Abstract
Mutations in the α-subunit of cardiac sodium channel gene SCN5A can lead to the overlapping phenotypes of both the Brugada and type 3 long QT syndromes. However, the combination of Brugada and a short QT phenotype resulting from mutation in SCN5A has not previously been described. A man with concomitant Brugada-like and short QT electrocardiogram (ECG) was identified and the SCN5A gene was sequenced. Whole-cell patch clamp analysis of human embryo kidney (HEK) 293 cells expressing a SCN5A channel with the patient's sequence was used to investigate the biophysical properties of the channel. The patient with the family history of sudden death showed Brugada-like and short QT interval ECG. Sequence anlaysis of the coding region of the SCN5A gene, identified a G to A heterozygous missense mutation at nucleotide site 2066 that resulted in a amino-acid substitution of arginine to histidine at amino-acid site 689 (R689H). Patch clamp analysis showed that the R689H failed to generate current when heterologously expressed in HEK293 cells, indicating it was a loss-of-function mutation. Our finding firstly showes that a heterozygous missense mutation R689H in SCN5A gene results in the loss of protein function and the coexistents of the Brugada-like and short QT interval ECG phenotypes.
Keywords: short QT, Brugada, mutation, SCN5A
Introduction
Brugada syndrome (BrS) is an electrical disorder of the heart that does not result from abnormal heart structure and is characterized by the coved ST segment elevation in the right precordial leads. Short QT syndrome (SQTS) clinically first described in 2000 has been associated with mutations in three potassium channel genes (KCNH2, KCNQ1, and KCNJ2) that increase the activity (gain-of-function) of the channels.1, 2, 3 In 2007, loss-of-function mutations in the CACNA1C (A39V and G490R) and CACNB2 (S481L) genes encoding the alpha1- and beta2b-subunits of the L-type calcium channel were reported in patients with short QT interval and Brugada-like electrocardiogram (ECG).4
The QT interval is determined by the cardiac action potential, which is balanced between inward and outward currents of myocardial cells. Genetic testing has shown that mutations in cardiac ion channels are responsible for an ever-increasing number and diversity of familial cardiac arrhythmia syndromes. For example, mutations in the α-subunit of sodium channel gene (SCN5A) have been found in BrS associated with type 3 long QT syndrome (LQTS),5 sick sinus syndrome,6 atrial standstill,7 cardiac conduction disturbance,8 and atrial fibrillation.9
In this study, we describe the overlapping phenotypes of short QT interval with Brugada-like ECG in a Chinese man. Furthermore, we identify a heterozygous missense mutation in the patient′s SCN5A sodium channel and show that this mutation (R689H) is a loss-of-function mutation possibly causing the cardiac defects.
Materials and Methods
The study was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and approved by the hospital review board. The patient gave written informed consent.
Clinical Evaluation
The patient underwent detailed clinical examinations including baseline of 12-lead ECG, echocardiography and laboratory test. The patient did not permit electrophysiological studies to be performed.
Molecular genetics
Genomic DNA was extracted from peripheral lymphocytes. All exon coding regions of disease-causing genes of SQTS (KCNQ1, KCNH2, KCNJ2, CACNA1C, CACNB2) and SCN5A were amplified by polymerase chain reaction (PCR) using primers that would amplify intronic flanking sequences. The PCR products were directly sequenced using a Bigdye Terminator Mix (Applied Biosystems, Carlsbad, CA, USA) in both strands and analyzed by cycle sequencing using an automated DNA sequencer (ABI Prism 3130XL; Applied Biosystems).
Mutagenesis
The mutation in SCN5A that altered an arginine to a histidine (R689H) was generated by site-directed mutagenesis of the plasmid pcDNA-SCN5A, which contained SCN5A cDNA cloned into pcDNA3.1+ (Invitrogen, Carlsbad, CA, USA). The following mutagenic sense and anti-sense primers were used to generate the R689H mutation: sense primer: 5′-TGCTGGAACCATCTCGCCCAGCGCTACCTG-3′ anti-sense primer: 5′-CTGGGCGAGATGGTTCCAGCATGGTGGAC-3′. The mutated plasmid was sequenced to ensure the presence of the R689H mutation as well as the absence of other substitutions introduced by PCR.
