Abstract
The nucleobase-ascorbate transporter or nucleobase-cation symporter-2 (NAT/NCS2) family is one of the five known families of transporters that use nucleobases as their principal substrates and the only one that is evolutionarily conserved and widespread in all major taxa of organisms. The family is a typical paradigm of a group of related transporters for which conservation in sequence and overall structure correlates with high functional variations between homologs. Strikingly, the human homologs fail to recognize nucleobases or related cytotoxic compounds. This fact allows important biomedical perspectives for translation of structure-function knowledge on this family to the rational design of targeted antimicrobial purine-related drugs. To date, very few homologs have been characterized experimentally in detail and only two, the xanthine permease XanQ and the uric acid/xanthine permease UapA, have been studied extensively with site-directed mutagenesis. Recently, the high-resolution structure of a related homolog, the uracil permease UraA, has been solved for the first time with crystallography. In this review, I summarize current knowledge and emphasize how the systematic Cys-scanning mutagenesis of XanQ, in conjunction with existing biochemical and genetic evidence for UapA and the x-ray structure of UraA, allow insight on the structure-function and evolutionary relationships of this important group of transporters. The review is organized in three parts referring to (I) the theory of use of Cys-scanning approaches in the study of membrane transporter families, (II) the state of the art with experimental knowledge and current research on the NAT/NCS2 family, (III) the perspectives derived from the Cys-scanning analysis of XanQ.
Keywords: Nucleobase uptake, xanthine permease, Cys-scanning analysis, evolution, specificity, binding site
Using Cys-scanning approaches in secondary active transporters: an overview
Current research challenges
Membrane transport proteins have crucial roles in either human physiology and disease or diverse environmental adaptations of microorganisms. On the other hand, inherent problems in the experimental study of such proteins outside the membrane have long hindered their analyses at high resolution and have limited availability of analytical structure-functional models. Recently, however, there is considerable progress with crystallography and structural modeling of membrane transport proteins. In conjunction with expanding information on new sequence entries from genome analyses, these developments have set the stage for more systematic approaches to link high-resolution structural data with functional evidence.
The above considerations apply more promptly to secondary (ion-gradient driven) transporters, for which the first crystal structures appeared less than a decade ago [1]. The current structural insight on secondary transporters and their classification raises new fundamental questions on the relationships of structure with function. A crucial aspect is that many of the functionally divergent homologs or even separate families of secondary transporters are evolutionarily and structurally related. The majority of known structures fall in only two common folds represented by the lactose permease (LacY) and the neurotransmitter-sodium symporter prototype (LeuT). Other transporters with different folds still display the typical feature of organization in structural repeats that coordinate to form the dynamic binding site [1]. The binding site operates through the commonly accepted alternating access mechanism which is explained as involving both rocking movements of domains pivoting at the binding site and local motions of outside and inside gates flanking the binding site, leading to alterations between outward-facing, inward-facing and intermediate substrate-occluded conformation states [2]. Apart from the structural knowledge, insight on the secondary transport mechanisms requires the concerted use of structural and functional approaches [3]. This is important, since the x-ray structures represent static snapshots of highly dynamic proteins outside their native membrane environment and, with few exceptions [4], interpretations have been based on compilations of different snapshots from different structural homologs.
In the context of the recent evidence, it is striking that conservation in sequence and overall structure in many evolutionarily broad transporter families often correlates with high functional variations between homologs. In most cases, however, very few of the members are characterized experimentally or studied with site-directed mutagenesis while high-resolution evidence from crystallography is scarce. To explore the spectrum of substrate selectivity determinants in such families, it is important to introduce effective mutagenesis designs for the comprehensive ab initio study of new homologs, based on the existing evidence from other, known members. Capitalization on data from Cys-scanning or other systematic mutagenesis analyses of a well-studied homolog, if available, offers a powerful approach to this end.
The Cys-scanning concepts in brief
Cys-scanning mutagenesis is a well-established strategy for structure-function analysis of proteins. It has proven particularly useful and provided valuable insight for the analysis of polytopic membrane proteins and, in particular, membrane transporters [5,6]. Cys-scanning protocols rely on the engineering and availability of functional protein variants which are devoid of all or part of the native Cys residues (Cys-less or Cys-depleted versions, respectively) and the use of these variants as a background for site-specific mutagenesis to introduce new single-Cys replacements at selected positions. The term scanning derives from the common application of this strategy to individually replace each amino acid residue in a contiguous sequence portion or even in the whole sequence of a protein with Cys and create an extensive library of single-Cys replacement mutants for this protein. A battery of different site-directed techniques can be applied to probe specific features of each Cys-substituted position (accessibility to solvent, relevance to substrate binding, sensitivity to the conformational changes of turnover, proximity to other sites in the protein) with appropriate sulfhydryl-specific reagents. Thus, Cys-scanning analysis often yields a wealth of data that are used to build comprehensive structure-mechanistic models for the protein under study, even in the absence of high-resolution crystallographic evidence [5-8].
