Abstract
Cytotoxic CD8 T cells constitute a subgroup of T cells that are capable of inducing the death of infected or malignant host cells1. These cells express a specialized receptor, called the T cell receptor (TCR), which can recognize a specific antigenic peptide bound to HLA class I molecules2. Engagement of infected cells or tumor cells through their HLA class I molecule results in production of lytic molecules such as granzymes and perforin resulting in target cell death. While it is useful to determine frequencies of antigen-specific CD8 T cells using assays such as the ELIspot or flow cytometry, it is also helpful to ascertain the strength of CD8 T cell responses using cytotoxicity assays3. The most recognizable assay for assessing cytotoxic function is the Chromium Release Assay (CRA), which is considered a standard assay 4. The CRA has several limitations, including exposure of cells to gamma radiation, lack of reproducibility, and a requirement for large numbers of cells. Over the past decade, there has been interest in adopting new strategies to overcome these limitations. Newer approaches include those that measure caspase release 4, BLT esterase activity 5 and surface expression of CD107 6. The impedance-based assay, using the Roche xCelligence system, was examined in the present paper for its potential as an alternative to the CRA. Impedance or opposition to an electric current occurs when adherent tumor cells bind to electrode plates. Tumor cells detach following killing and electrical impedance is reduced which can be measured by the xCelligence system. The ability to adapt the impedance-based approach to assess cell-mediated killing rests on the observation that T cells do not adhere tightly to most surfaces and do not appear to have much impact on impedance thus diminishing any concern of direct interference of the T cells with the measurement. Results show that the impedance-based assay can detect changes in the levels of antigen-specific cytotoxic CD8 T cells with increased sensitivity relative to the standard CRA. Based on these results, impedance-based approaches may be good alternatives to CRAs or other approaches that aim to measure cytotoxic CD8 T cell functionality.
Protocol
1. Generation of Short-term Human Antigen-specific CD8 T cell Lines7
Separate peripheral blood mononuclear cells (PBMCs) from the blood of normal healthy HLA-A2 positive donors using Ficoll-paque plus.
Add 2 ml/well of PBMCs to 6-well plates at 2 x 106 cells/ml.
Add HLA-A2-binding HER-2/neu p369-377 peptide (amino acid sequence KIFGSLAFL) or HLA-A2-binding influenza p58-66 peptide (GILGFVFTL) directly to the wells at a final concentration of 10 μg/ml and place in an incubator at 37 °C with 5% CO2.
To help stimulate and expand T cells, add, every 2 days, 250 μl/well fresh media supplemented with human recombinant IL-2 (stock: 50 units/μl) to achieve a final concentration of 50 units/ml in the culture media.
Re-stimulate the culture cells with 2 x 106 cells/ml irradiated autologous PBMCs pulsed for 2 hours with 10 μg/ml of the same peptides used in Step 1.3 and add these cells to cultures at 1 ml/well one week after the first peptide stimulation.
Add human recombinant IL-2 and fresh media, as described in Step 1.4, into the existing cultures every 2 days.
Re-stimulate the cells with peptide and irradiated autologous PBMCs as described in Step 1.5 one week after the second peptide stimulation.
Prepare to use cells six days after the last re-stimulation at the earliest.
2. Preparation of Breast Cancer Cells
Place the xCelligence impedance measuring station (xCelligence station) in an incubator at 37 °C with 5% CO2.
Harvest human tumor cells (SKBR3, BT20, MCF7 or HCC1419) that are 75% confluent using 0.25% trypsin and centrifugation (1,000 rpm for 5 minutes).
Count tumor cells using a hemocytometer and reconstitute them in RPMI tumor media (supplemented with 10% fetal bovine serum (FBS) and 500 units/ml IFN-gamma) at a concentration of 7.5 x 103 tumor cells/100 μl.
