gel electrophoresis of Enterobacteriaceae isolates. *Salmonella braenderup molecular mass marker; †Isolate was obtained from same patient (lane 3) four days later; §Isolate was obtained from same patient (lane 1) 72 days later; ¶Isolate was obtained from same patient (lane 10) seven days later. E Escherichia; K Klebsiella; S Serratia
RESULTS
Between December 2009 and July 2010, 10 unique MHT-positive Enterobacteriaceae isolates from nine ICU-hospitalized patients were identified. A summary of the clinical characteristics of the case patients is shown in Table 1. Although not all patients were in the ICU simultaneously, each case patient’s ICU hospitalization overlapped with several other case patients (Figure 1). A summary of the isolates, types of infection and antibiotic susceptibilities are shown in Table 2. The complete DNA sequences of the blaKPC gene and deduced amino acid sequences were 99.9% to 100% identical to that of K pneumonia CL-5761 containing the blaKPC-3 allele. Clonal diversity was interpreted using published criteria for PFGE and results showed four E coli, one Klebsiella oxytoca and one Serratia marcescens clonal group(s) (12).
TABLE 1.
Demographic and clinical characteristics of patients infected or colonized with Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae
| Variable | Result |
|---|---|
| Age, years, mean (range) | 65 (40–88) |
| Male sex | 6 |
| Patients admitted to the intensive care unit from: | |
| Another hospital | 4 |
| Another ward | 1 |
| Community | 4 |
| Charlson comorbidity index, mean ± SD | 6.8±2.8 |
| Age-adjusted Charlson comorbidity index, mean ± SD | 8.8±2.1 |
| Length of hospital stay before isolation of KPC-Ent, days, mean ± SD | 100.7±84.2 |
| Length of ICU stay before first isolation of KPC-Ent, days, mean ± SD | 58.2±52.4 |
| Patients colonized or infected with other multidrug-resistant organisms | |
| Methicillin-resistant Stayphlococcus aureus | 4 |
| Vancomycin-resistant Enterococcus species | 3 |
| Clostridium difficile | 1 |
| Patients who had surgeries before isolation of KPC-Ent | 7 |
| Patients with invasive devices | |
| Central venous catheters | 9 |
| Hemodialysis catheters | 7 |
| Endotracheal tubes | 9 |
| Foley catheters | 7 |
| Intra-abdominal drains | 6 |
| Intrathroracic drains | 2 |
| Patients who received antibiotics within 12 weeks before isolation of KPC-Ent | |
| None | 0 |
| Aminoglycosides | 3 |
| Cephalosporins | 3 |
| Carbapenems | 7 |
| Penicillins | 3 |
| Penicillins/β-lactam inhibitors | 8 |
| Quinolones | 7 |
| Trimethroprim/sulfamethoxazole | 1 |
| Colistin | 0 |
| Tigecycline | 1 |
| Patients who died, n (crude mortality %) | 4 (44.4) |
Data presented as n unless otherwise indicated. KPC-Ent Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. ICU Intensive care unit
TABLE 2.
Characteristics of Enterobacteriaceae isolates producing Klebsiella pneumoniae carbapenemase type 3 isolated from an intensive care unit outbreak
| Organism | Source | Infection |
Etest*
|
Vitek 2*
|
||||
|---|---|---|---|---|---|---|---|---|
| TC | CO | MP | IP | IP | Susceptible antibiotics | |||
| Escherichia coli | ETT | Pneumonia | 0.75 | 1.5 | 0.5 | 0.75 | 2 | Amikacin, nitrofurantoin |
| Escherichia coli | Jejunum | Surgical site infection | 0.38 | 1.5 | 0.5 | 0.75 | 4 | Amikacin, nitrofurantoin |
| Citrobacter freundii | ETT | Colonization | 0.5 | 1 | 0.5 | 0.75 | 8 | Ceftriaxone, cefepime, amikacin, tobramycin, nitrofurantoin |
| Klebsiella oxytoca | ETT | Colonization | 0.5 | 1 | 0.75 | 1 | ≥16 | Cefoxitin, amikacin, gentamicin, tobramycin, trimethoprim/sulfamethoxazole |
| Klebsiella oxytoca | ETT | Pneumonia | 1.5 | 1.5 | 0.75 | 0.75 | ≥16 | Cefoxitin, amikacin, gentamicin, tobramycin, trimethoprim/sulfamethoxazole |
| Escherichia coli | Urine | Colonization | 0.75 | 2 | 0.5 | 0.75 | ≥1 | Amikacin, nitrofurantoin |
| Escherichia coli | Urine | Colonization | 0.5 | 1.5 | 1.5 | 3 | 4 | Amikacin, nitrofurantoin, trimethoprim/sulfamethoxazole |
| Serratia marcescens | Trachea | Pneumonia | 1 | 256 | >32 | >32 | ≥16 | Amikacin, gentamicin, ceftriaxone |
| Serratia marcescens | Urine | Urinary tract infection | 3 | 256 | >32 | >32 | ≥16 | Amikacin, gentamicin |
| Escherichia coli | ETT | Colonization | 0.38 | 1.5 | 1 | 0.38 | 2 | Amikacin, gentamicin, tobramycin, nitrofurantoin |