Cys-scanning mutagenesis and site-directed Cys modification have been widely used to elucidate structure-function relationships in membrane transport proteins. The reasons for this broad application can be outlined as follows:
Cys-less background: Engineering of many bacterial transporters devoid of Cys residues that are functionally equivalent to wild type [9-14] has proven to be feasible. The majority of single-Cys mutants constructed on such a Cysless background do not affect dramatically the transporter expression, structural integrity or function [5,8].
Cys-specific reagents: A diverse compendium of thiol-specific reagents from established companies such as Molecular Probes or Toronto Research Chemicals have been made widely available.
Cys-scanning strategies: A range of thiol-specific reagents and strategies has been developed for membrane proteins, such as substituted Cys accessibility method (SCAM) using hydrophilic methanethiosulfonate (MTS) derivatives [15] or other reagents [16], Cys-Cys cross linking [3,17], site-directed fluorescence spectroscopy [18,19], site-directed spin labeling (SDSL) [20], and site-directed alkylation in situ with radioactive or fluorescent probes [21-23].
Other scanning strategies: The Cys-scanning technologies offer obvious advantage over strategies like Ala-scanning mutagenesis which do not allow further site-specific derivatization of the substituted amino acid.
Lag of appearance of x-ray structures: High-resolution crystallographic models did not appear for membrane transport proteins until the last two decades, due to inherent difficulties with these hydrophobic, integral in the membrane and conformationally dynamic proteins. This fact allowed sufficient time for Cys-scanning applications to expand and provide alternative approaches to the study of membrane transporters.
Delineation of important residues: Cys-scanning methods have succeeded in revealing important residues of membrane transport proteins, including irreplaceable residues, bindingsite residues or residues that are essential for the mechanism of energy coupling. This success is related to the fact that single-Cys mutants are very useful in indicating positions of low significance (active and alkylation-insensitive Cys mutants) and, at the same time, delineating the relatively few residues of potentially major significance (inactive or alkylationsensitive Cys mutants) for more extensive mutagenesis study. It is also based on the diverse array of specific Cys modification reagents and protocols developed and used to probe accessibility to solvent, relevance to substrate binding, sensitivity to the conformational changes of turnover, proximity to other sites or other functional properties for each Cys-substituted position.
Validity of the scanning data: Low-resolution models derived for membrane transport proteins with Cys-scanning approaches continue to provide insight even in the post-crystallization era of research for this class of proteins [3,6]. This is particularly due to the fact that they provide information on the conformational dynamics that is always needed for an integrated insight on the structure-function mechanism.
The research potential of Cys-scanning analysis is not limited to the systematic study of structure-function relationships of individual proteins. The data derived from Cys-scanning analysis of a study prototype can serve as a basis for rapid and effective mutagenesis designs in new, previously unknown or unstudied proteins which are related to the study prototype by sequence or structure homology. In this way, evolutionarily broad families of related proteins can be studied with respect to their active site consensus architecture and function, the spectrum of different specificity trends and mechanistic deviations, and the molecular determinants undermining these differences.
The data derived from the library of mutants produced for a particular transporter in the course of a Cys-scanning mutagenesis study can be used to design new approaches for the analysis of other homologs that might be poorly studied or even not characterized previously with respect to function. In many cases, availability of at least one high-resolution structural prototype (from a solved x-ray structure for one homolog representing an evolutionarily broad family or group of families with structurally related transporters) might provide valuable additional information and allow the formulation of preliminary structural models. However, even in the absence of such models, the information from Cys-scanning analysis of a prototypic homolog per se is sufficient to guide selection of new homologs for study and of amino acid targets for effective site-directed mutagenesis designs in these homologs.