Program the xCelligence software by setting up the layout page to determine well layout and by setting up the schedule page to take impedance readings every 5 minutes for a 40 hour period. For the first 18 hours, the xCelligence system will take impedance readings of the tumor cells only.
Add 100 μl of RPMI tumor media to the xCelligence E-plates and perform a background reading by measuring impedance in the absence of cells.
Add tumor cells to the E-plates at 100 μl per well and place in the xCelligence station.
Press the start button on the xCelligence software to begin taking impedance readings of the tumor cells, which immediately start adhering to the E-plates.
3. Co-culture of Tumor with CD8 T cells
Once the tumor cells have attached and doubled to 1.5 x 104 tumor cells per well (approximately 18 hours after initially being plated), harvest T cells prepared in Section 1 by gentle scraping and agitation
Centrifuge at 1,000 rpm for 5 minutes and count T cells using a hemocytometer and reconstitute them in RPMI media with 10% FBS at various ratios, starting at a maximum ratio of 1 tumor cell to 40 T cells and dilute 2-fold until a ratio of 1 tumor cell to 1.25 T cells is reached. For example, there are 1.5 x 104 tumor cells per well. To achieve a 1:40 ratio, add 6 x 105 T cells per well and dilute the T cells 2-fold until a ratio of 1.5 x 104 tumor cells to 1.875 x 104 T cells per well (1:1.25 ratio) is achieved.
Pause the xCelligence station and remove the E-plate.
Remove the media in the wells with a pipet and add 200 μl of media alone or the appropriate ratio of T cells to the wells.
Place the E-plate back in the xCelligence station and continue the xCelligence software program so impedance readings are taken every 5 minutes for approximately 20 hours after T cell addition.
Normalize the results at the end of the assay.
4. Chromium Release Assay (CRA)7
Follow institutional radiation safety procedures when working with 51Cr and 51Cr-labeled target cells. Always use appropriate shielding to reduce radiation exposure.
Pulse tumor cells for 2 hours at 1 x 106 tumor cells/ml with 10 μl/ml fresh 51Cr (0.005 Ci/ml).
Wash tumor cells in 10 ml of RPMI media with 10% FBS and centrifuge at 1,000 rpm for 5 minutes. Add tumor cells to a 96-well round-bottom plate at 1 x 105 tumor cells in 100 μl /well.
Add T cells at various ratios, starting at 1 tumor cell to 40 T cells and dilute 2-fold until a ratio of 1 tumor cell to 1.25 T cells is achieved. For example, there are 1 x 105 tumor cells per well. To achieve a 1:40 ratio, add 4 x 106 T cells per well and dilute the T cells 2-fold until a ratio of 1 x 105 tumor cells to 1.25 x 105 T cells per well (1:1.25 ratio) is achieved.
Add spontaneous and maximum tumor cell controls to the plate. In order to calculate the spontaneous release of 51Cr from the tumor cells, add six wells each containing 1 x 105 tumor cells in 100 μl to 100 μl of RPMI media with 10% FBS. To calculate maximum release of 51Cr from the tumor cells, add six wells each containing 1 x 105 tumor cells in 100 μl to 100 μl 0.1% Triton X-100 in PBS, which lyses all cells.
Incubate the plates for 5 hours at 37 °C with 5% CO2.
After incubation, remove 50 μl of the supernatant from each well and add it to a Luma plate.
Dry the plates overnight and measure CPM using a Gamma Counter.
5. Percent Lysis Calculation
Impedance assay: Take the impedance reading, reported as the cell index (CI), at various time points and use the following formula to determine percent lysis: (CI tumor only - (CI tumor + T cells)) / (CI tumor only) *100.
CRA: Use the following formula to determine percent lysis: (CPM experimental - CPM spontaneous) / (CPM maximum - CPM spontaneous)*100.