Patch clamp
The R689H mutation was expressed in the human embryo kidney (HEK) 293 cells and characterized by whole-cell patch clamp recordings as previously described.1 Membrane currents were measured with the EPC 10 amplifier (HEKA Instruments Inc., Bellmore, NY, USA).The pipette solution contained (mmol/l): CsF 110, CsCl 20, NaF 10, EGTA 10, and HEPES 10 (pH 7.35 adjusted with CsOH). The bath solution consisted of (mmol/l): NaCl 145, KCl 4, MgCl2 1.0, CaCl2 1.8, glucose 10, and HEPES 10 (pH 7.35 adjusted with NaOH). Recordings were made at room temperature.
Voltage-dependent sodium currents were recorded at various membrane potentials from −80 mV to 60 mV in 10 mV increment for 50 ms, holding potential was −120 mV. Steady-state activation was estimated by measuring the peak sodium currents. Conductance G(v) was calculated by the equation: G(v)=I/(Vm−Erev) (I: peak currents, Vm: membrane potentials), Eres: the measured reversal potentials). Steady-state inactivation was estimated by a prepulse protocol that was a various membrane potentials from −140 mV to −50 mV for 500 ms followed by depolarizing to −20 mV for 20 ms. Data for steady-state activation and steady-state inactivation were fitted with the Boltzmann equation.
Data Acquisition and Analysis
All signals were acquired at 20 kHz with Patchmaster software (HEKA Instruments Inc., Bellmore, NY, USA). The data were analyzed with Fitmaster, Sigmaplot 10.0 and EXCEL data handling. Results were presented as mean±SE. Statistical analysis was performed with t-test and P<0.05 was considered statistically significant.
Results
Clinic and genetic data
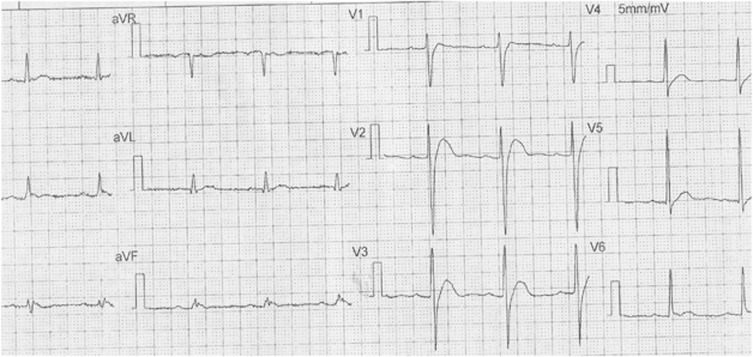
A 40-year-old man was admitted to our hospital with a ruptured quadriceps femoris tendon of the right knee. Cardiac analysis (12-lead ECG) indicated the patient had Brugada-like ECG (Figure 1) and was diagnosed by a cardiologist as also having a short QT interval (QT=320 ms, HR=71 bpm). Echocardiography and laboratory analysis including serum electrolytes were normal. There was a history of sudden death in the man's family (his father encountered sudden death at the age of 39 years from unknown cause) consistent with the Brugada-like and short QT interval ECG phenotypes resulting from a genetic mutation. All medicines were withdrawn.
Figure 1.
12-lead ECG (5 mm/mV, 25 mm/s) showed Brugada-like ST-T changes and short QT interval (QT=320 ms, HR=71bpm).