Cys-scanning data as a basis for the study of new homologs
In principle, the Cys-scanning data refer to the functional properties of single-Cys, paired-Cys and other site-directed mutants engineered in the course of the relevant studies. A scanning experiment has two parts at minimum:
Analysis of Cys-replacement mutants: the scope is to delineate positions where a mutant is inactive, of very low activity or sensitive to inactivation by specific alkylating agents [such as the relatively small and membrane permeant N-ethylmaleimide (NEM) which is commonly used to scan for alkylation-sensitive cysteines] and analyze selected mutants for site-directed alkylation in the presence or absence of substrate or other conditions pertinent to appropriate mechanistic questions.
Further site-directed mutagenesis at positions where a single-Cys mutant presents with very low or negligible activity or high sensitivity to inactivation by NEM: the scope is to delineate positions where several replacements yield very low activity or different kinetics or specificity than wild type, and define a pattern of permissive and non-permissive replacements (taking into account the bulk, hydrophobicity, polarity, geometry or other properties of the side chain changes).
Overall, the two lines of experiments are expected to delineate a set of residues that are crucial for the transport mechanism of the study prototype in various respects. Positions where bulky replacements or alkylation of a substituted Cys with the maleimidyl adduct lead to inactivation may reflect important conformational constraints and interactions with other parts of the protein that are essential for the permease turnover [23,24]. Residues which are replaceable with few or no other side chains and, at the same time, accommodate site-specific mutants of impaired affinity or distorted specificity for substrate may be crucial for substrate recognition and binding, while positions of Cys replacements which are protected from alkylation in the presence of substrate may be at the vicinity of the binding site.
It is generally true that such important residues fall in one of the following three categories: (a) irreplaceable; (b) replaceable with few other side chains; (c) sensitive to inactivation of the Cys replacement by NEM. These three potential properties can be used to define the set of important residues of the study prototype as deduced from Cys-scanning analysis data [24-26]. It is also true that this set of residues represents positions with higher degree of side chain conservation than the rest of the protein and, in cases of transporters that have been studied thoroughly, correspond to only 10-15% of the total amino acids in the sequence [5,8,25]. Based on these features, this clearly defined set of residues can be used as a basis to select (i) new homologs for ab initio study and (ii) amino acid targets for site-specific mutagenesis in these homologs. Homology modeling is also of great value in selecting mutagenesis targets for the new homologs provided that a prototypic crystal structure is available; in addition, the Cys-scanning approaches yield dynamic information on the role of specific residues that cannot be provided by the structural models per se.
New homologs to study (from the unknown or poorly characterized pool of sequence entries) can be selected through homology search referring not to the whole coding sequence but to the set of the important residues of the study prototype (as defined above). The aim of this search is to select new homologs on the basis of specific differences in sequence implying distinct conservation patterns that might correlate with shifts in specificity. This process resembles the search for characteristic sequence motifs that are conserved as a consensus throughout a transporter family but is more effective in practice as it is reinforced with laboratory experimental data [26]. In parallel, provided that the homolog under study is characterized functionally and turns out to be different in specificity from the prototype in an assayable manner, amino acid targets for sitedirected mutagenesis of the new homolog are selected from the set of important residues of the prototype, as follows:
Residues that are invariant between the new homolog and the study prototype are used as targets of conservative replacements or more extensive mutagenesis, to provide a measure of the functional conservation for key conserved residues between the two transporters.
Residues that differ between the new homolog and the study prototype are used as targets of replacements of each relevant position of the new homolog with the corresponding amino acid found in the prototype, in an attempt to modulate specificity or other substrate-recognition properties and draw comprehensive conclusions on the determinants of different transporter preferences. Such mutagenesis designs may need to include combinatorial replacements and/or construction of cross-homolog chimeras.
The nucleobase-ascorbate transporter family (NAT/NCS2): state of the art
Nucleobase and nucleobase-related transporters
Purine and pyrimidine nucleobase transporters that are responsible for the cellular uptake of nucleobases and nucleobase-related compounds fall in five distinct families of homologs. All these families are classified as secondary active transporters (ion gradient-driven active transporters) and functionally known members act as H+ or Na+ co-transporters at the plasma membrane of microbial, plant or animal cells. Two of them (the UPS/NBUT family of ureide, uracil, xanthine and/or uric acid transporters and the PUP/POP family of adenine, hypoxanthine, cytosine, cytokinin and/or alkaloid transporters) are specific for the plant kingdom [27] and classified in the Drug/Metabolite Transporter Superfamily (DMT) and the structural prototype of the small multidrug transporter EmrE. A third one (the NCS1 family of cytosine, hypoxanthine, adenine and/or guanine transporters, of uracil/thymine and/or uridine transporters and of allantoin/hydantoin transporters) is confined to archaea, bacteria, fungi, plants and few photosynthetic protists and conforms to the structural prototype of the single crystallographically known homolog Mhp1 which shares common binding site architecture with a range of distantly related families including neurotransmitter transporters, amino acid transporters and sugar transporters, as well [4,28-30]. The remaining two families (the NCS2 or NAT family of uracil and/or purine, xanthine, uric acid, and L-ascorbic acid transporters which is distributed in all major taxa of organisms except protozoa and the AzgA-like family of adenine, hypoxanthine and/or guanine transporters which is confined to archaea, bacteria, fungi, plants and few photosynthetic protists) model on the recently described structure of the single structurally known homolog of NAT/NCS2 family, the uracil permease UraA, which is distinct from any other common fold of secondary active transporters [31].