6. Representative Results
T cells alone do not increase impedance
T cells, in general, do not adhere to most solid surfaces and do not require adherence for activation and proliferation. Therefore, it was hypothesized that T cells should not contribute to impedance on the xCelligence E-plates. To test this, wells were loaded with either SKBR3 breast cancer cells or freshly purified human CD8 T cells. Impedance was measured over the course of 40 hours and a representative example demonstrating essentially little effect of T cells is shown in Figure 1A. In contrast, it could be argued that T cells may diminish background impedance, albeit only slightly. Despite that, co-culture revealed that the process of adding T cells at 18 hours following initiation of impedance measurements resulted in an impedance disruption (Figure 1B). Other studies revealed that disruptions were due in part to handling since the diminution in signal could be reduced by ensuring that temperature of the T cell-containing solution was the same as the interior of the incubator.
Reductions in impedance mediated by T cells are dose-dependent.
The utility of an impedance-based assay to compare samples for cytotoxic T cell activity requires the ability to distinguish between different concentrations of antigen-specific T cells. To test whether this is possible, SKBR3 cells were co-cultured with varying concentrations of T cells derived from a HER-2/neu p369-specific T cell line. As shown in Figure 2A, reductions in impedance were dependent on the dose of T cells. Figure 2B shows that the cell indices at 10 hours follow a simple second order polynomial over the range of T cells. Thus, the xCelligence station monitors CD8 T cell mediated death of SKBR3 tumors in real time as a drop in cell index.
The impedance-based assay detects antigen-specific T cells
To determine if reductions in impedance of SKBR3, which is a HER-2/neu and HLA-A2 positive breast cancer tumor cell line, by HER-2/neu p369-specific T cells were antigen specific, separate wells containing SKBR3 were also incubated with T cells derived from an influenza (FLU)-specific T cell line. The FLU specific-T cells served as a non-specific control, being HLA-A2 positive, but not having any ability to recognize HER-2/neu. As shown in Figure 3, there is a significant difference between the lytic activity of HER-2/neu p369-specific T cells as compared to FLU-specific T cells (p<0.0001). In some cases, T cells not specific for tumor (e.g. FLU-specific or anti-CD3/anti-CD28 stimulated) demonstrated a low level of lytic activity (Figures 3A-B). T cells can kill in one of two ways, either non-specifically through Fas:Fas ligand interactions or antigen-specifically through the release of granzymes and perforins. To further corroborate the notion that HER-2/neu-specific T cells were killing in an antigen-specific fashion, we incorporated a blocking antibody to Fas ligand. As shown in Figure 3B, the antibody did not reduce the lytic activity of the HER-2/neu p369-specific T cells. The antibody could inhibit interactions between Fas and Fas Ligand, as demonstrated with T cells that were non-specifically activated with anti-CD3/CD28 beads. These results suggest that when tumor-specific T cells are tested with non-tumor-specific T cells, Fas ligand blockade may be needed to reduce non-TCR mediated interactions to a minimum. Alternatively, when using short-term cultured T cells lines in which only a fraction of the T cells are specific for the antigen used for ex vivo generation, the possibility of MHC mismatch-mediated T cell activation could occur leading to non-specific lysis. In this case, the addition of antibodies that block the irrelevant HLAs may be warranted. To determine if the p369-specific T cells were killing tumor cells in an HLA class I dependent manner, either anti-HLA-class I antibody or isotype control antibody were added to the co-cultures. As shown in Figure 3C, lysis by p369-specific T cells was suppressed by inclusion of antibody demonstrating HLA class I restriction. Lastly, since the development of this assay was largely done using SKBR3, it was important to ascertain if other adherent HLA-A2 and HER-2/neu expressing tumor cells could be killed by p369-specific T cells. Thus, T cells were prepared and added at a 1:5 ratio to the HLA-A2 and HER-2/neu-expressing tumor cells, SKBR3, MCF-7, and HCC1419 cells. The HLA-A2 negative tumor cell line, BT20 was used as a negative control. As shown in Figure 3D, all of these additional tumor cells, except BT20 were also killed as assessed by impedance. Thus, the method appears to be useful for multiple target adherent tumor cells.