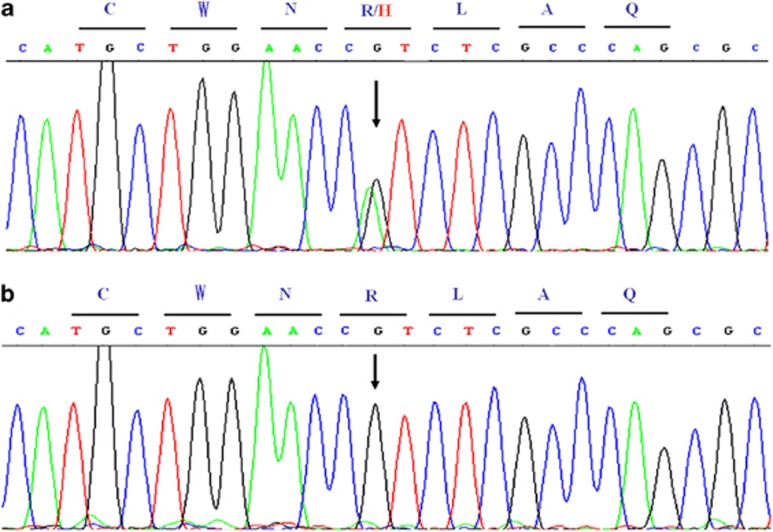
To explore the possibility that the cardiac phenotypes may represent a mutation in the SCN5A gene, we sequenced the coding regions of SCN5A and identified a G to A substitution at nucleotide site 2066 that resulted in heterozygous missense mutation R689H (amino-acid change of arginine to histidine, Figure 2a). The same mutation was not detected in the 200 control chromosomes of the same ethnic background (Figure 2b). There was no mutation found in KCNQ1, KCNH2, KCNJ2, CACNA1C, CACNB2 genes. More indepth analysis was not possible, as the patient refused to participate in a cardiac electrophysiological study and family members would not allow their DNA to be analyzed.
Figure 2.
DNA sequence analysis of the patient with the SCN5A R689H heterozygous mutation (a) and a WT normal SCN5A sequence (b). The capital letters on the lines stand for amino-acids abbreviations.
Biophysical properties
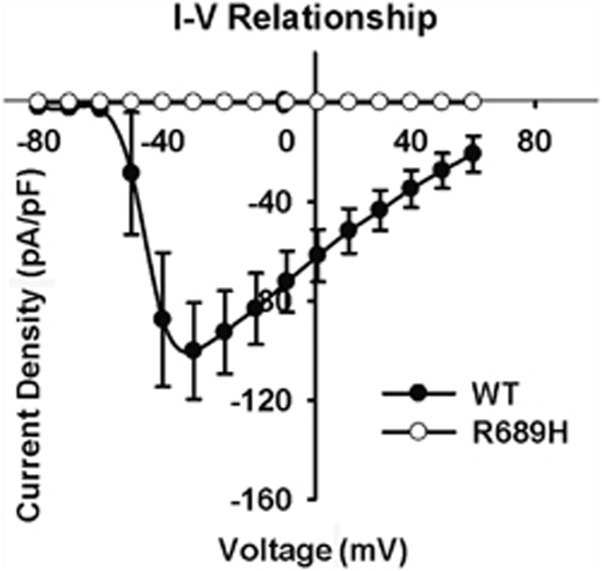
Patch clamp analysis was used to further explore how the SCN5A R689H mutation resulted in Brugada-like and short QT interval ECG. HEK293 cells were transfected with wild-type (WT) and mutant (R689H) SCN5A cDNA. Figure 3 showed the current–voltage relationships in WT and R689H mutant sodium channel. There was no sodium current with the R689H mutant sodium channel indicating loss-of-function of the mutation.
Figure 3.
Current–voltage relationships for WT and R689H mutant sodium channel expressed in HEK293 cells. The peak value of I–V curve for WT occurred at −30mV, and the current density for R689H was zero.