Strikingly, only one of the above five nucleobase-related transporter families (namely the NAT/NCS2 family) contains homologs in mammals and other animals and, even in this family, the human homologs fail to recognize nucleobases or related cytotoxic compounds [32]. In addition, the human transport systems that engage in the uptake of nucleobases and related drugs are typically very different in topology or predicted structure and belong to different families, mostly to the Equilibrative Nucleoside Transporter (ENT) family, a group of facilitators that recognize nucleosides as their principal substrates [32-34]. In view of the fact that nucleobase-specific transporter homologs are absent from the human or other animal hosts, studies on the structure-function-specificity relations and mechanism of the microbial nucleobase uptake systems might eventually lead to the development of novel antimicrobial drugs targeted specifically at the relevant transporters of pathogens. Such structure-function relationship studies are available to a significant extent for representative members of the NAT/NCS2 family, as described below. On the other hand, considerable progress towards the development of novel therapeutic drugs has been made with targeting the ENT transporters of parasitic protozoa, in particular of kinetoplastid protists [35,36]. The studies with protozoan ENTs (which represent the only family responsible for the uptake of purine nucleobases in parasitic protozoa) have two drawbacks that are absent from the case of the potential relevant research in bacterial or fungal members of the NAT/NCS2 family. These drawbacks are the lack of a robust high-resolution structural model for the ENT family [37,38] and the need to consider for selective inhibitors distinguishing the homologous nucleobase-transporting ENTs of humans or other animal hosts [35,39].
Functional diversification of the NAT/NCS2 family
The Nucleobase-Ascorbate Transporter (NAT) or Nucleobase-Cation Symporter-2 (NCS2) family is unique among the other nucleobase transporter families as it is evolutionarily ubiquitous and includes more than 2,000 putative members in all major taxa of organisms (Figure 1). Among the few exceptions, it is notable that NAT/NCS2 genes are absent from the genomes of most protozoa and of the yeast Saccharomyces cerevisiae. On the other hand, less than 20 members of the NAT/NCS2 family have been characterized experimentally to date; these are specific for the cellular uptake of uracil, xanthine or uric acid (microbial, plant and non-primate mammalian genomes) or vitamin C (mammalian genomes) [26,32,40]. Understanding the mechanism and evolution of different substrate profiles within NAT/NCS2 family is of particular interest in both an evolutionary and a biomedical respect. From the evolutionary perspective, transporters of this family have largely different substrate profiles but share highly conserved short sequence motifs that are functionally relevant [25,26,40-42] and present the same overall topology and predicted tertiary structure [31]. In addition, members of a separate phylogenetic cluster (the AzgA-like family) with clearly distinct substrate profiles [43-45] or sequence conservation motifs are distant structural homologs of the NAT/NCS2 family and model on the same x-ray structure (UraA, 3QE7). From the biomedical perspective, analysis of the NAT/NCS2 mechanism at the molecular level offers important possibilities for translation of knowledge to the rational design of targeted antibacterial or antifungal drugs, based on the fact that the human homologs do not recognize nucleobases or nucleobase-related drugs [32]. In the above context, it is challenging that only 1% (less than 20) of the 2,000 or more predicted members of the family have been identified experimentally with respect to function and only 0.1% (two members, the xanthine permease XanQ and the uric acid/xanthine permease UapA) have been studied rigorously with respect to structure-function relationships [25,40-42,46,47].
Figure 1.