The impedance-based assay is more sensitive than the standard chromium release assay in detecting cytotoxic T cell activity
The chromium (51Cr) release assay (CRA) that measures cytolytic activity has long been the gold standard by which to monitor CD8 T cell responses. In this assay, target cells (e.g. tumor cells) are labeled with 51Cr. In most cases, healthy target cells can retain the majority of the radiolabel over the course of the assay. CD8 T cells are added to the target cells, usually at varying concentrations, and killing of the target cells is detected by release of the 51Cr into the medium. Aside from the fact that it uses hazardous radiation, two major problems with the CRA are a lack of sensitivity and that the assay typically requires large amounts of CD8 T cells, limiting its utility. To determine if the impedance-based assay was more sensitive than the CRA, HER-2/neu p369-specific T cells were generated in vitro and applied, in varying amounts, to either E-plates containing unlabeled tumor cells or standard polypropylene plates containing 51Cr-labeled target cells. Measurements, either impedance or free chromium, were measured at 5 hours following T cell addition. Figure 4, a representative example, shows the impedance assay outperforms the chromium release.
The intra-assay variation of the impedance-based assay is typically below 20%.
Intra-assay variation was examined, since wider use of this assay will be largely determined by its reproducibly and variation (Figure 5). Two p369-specific T cell lines were generated independently 30 days apart and added in triplicate over a range of tumor cell to T cell ratios. The inset graph in Figure 5 shows that the lines were largely equivalent in terms of lysing SKBR3 cells. The % coefficient of variation was calculated at each cell ratio and plotted. As shown in Figure 5, between 1:40 and 1:5 ratios the % coefficients of variation (CV) were below 15%. At the 1:2.5 and 1:1.25, however, the CVs were high or unpredictable. Because of extensive biologic variation, it is not possible to measure inter-assay variability with primary T cell lines.
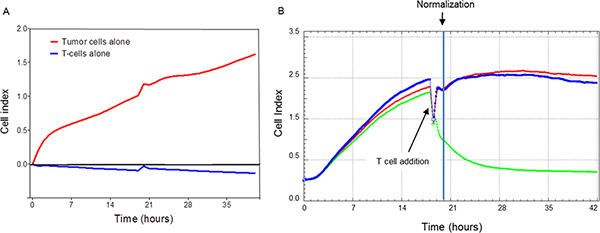 Figure 1. Impedance needs to be normalized following the addition of the T cells.
Panel A: demonstrates that tumor cells (red line) and not T cells (blue line)
are responsible for the impedance. Shown are the impedance tracings over 40 hours for
either tumor or T cells alone. Similar results were obtained from a second experiment.
Note that the signal perturbation at approximately 20 hours represents the point of
addition of T cells to the wells not containing tumor cells. Panel B: shows
that impedance measurements are transiently disrupted with the addition of T cells to
tumor cells. This requires normalization at some time point following the addition of T
cells (See point of normalization marked in graph). Traces are composed from duplicate
data points. The trace for tumor cells alone is shown in red. The other traces represent
tumor cells co-cultured with HER-2/neu p369-specific T cells (Blue: ratio of 1.25 T
cells/tumor cell, Green: ratio of 40 T cells/tumor cell). Each trace is calculated from
duplicate data points collected every 5 minutes through the duration of the culture.
Results are representative of 3 separate experiments. Click here to
view larger figure.
Figure 1. Impedance needs to be normalized following the addition of the T cells.
Panel A: demonstrates that tumor cells (red line) and not T cells (blue line)
are responsible for the impedance. Shown are the impedance tracings over 40 hours for
either tumor or T cells alone. Similar results were obtained from a second experiment.