Discussion
The patient in this study had both Brugada-like and short QT interval ECG. However, to date he had no symptoms of heart disease, although there was a family history of sudden death of unknown origin. The lack of cardiac symptoms may result from the phenotypes of BrS and SQTS being only partially penetrant. These phenotypes often become more apparent in the presence of external factors, such as medication (sodium channel blocker), fever,10 or electrolyte disorder. Because the ECG of this patient was not the typical type 1 Brugada ECG pattern, it would of course have been very interesting to see the response to a sodium channel blocker. Unfortunately, the patient refused to do this test. There was no mutation in the KCNQ1, KCNH2, KCNJ2, CACNA1C and CACNB2 genes that had been confirmed to cause SQTS. In this patient, the Brugada-like and short QT phenotypes appears to result from a single heterozygous missense mutation in the SCN5A protein.
It is clear that SCN5A protein carries a large inward depolarizing current (INa) during phase 0 of the cardiac action potential. The importance of INa for the cardiac electrical activity is reflected by the high incidence of arrhythmias in cardiac sodium channelopathies. It is now recognized that reduced INa decreases cardiac excitability and electrical conduction velocity, and induces BrS, progressive cardiac conduction disturbance, sick sinus syndrome, dilated cardiomyopathy, atrial fibrillation, sudden infant death syndrome, or combinations thereof.11 In our study, the R689H mutation induced INa dysfunction (zero current) that might result in the reduction of cardiac excitability and electrical conduction velocity, and therefore induce Brugada-like type ECG. It could also cause the abbreviation of the ventricular action potential resulting in a short QT interval. This finding is similar to another study in which blocking of the sodium channel shortened the QT but exacerbated the ST segment elevation of BrS phenotype causing enhanced risk for arrhythmia.12
The R689H mutation has been described previously in patients with isolated LQTS13 or BrS.15, 16 Napolitano C13 found R689H mutation in isolated LQTS probands (97% White). Kapplinger JD15 and Nakajima T16 found R689H mutation in isolated BrS patients (Hispanic and Japanese), respectively. The functional analysis of R689H mutation was not done in those three study. Although we found R689H mutation associated with short QT interval and Brugada-like ECG in a Chinese patient with the family history of sudden death and the functional analysis in HEK293 cell system did show the loss of function (no current). It is known that some genetic defects can lead to the overlapping syndromes in a single patient.14 This is the first report that a mutation in SCN5A causes a subclinical phenotype – Brugada-like and short QT interval ECG. The same mutation causing different manifestations suggested the complexity of genotype–phenotype relationship. The mechanism of this phenomenon was not well understood to date and it is commonly held that environmental and genetic modifiers presumably have a role in phenotype.17 The R689H mutation caused LQTS in White race and caused the other different phenotypes in other race,which suggested that the ethnic genetic background have an important role in the arrhythmogenic phenotype susceptibility.18 The results of R689H mutation in HEK293 cells expression system only reflected the changes in HEK293 cells, which could not faithfully represent the function changes of R689H heterozygous mutation. So the heterologous cell system had limited value in explaining the pathophysiological mechanism of the final comprehensive manifestation from a global patient. More sophisticated cell systems such as human stem cell-derived cell lines may be helpful to resolve these issues.
In conclusion, our finding is the first to show that a heterozygous missense mutation R689H in SCN5A gene results in the loss of protein function and the coexistents of the Brugada-like and short QT interval ECG phenotypes.
Acknowledgments
We would like to thank Xiuxia Liu and Xin Yu for DNA extraction. This work was supported in part by the National Basic Research Program of China (973 Program: 2007CB512002 and 2008CB517305), the National Natural Science Foundation of China (30760076 and 81070148), and the JiangXi Scientific Foundation (2007BS02346).
The authors declare no conflict of interest.