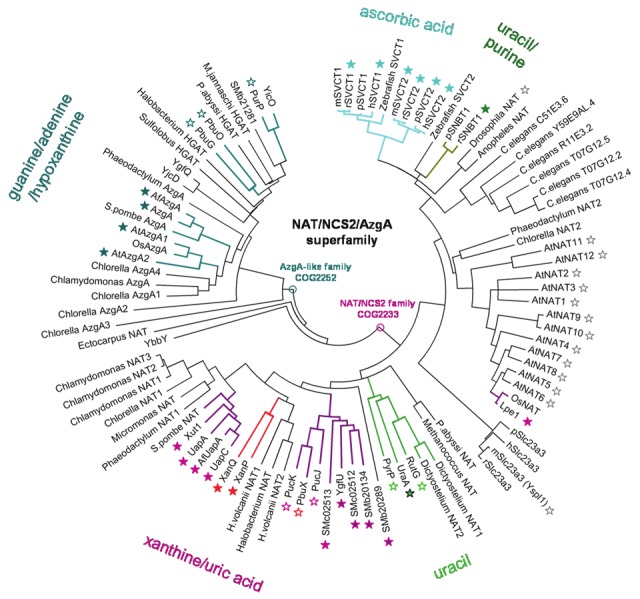
Phylogenetic tree of known members of the NAT/NCS2 family and their relationship with AzgA-like transporters. The NAT/NCS2 family (phylogenetic cluster COG2233) is distinct from the AzgA-like family (phylogenetic cluster COG2252) but they both constitute a superfamily of transporters that conform to the same structural fold [31]. The phylogenetic tree shown has been constructed taking into account all characterized homologs and selected non-characterized ones to represent key organisms from all major taxa [for a complete list of homologs with full names of species and accession numbers, see Supplemental Table S1]. Functionally known homologs are denoted by a colored asterisk that is either filled (information derived directly from transport assays) or empty (information deduced from genetic or genomic studies). Different colors indicate xanthine/uric acid transporters (purple), xanthine-specific transporters (red), uracil transporters (green), uracil/purine transporters (SNBT1; smudge/green), L-ascorbate transporters (SVCT1 or SVCT2; light blue) and adenine/guanine/hypoxanthine transporters of the AzgAlike family (teal). Empty grey asterisks indicate homologs from Drosophila melanogaster [59] or Arabidopsis thaliana [40] that have been studied but their function remains unknown due to inefficient heterologous expression or lack of detectable phenotypes of mutants. Multiple protein sequence alignments were performed with ClustalW and the phylogenetic unrooted tree was constructed by neighbor-joining based on the amino acid pairwise distance with the Poisson correction method and bootstrap test of inferred phylogeny, using the Mega 4.1 software [92].
The first NAT/NCS2 members to be studied and characterized experimentally were the purine permeases UapA and UapC of Aspergillus nidulans [48-50] and the uracil permeases UraA and PyrP of Escherichia coli and Bacillus subtilis, respectively [51,52]. Since then, several other purine (xanthine and/or uric acid)-transporting members have been identified in bacteria (E. coli and B. subtilis) [26,53-55], pathogenic fungi (Aspergillus fumigatus and Candida albicans) [56,57] and plants (Zea mays) [58]. Interestingly, attempts to demonstrate transporter function were unsuccessful for putative homologs of the model angiosperm Arabidopsis thaliana, even though this organism contains 12 members [59]. In contrast to the studied microbial and plant homologs, mammalian members of the family were found to include both nucleobase transporters and L-ascorbate (vitamin C) transporters [32,60-63]. The mammalian genomes contain four homologs, of which two (SLC23A1 and 2, or SVCT1 and 2) are Na+-dependent L-ascorbic acid transporters [61-63], one (SLC23A3) remains functionally unidentified to date [64], and one (SLC23A4, or SNBT1) functions as a Na+-dependent nucleobase transporter of non-primate mammals, responsible for the high-affinity uptake of uracil, but also thymine, hypoxanthine, guanine, and xanthine, as identified recently [32]. It is notable that humans and other primates have lost the corresponding nucleobase uptake system (rSNBT1), for which they contain a genetically defective ortholog (pseudogene) [32]. They retain only two known functional homologs (hSVCT1 and hSVCT2) that function as specific L-ascorbate transporters and do not recognize nucleobase ligands.