Note that the signal perturbation at approximately 20 hours represents the point of
addition of T cells to the wells not containing tumor cells. Panel B: shows
that impedance measurements are transiently disrupted with the addition of T cells to
tumor cells. This requires normalization at some time point following the addition of T
cells (See point of normalization marked in graph). Traces are composed from duplicate
data points. The trace for tumor cells alone is shown in red. The other traces represent
tumor cells co-cultured with HER-2/neu p369-specific T cells (Blue: ratio of 1.25 T
cells/tumor cell, Green: ratio of 40 T cells/tumor cell). Each trace is calculated from
duplicate data points collected every 5 minutes through the duration of the culture.
Results are representative of 3 separate experiments. Click here to
view larger figure.
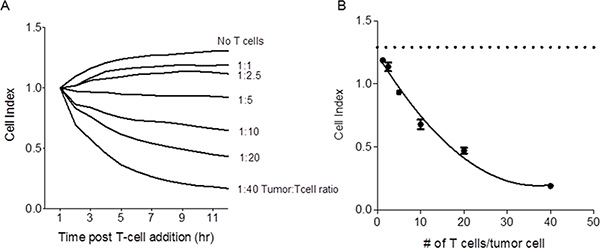 Figure 2. Reduction in impedance mediated by T cells is dose dependent. Panel A:
Shown are the impedance traces of SKBR3 tumor cells incubated alone or with
varying concentrations of tumor-specific T cells. Each trace is calculated from triplicate
data points collected every 5 minutes through the duration of the culture. Results are
representative of 3 separate experiments. Panel B: Shown are the cell indices
at 10 hours across all T cell/tumor cell concentrations. The dashed line represents the
cell index for tumor cells alone. Each symbol is the mean (±s.e.m.) of triplicate
determinations. Results are representative of 3 separate experiments. Click here to
view larger figure.
Figure 2. Reduction in impedance mediated by T cells is dose dependent. Panel A:
Shown are the impedance traces of SKBR3 tumor cells incubated alone or with
varying concentrations of tumor-specific T cells. Each trace is calculated from triplicate
data points collected every 5 minutes through the duration of the culture. Results are
representative of 3 separate experiments. Panel B: Shown are the cell indices
at 10 hours across all T cell/tumor cell concentrations. The dashed line represents the
cell index for tumor cells alone. Each symbol is the mean (±s.e.m.) of triplicate
determinations. Results are representative of 3 separate experiments. Click here to
view larger figure.
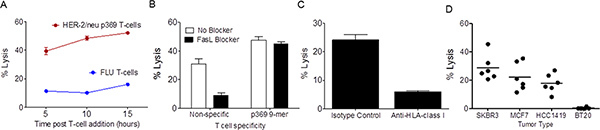 Figure 3. The impedance based assay detects antigen-specific cytotoxic T cell
responses. Panel A shows the level of lysis of SKBR3 cells by HER-2/neu
p369-specific T cells (Red) or FLU-specific T cells (Blue) at 5, 10, and 15 hours
following the co-culture. Each data point is the mean (±s.e.m.) of triplicate
measurements. Results are representative of 3 separate experiments. Panel B
shows lysis of SKBR3 by p369-specific T cells or non-specifically activated T
cells, demonstrating that antigen-specific lysis is not due to Fas-Fas ligand
interactions. Black bars represent co-cultures that contained a Fas ligand antibody. Each
bar is the mean (±s.e.m.) lysis in triplicate at 5 hours following the addition of T
cells. Results are representative of 3 separate experiments. Panel C shows
lysis 10 hours after T cell addition of SKBR3 by p369-specific T cells is HLA-class I
dependent. Either HLA-class I blocking or isotype control was added during co-culture.
Each bar is the mean (±s.e.m.) lysis in triplicate. This experiment was repeated twice
with similar results. Panel D shows % lysis 10 hours after T cell addition of
several HLA-A2+, HER-2/neu-expressing tumor cell lines by p369-specific T cells. BT20, a
HLA-A2-negative breast cancer cell line was used as a negative control. Each spot
represents a unique experiment calculated from triplicate determinations. Click here to
view larger figure.