References
- Brugada R, Hong K, Dumaine R, et al. Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation. 2004;109:30–35. doi: 10.1161/01.CIR.0000109482.92774.3A. [DOI] [PubMed] [Google Scholar]
- Bellocq C, van Ginneken AC, Bezzina CR, et al. Mutation in the KCNQ1 gene leading to the short QT-interval syndrome. Circulation. 2004;109:2394–2397. doi: 10.1161/01.CIR.0000130409.72142.FE. [DOI] [PubMed] [Google Scholar]
- Priori SG, Pandit SV, Rivolta I, et al. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ Res. 2005;96:800–807. doi: 10.1161/01.RES.0000162101.76263.8c. [DOI] [PubMed] [Google Scholar]
- Antzelevitch C, Pollevick GD, Cordeiro JM, et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation. 2007;115:442–449. doi: 10.1161/CIRCULATIONAHA.106.668392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita N, Behr E, Shimizu W, et al. The E1784K mutation in SCN5A is associated with mixed clinical phenotype of type 3 long QT syndrome. J Clin Invest. 2008;118:2219–2229. doi: 10.1172/JCI34057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiyama T, Akao M, Tsuji K, et al. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol. 2005;46:2100–2106. doi: 10.1016/j.jacc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Takehara N, Makita N, Kawabe J, et al. A cardiac sodium channel mutation identified in Brugada syndrome associated with atrial standstill. J Intern Med. 2004;255:137–142. doi: 10.1046/j.0954-6820.2003.01247.x. [DOI] [PubMed] [Google Scholar]
- Cordeiro JM, Barajas-Martinez H, Hong K, et al. Compound heterozygous mutations P336L and I1660V in the human cardiac sodium channel associated with Brugada syndrome. Circulation. 2006;114:2026–2033. doi: 10.1161/CIRCULATIONAHA.106.627489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano KF, Taniyama M, Nakamura K, et al. Atrial fibrillation in patients with Brugada syndrome relationships of gene mutation, electrophysiology, and clinical backgrounds. J Am Coll Cardiol. 2008;51:1169–1175. doi: 10.1016/j.jacc.2007.10.060. [DOI] [PubMed] [Google Scholar]
- Patruno N, Pontillo D, Achilli A, Ruggeri G, Critelli G. Electrocardiographic pattern of Brugada syndrome disclosed by a febrile illness: clinical and therapeutic implications. Europace. 2003;5:251–255. doi: 10.1016/s1099-5129(03)00029-1. [DOI] [PubMed] [Google Scholar]
- Amin AS, Asghari-Roodsari A, Tan HL. Cardiac sodium channelopathies. Pflugers Arch. 2010;460:223–237. doi: 10.1007/s00424-009-0761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priori SG, Napolitano C, Schwartz PJ, Bloise R, Crotti L, Ronchetti E. The elusive link between LQT3 and Brugada syndrome: the role of flecainide challenge. Circulation. 2000;102:945–947. doi: 10.1161/01.cir.102.9.945. [DOI] [PubMed] [Google Scholar]
- Napolitano C, Priori SG, Schwartz PJ, et al. Genetic testing in the long QT syndrome:development and validation of an efficient approach to genotyping in clinical practice. JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- Makita N. Phenotypic overlap of cardiac sodium channelopathies: individual-specific or mutation-specific. Circ J. 2009;73:810–817. doi: 10.1253/circj.cj-09-0014. [DOI] [PubMed] [Google Scholar]
- Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Kaneko Y, Saito A, et al. Identification of six novel SCN5A mutations in Japanese patients with Brugada syndrome. Int Heart J. 2011;52:27–31. doi: 10.1536/ihj.52.27. [DOI] [PubMed] [Google Scholar]
- Scicluna BP, Wilde AA, Bezzina CR. The primary arrhythmia syndromes: same mutation, different manifestations. Are we starting to understand why. J Cardiovasc Electrophysiol. 2008;19:445–452. doi: 10.1111/j.1540-8167.2007.01073.x. [DOI] [PubMed] [Google Scholar]
- Ackerman MJ, Splawski I, Makielski JC, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1:600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]