Investigating the substrate selectivity differences between closely related members of the NAT/NCS2 family poses important specific questions. For example, what is the basis of the striking difference in substrate preference between rSNBT1 (nucleobases) and hSVCT1 or 2 (L-ascorbic acid) which share a stunning 50% identity (70% similarity) in sequence? A prerequisite to address such questions on the structure-functional deviation between the ascorbate-specific members and their closely related uracil/nucleobase-transporting homologs would be a systematic mutagenesis study of the relevant homologs. It is noteworthy, however, that human or other mammalian NAT/NCS2 transporters have not been studied systematically with mutagenesis to date, albeit scarcely [65,66]. Another question refers to the spectrum of NAT/NCS2 specificities with respect to the recognition of purine substrates. It is true that most of the experimentally known members of the NAT/NCS2 family have been characterized as purine nucleobase transporters which are specific for the proton gradient-driven uptake of xanthine, uric acid (8-oxy-xanthine) or both. This group of related transporters include 11 bacterial, fungal or plant homologs, namely the xanthine transporters XanQ and XanP from E. coli [55] and PbuX from B. subtilis [53], the uric-acid transporters UacT (or YgfU) from E. coli [26] and PucK and PucJ from B. subtilis [54], and the dual-selectivity uric-acid/xanthine transporters UapA [49] and UapC [50] from the filamentous fungus Aspergillus nidulans, AfUapA from its pathogenic relative Aspergillus fumigatus [57], Xut1 from the yeast Candida albicans [56], and Lpe1 from maize (Zea mays) [58]. Based on the known spectrum of these specificities, a major research challenge is to understand the mechanism of differential recognition between xanthine and uric acid (8-oxy-xanthine) and between different binding-site preferences for xanthine analogues with variations at the imidazole moiety (8-methylxanthine, 8-azaxanthine, oxypurinol) [55,56]. In this vein, it is promising that two paradigmatic purine-transporting homologs, the xanthine permease XanQ of E. coli [25,41,42] and the uric acid/xanthine permease of A. nidulans [40,46,47], have been studied extensively with site-directed mutagenesis at motif sequence regions and these studies have reinforced the idea that few residues conserved throughout the family may be invariably critical for function and underlie specificity differences. In further support, most of the conserved residues delineated as important in XanQ or UapA are highlighted as active-site relevant in models built on the recently released x-ray structure of the uracil permease homolog UraA (Protein Data Bank ID: 3QE7) [31]. The relevant studies and related research perspectives are described in more depth below.
Structure and structure-function relationship studies
Of the five known transporter families that specialize in the uptake of nucleobases [27-31], only the NAT/NCS2 family has been studied with mutagenesis to a significant extent. Studies of NAT/NCS2 transporters have included three major approaches, the forward and reverse genetic analysis of the uric acid/xanthine permease UapA from Aspergillus nidulans [47,56,67-74], the systematic Cys-scanning and site-directed mutagenesis of the xanthine permease XanQ from Escherichia coli [25,41,42,75-77] and mutagenesis of selected residues of the uracil permease UraA from E. coli on the basis of its recently solved x-ray structure, the first one elucidated for a NAT homolog [31]. The human ascorbate-transporting members SVCT1 and SVCT2 have also been studied with mutagenesis to some extent, but these studies were rather sporadic and limited to interrogation of the role of conserved His residues [65,66] and of membrane-domain targeting [78,79] or N-glycosylation sites [80]. Briefly, the key methods and findings derived from the structure-function analyses of microbial NAT/NCS2 transporters are as follows:
Studies on the dual-selectivity uric acid/xanthine transporter UapA: The fungal uric acid/xanthine permease UapA from Aspergillus nidulans represents the first member of NAT/NCS2 family to be identified functionally [48] and the only eukaryotic member that has been studied extensively with mutagenesis. These studies began in 1998 with analysis of the so called NAT-signature motif [67-70]. The NAT-signature motif, a conserved 11-amino acid sequence present in all members of the NAT/NCS2 family [41] but not in the structurally related AzgA-like family, was proposed to be associated with determining the purine substrate specificity, based originally on in vivo complementation assays between UapA and its broader-specificity paralog UapC [67]. Subsequently, mutagenesis of the five most highly conserved residues of this motif revealed that Gln-408 and Asn-409 are critical for the UapA function while Gly-411, Thr-416 and Arg-417 are important contributors to the specificity profile [70].
Based on a series of second-site suppressor analyses and targeted mutagenesis of other regions of the protein, the specificity of UapA for uric acid was proposed to derive from conformational interaction between key residues at three distinct regions or “domains” of this transporter [73], namely the first transmembrane segment [47,71], the last transmembrane segment [69,72-74] and an extended sequence region including and flanking the NAT-signature motif [67,73]. More recently, specific interactions between residues of different domains not directly involved in binding were proposed to control accessibility of substrates to the UapA binding site and regulate selection of xanthine/uric acid versus adenine and other purines by a dynamic gating mechanism [74]. Finally, a residue important for optimizing specificity was also found at the third transmembrane segment of UapA (Ser-154) [47].