Figure 3. The impedance based assay detects antigen-specific cytotoxic T cell
responses. Panel A shows the level of lysis of SKBR3 cells by HER-2/neu
p369-specific T cells (Red) or FLU-specific T cells (Blue) at 5, 10, and 15 hours
following the co-culture. Each data point is the mean (±s.e.m.) of triplicate
measurements. Results are representative of 3 separate experiments. Panel B
shows lysis of SKBR3 by p369-specific T cells or non-specifically activated T
cells, demonstrating that antigen-specific lysis is not due to Fas-Fas ligand
interactions. Black bars represent co-cultures that contained a Fas ligand antibody. Each
bar is the mean (±s.e.m.) lysis in triplicate at 5 hours following the addition of T
cells. Results are representative of 3 separate experiments. Panel C shows
lysis 10 hours after T cell addition of SKBR3 by p369-specific T cells is HLA-class I
dependent. Either HLA-class I blocking or isotype control was added during co-culture.
Each bar is the mean (±s.e.m.) lysis in triplicate. This experiment was repeated twice
with similar results. Panel D shows % lysis 10 hours after T cell addition of
several HLA-A2+, HER-2/neu-expressing tumor cell lines by p369-specific T cells. BT20, a
HLA-A2-negative breast cancer cell line was used as a negative control. Each spot
represents a unique experiment calculated from triplicate determinations. Click here to
view larger figure.
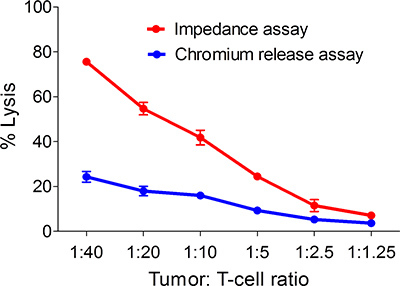 Figure 4. The impedance based assay is more sensitive than the chromium release
assay for the detection of tumor antigen-specific T cells. Shown are % lysis of
SKBR3 in response to varying concentrations of p369-specific T cells as measured at 5
hours following tumor and T cell mixing using both the impedance-based assay and the
chromium release assay. Each symbol is the mean (±s.e.m) of three replicates. Results are
representative of 3 separate experiments.
Figure 4. The impedance based assay is more sensitive than the chromium release
assay for the detection of tumor antigen-specific T cells. Shown are % lysis of
SKBR3 in response to varying concentrations of p369-specific T cells as measured at 5
hours following tumor and T cell mixing using both the impedance-based assay and the
chromium release assay. Each symbol is the mean (±s.e.m) of three replicates. Results are
representative of 3 separate experiments.
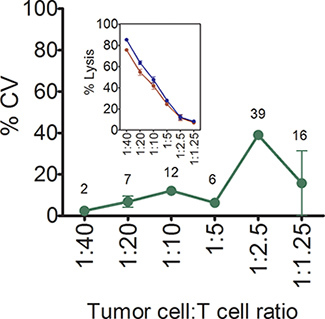 Figure 5. % intra-assay coefficients of variation of the impedance-based T cell
cytotoxicity assay. Shown are mean (±SD) % intra-assay coefficients of variation
in lysis of SKBR3 over a range of concentrations of antigen-specific T cells. Each symbol
is the mean of three replicates. Numbers shown in the graph are the mean % CVs.
Inset panel shows the mean (±s.e.m, n=3) % lysis of SKBR3, at varying
concentrations of p369-377-specific T cells. Two independently generated p369-specific T
cell lines using T cells from two separate donors are shown.