bioMérieux, France. CO Colistin; ETT Endotracheal tube aspirate; IP Imipenem; MP Meropenem; TC Tigeycline
DISCUSSION
To the best of our knowledge, this is the first outbreak of KPC-Ent in Canada. To have multiple genera involved in this outbreak is concerning. Horizontal transmission is suggested by the epidemiological link (Figure 1) and clonal clusters in four pairs of patients (Figure 2). Although the outbreak involved multiple genera, PFGE was the only molecular typing method used. Interestingly, the E coli isolates in lanes 1 and 6 belong to a different pulsovar (Figure 2) and were isolated from the same patient 72 days apart. This strongly suggests in vivo intra-species transfer of the blaKPC gene. In vivo transfer of the blaKPC gene between K pneumoniae isolates from the same patient has been recently documented (13). There is also evidence for interspecies conjugative transfer of the blaKPC gene (14). We hypothesize that this outbreak has resulted from ongoing horizontal transfer of Enterobacteriaceae with subsequent in vivo interspecies transfer of the blaKPC-3 gene. Plasmid and transposon characterization and multilocus sequence typing were not performed to further characterize genetic relatedness. Previous studies have demonstrated remarkable diversity of the molecular features of KPC genes with plasmids differing in size and incompatibility groups. Future epidemiological investigations of KPC producers should use different molecular epidemiological approaches to characterize transmission with more certainty.
The antibiotic susceptibility patterns of the 10 isolates are shown in Figure 2. Vitek 2 susceptibilities were different than the Etest results for imipenem. We would have misclassified eight of the isolates as susceptible using Etest, whereas five isolates would have been misclassified as susceptible using Vitek 2 using the Clinical and Laboratory Standards Institute (CLSI) 2009 breakpoints for Enterobacteriaceae (15). In June 2010, the CLSI revised the carbapenem breakpoints for Enterobacteriaceae (16). The CLSI revisions are based on evaluation of pharmokinetic and pharmacodynamic data, clinical data and MIC distributions of carbapenemase-producing strains. The breakpoints were lowered by two doubling dilutions for meropenem, imipenem and doripenem, and by three doubling dilutions for ertapenem. Although the use of lower breakpoints decreases the chance of falsely misclassifying Enterobacteriaceae isolates, it still has the potential to result in major errors. Using these new breakpoints, the imipenem and meropenem Etest would have misclassified seven of the isolates as susceptible, while Vitek 2 would have misclassified only one isolate as susceptible. However, this observation is based on the Vitek 2 AST085 card, which only has imipenem.
The CLSI 2010 supplemental update for the Performance Standards for Antimicrobial Susceptibility Testing (16) states that if a laboratory uses new interpretive criteria, the MHT does not need to be performed other than for epidemiological or infection control purposes. If we had only relied on the revised breakpoints recommended by the most recent CLSI, we would have missed one or seven cases depending on the method and antibiotics used for susceptibility testing. This would have prevented the early implementation of infection control measures. Based on our limited experience and the general availability of tests in hospital-based microbiology laboratories, we recommend that laboratories in Canada continue to use the MHT on Enterobacteriaceae isolates with elevated breakpoints to carbapenems. If positive by MHT, further testing at a reference laboratory for the underlying resistance mechanism is warranted because the prevalence of KPCs in Canada is believed to be low.
The possibility of major errors in detecting KPCs using automated susceptibility systems including Vitek 2 has previously been reported and is believed to be a result of an inoculum effect and porin changes affecting carbapenem permeability (17). There are little data on the ability of Etest to consistently detect KPC-Ent. Our isolates also contained other β-lactamases (data not shown), which may have contributed to the variable susceptibilty profile. Data suggest that ertapenem MICs may be the most sensitive, but meropenem and imipenem MICs are more specific for KPC detection (18,19).