The studies on UapA have made extensive use of forward and reverse genetics approaches in the model ascomycote A. nidulans and capitalized on the fact that the regulation and functional profiles of all nucleobase transporters and corresponding knock-out strains of this organism as well as the enzymes of purine catabolism are thoroughly known [81-83]. In addition, attempts are under way for the crystallization of UapA that has been overexpressed, purified and characterized biophysically from either the native ascomycote [84] or a yeast host [85,86]. On the other hand, the structure-function analyses of UapA permease are technically limited from the lack of a robust western blotting method to monitor the concentration of UapA mutants in the plasma membrane or a systematic scanning approach, such as Cys-scanning, to rapidly evaluate large contiguous regions of the transporter and assess the role of a more significant fraction of residues [46].
Studies on the xanthine-specific transporter XanQ: A systematic mutagenic analysis of XanQ was launched in 2005, right after the functional identification of this specific, high-affinity xanthine: H+ symporter of Escherichia coli K-12, representing the first purine-transporting NAT/NCS2 member to be characterized in enterobacteria [55]. To facilitate further analysis, a number of experimental tools were developed, including a robust IPTG-inducible expression system to achieve highly controllable overexpression of XanQ [55], a variety of different C-terminal tags to allow quantitation of the XanQ levels in the plasma membrane and purification [55,75] and a fully functional Cys-less version in which the five native Cys residues had been replaced with Ser to allow application of Cys-scanning mutagenesis and Cys derivatization protocols [75].
Cys-scanning analysis was applied to several contiguous regions of XanQ, including the NAT-signature motif [41,75], transmembrane segments and loops flanking the NAT-signature motif [42], the last transmembrane segment [76], the third transmembrane segment [25], as well as amino acid positions that represent highly conserved short sequence motifs [87] or highly polar residues [77] and are scattered throughout the protein. Overall, nearly 50% of the XanQ sequence (220 residues) has been subjected to Cys-scanning mutagenesis to date. The corresponding native amino acids are replaced individually with Cys in the Cys-less background and are initially assayed for expression, activity and sensitivity to site-directed alkylation of the relevant single-Cys mutant. Positions of very low or negligible mutant activity are further subjected to extensive mutagenesis in the wild-type transporter background and selected mutants are subjected to full analysis of the substrate specificity profile using a set of purines and purine analogues as putative competitors of the xanthine uptake [56].
Important residues of the XanQ transporter delineated in this experimental screen are located in transmembrane segments 1, 3, 8, 9, 10 and 14 (Figure 2A). They include four functionally irreplaceable residues (Asp-272, Asp-304, Gln-324, Asn-325), the invariably conserved His-31 where hydrogen bonding is needed for optimal substrate affinity, Asp-276 where a carboxyl group is essential for activity, the specificity-related Asn-93, Gly-333 and Asn-430, the membrane insertion/stabilityrelated Gly-83 and Pro-318, and a number of other positions that display sensitivity to inactivation by a set of site-directed replacements or Cys modification reagents [25,26]. Comparison of the data derived for XanQ with the ones on UapA [47,70-74] has revealed striking similarities between key NAT determinants of the two transporters. Based on homology modeling on the structure of the uracil permease UraA [31], the majority of these determinants are found at the vicinity or at the periphery of the binding site in transmembrane segments 1, 3, 8 and 10 (Figure 2B). As with the fungal UapA, crystallization studies have been performed with XanQ and other closely related bacterial homologs [88] but high-resolution diffracting crystals for a purine-transporting homolog are not yet available.
Figure 2.