Figure 5. % intra-assay coefficients of variation of the impedance-based T cell
cytotoxicity assay. Shown are mean (±SD) % intra-assay coefficients of variation
in lysis of SKBR3 over a range of concentrations of antigen-specific T cells. Each symbol
is the mean of three replicates. Numbers shown in the graph are the mean % CVs.
Inset panel shows the mean (±s.e.m, n=3) % lysis of SKBR3, at varying
concentrations of p369-377-specific T cells. Two independently generated p369-specific T
cell lines using T cells from two separate donors are shown.
Discussion
CD8 T cells are important to the adaptive immune response, being able to directly kill infected or malignant cells4. While there are several ways to measure the numbers of antigen-specific CD8 T cells (e.g. ELIspot, tetramer, flow cytometry), often it is helpful to know whether these cells are functionally cytotoxic3. The most recognizable assay for assessing cytotoxic function is the CRA, which is considered the gold standard assay4. Major limitations of the CRA are (1) the requirement of radioactivity, (2) lack of sensitivity, (3) lack of mechanistic information, (4) requirement for large amounts of cells, and (5) laborious set up. Thus, over the past decade, there has been interest in adopting new strategies to minimize these limitations. Newer approaches include those that measure caspase release4, BLT esterase activity5, and surface expression of CD1076. The present manuscript suggests that the impedance-based assay, performed on the Roche xCelligence system, may also be useful as an alternative to the CRA. The ability to adapt the impedance-based approach rests on the observation that T cells, which do not adhere tightly to surfaces, do not appear to have much impact on impedance thus diminishing any concern of direct interference of the T cells with the measurement. The labor required for the use of the impedance assay system is expected to be significantly less primarily due to the elimination of the regulatory concerns for the use of radioactivity. The only notable drawback of the impedance-based system is that an outlay of $50,000 to $60,000 is required for purchase of the xCelligence unit. However, the xCelligence unit eliminates the need for a radiation counter and is far more versatile, being useful for assessing cellular response to drugs and migration in addition to T cell cytotoxicity 8.
The observations that the diminution in tumor cell impedance is dose-dependent, as depicted in Figure 2, with a reasonably steep slope following a simple secondary quadratic function suggests that the assay could be used in limiting dilution assays (LDAs) to estimate the frequency of cytotoxic cells9. One advantage with the impedance-based approach is that the number of T cells and tumor cells required is less than the CRA, making LDA potentially more feasible. For example, in the CRA, at a 1:40 ratio, 1 x 105 tumor cells per well are plated which requires 4 x 106 total T cells per well. However, in the impedance-based assay, only 1.5 x 104 tumor cells per well are needed, which consumes only 6 x 105 total T cells per well. Thus, the impedance-based assay uses 85% less T cells and tumor cells. This reduction suggests that the impedance-based system could be adapted to immunologic monitoring in human clinical trials where the volumes of blood draws may be limited. Also, while the CRA is limited to a single time point, the impedance-based assay collects continuous real-time data, allowing the user to compare multiple lysis time points on the same experiment with minimal additional labor and cells. Frequency calculations could potentially be combined with other strategies that do not measure function such as tetramer analyses to establish the ratio of functional cytotoxic T cells to total antigen-specific CD8 T cells10. One of the most appealing features of the impedance-based approach is that, unlike most other assays, cytotoxicity can be monitored in real time, potentially making it easier to study cellular interactions with the use of inhibitors or antibodies. Another is that the method may be suitable for tumor cells that resist labeling. Lastly, since the non-killed tumor cells are not labeled and remain sterile, further harvest and study of the T cell-resistant cells is possible.
In summary, while both systems provide tumor killing data in response to specific T cells, the impedance-based assay is more sensitive than the CRA and provides a more flexible platform.
Disclosures
Production and Free Access of this video-article is sponsored by Roche Diagnostics.
Acknowledgments
This work was supported by grants to Keith L. Knutson from the Susan G. Komen for the Cure Foundation and NIH/NCI 9R01 CA152045 and P50-CA116201.