Only one patient in our outbreak received tigecycline for a VRE infection before the KPC-Ent isolation. Interestingly, S marcescens colonizing the urinary tract had an MIC of 3 μg/mL. The lower levels of tigecycyline in the urinary tract may have had a selective pressure effect. None of our patients were treated with colistin before or after identification of KPC-Ent. Patients who were treated for infections continued to receive a combination of a carbapenem and aminoglycoside. The efficacy of this treatment was difficult to discern because there were many other factors contributing to patient status. Overall, there were four deaths and none could be attributed entirely to KPC-Ent infection. However, we did not follow the status of patients who were discharged. Two of the case patients remained hospitalized in the ICU at the end of the study.
CONCLUSIONS
We described the first Canadian outbreak of KPC-3-producing Enterobacteriaceae isolates from patients in an ICU. After conducting two point prevalence studies in the ICU, using rectal swabs and the recommended CDC method followed by polymerase chain reaction confirmation, two new isolates (E coli and S marcescens) expressing KPC-3 from different patients have been found. The extent of this current outbreak is not fully realized, but further point prevalence studies will be performed. Infection control measures, including isolation of patients, contact precautions and increased hand hygiene education, have been implemented but the impacts of these measures and further active surveillance using rectal swabs will need to be evaluated. The ideal screening methods and optimal treatment for patients with KPC-Ent have not been determined. The potential for dissemination of KPC enzymes is a significant public health concern.
REFERENCES
- 1.Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–36. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 3.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob Agents Chemother. 2008;52:1257–63. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother. 2007;51:1553–5. doi: 10.1128/AAC.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldfarb D, Harvey SB, Jessamine K, Jessamine P, Toye B, Desjardins M. Detection of plasmid-mediated KPC-producing Klebsiella pneumoniae in Ottawa, Canada: Evidence of intrahospital transmission. J Clin Microbiol. 2009;47:1920–2. doi: 10.1128/JCM.00098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillai DR, Melano R, Rawte P, et al. Klebsiella pneumoniae carbapenemase, Canada. Emerg Infect Dis. 2009;15:827–9. doi: 10.3201/eid1505.081536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility Testing. Twentieth informational supplement. M100-S20. Wayne: The Institute; 2010. [Google Scholar]
- 8.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–32. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Guyatt G, Gutterman D, Baumann MH, et al. Grading strength of recommendations and quality of evidence in clinical guidelines: Report from an American college of chest physicians task force. Chest. 2006;129:174–81. doi: 10.1378/chest.129.1.174. [DOI] [PubMed] [Google Scholar]
- 10.Bradford PA, Bratu S, Urban C, et al. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York city. Clin Infect Dis. 2004;39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 11.Bratu S, Landman D, Alam M, Tolentino E, Quale J. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob Agents Chemother. 2005;49:776–8. doi: 10.1128/AAC.49.2.776-778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbier F, Ruppe E, Giakkoupi P, et al. Genesis of a KPC-producing Klebsiella pneumoniae after in vivo transfer from an imported Greek strain. Euro Surveill. 2010;15 doi: 10.2807/ese.15.01.19457-en. pii:19457. [DOI] [PubMed] [Google Scholar]
- 14.Rasheed JK, Biddle JW, Anderson KF, et al. Detection of the Klebsiella pneumoniae carbapenemase type 2 Carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K oxytoca carrying a common plasmid. J Clin Microbiol. 2008;46:2066–9. doi: 10.1128/JCM.02038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility Testing. Nineteenth informational supplement. M100-S19. Wayne: The Institute; 2009. [Google Scholar]
- 16.Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility Testing Twentieth informational supplement update M100-S20-U. Wayne: The Institute; 2010. [Google Scholar]
- 17.Tenover FC, Kalsi RK, Williams PP, et al. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg Infect Dis. 2006;12:1209–13. doi: 10.3201/eid1208.060291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doern CD, Dunne WM, Jr, Burnham CA. Detection of Klebsiella pneumoniae carbapenemase (KPC) production in non-Klebsiella pneumoniae Enterobacteriaceae isolates by use of the Phoenix, Vitek 2, and disk diffusion methods. J Clin Microbiol. 2011;49:1143–7. doi: 10.1128/JCM.02163-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson KF, Lonsway DR, Rasheed JK, et al. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007;45:2723–5. doi: 10.1128/JCM.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]