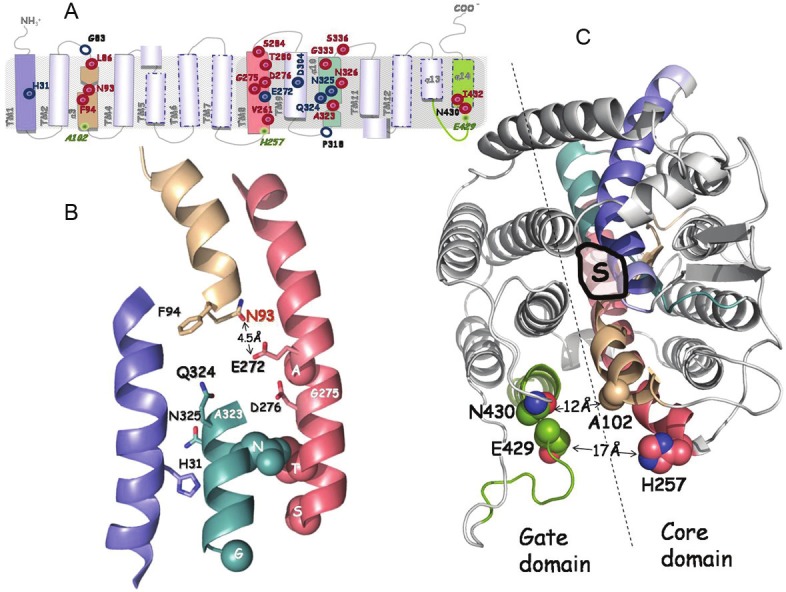
Synopsis of current knowledge on the structure-function analysis of xanthine permease XanQ. The positions of key residues of XanQ are shown in models depicting topology (A), arrangement of the four helices forming the substrate shelter (B) and the overall two-domain structure of the permease (C), as deduced from homology threading on the template of UraA [25,31]. Different colors indicate transmembrane segments TM1 (blue), TM3 (wheat), TM8 (salmon), TM10 (teal) and TM14 (split pea green). In panel A, important residues are indicated with empty blue circles (irreplaceable for expression in the membrane), blue targets (irreplaceable for uptake activity as well as the affinity-related H31) or red targets (sensitive to alkylation or replacement with several side chains). Positions studied for intramolecular interactions are indicated as miniscule green targets. In panel B, the side chains of important residues are shown as thick sticks (irreplaceable and/or substrate binding-relevant) or spheres (sensitive to alkylation or bulky replacement). Residues G275 and A323 where sensitivity to alkylation is enhanced by substrate are indicated with a white label. The specificity-related N93 is shown with red label. In panel C, shown are the coordination site of substrate (S) and distances between residues of the core and the gate domains at the periplasmic side. Further details can be found in the text.
X-ray structure of the uracil transporter UraA and mutagenesis thereof: The x-ray structure of the uracil/5-fluorouracil transporter UraA from E. coli [51] complexed with substrate (uracil) was solved at a resolution of 2.8 Å and released in 2011 (Protein Data Bank ID: 3QE7). Interestingly, uracil was essential for generation of usable UraA crystals and the uracilbound structure was stabilized further by n-nonyl-β, D-glucopyranoside (β-NG) which was included throughout crystallization [31]. The structure of UraA represents a novel fold, different from all other known structural prototypes for membrane transport proteins. The most unusual feature is that two of the 14 transmembrane segments, TM3 and TM10, are only halfway α-helical and contain extended unwound regions followed by short, antiparallel β-strands which provide a shelter for substrate binding at the center of the structure [31]. Overall, the 14 transmembrane segments are arranged in two structural inverted repeats and are spatially organized in two domains, a core domain (TM1-4 and 8-11) and a gate domain (TM5-7 and 12-14). The interface between the two domains is populated mainly with hydrophobic interactions and further stabilized by a bound β-NG molecule [31]. The unusual structure of TM3/TM10 in the core domain is stabilized by a multitude of buried hydrogen bonds [31]. The core domain contains all residues that are crucial in defining the uracil binding site, while the gate domain is proposed to contain more dynamic structural elements that contribute to the mechanism by controlling access of substrates to the binding site and implementing the appropriate conformational changes [31].
The core domain of UraA is almost exclusively responsible for the coordination of substrate at the binding site, which is achieved primarily through direct hydrogen bonds with side chains from TM8 (Glu-241) and TM10 (Glu-290) and nitrogen atoms of the main chain from TM3 (Phe-73) and TM10 (Gly-289), a β-NG-mediated hydrogen bond involving the side chain of His-245 (TM8) and van der Waals interactions involving the side chains of Ala-31 (TM1), Phe-73 (TM3), Tyr-288 (TM10) and Tyr-342 (TM12). Strikingly, residues of XanQ found at corresponding positions of TM3 (Phe-94), TM8 (Glu-272, Asp-276) and TM10 (Ala-323, Gln-324) have been shown to play important roles (Figure 2A and 2B) and Gln-324, at the NAT-signature motif, is irreplaceable for high-affinity substrate binding [41]. In addition to Gln-324, the neighboring Asn-325 is irreplaceable and linked with purine substrate binding [41], while similar observations have been made for Gln-408 and Asn-409 at the NAT-signature motif of UapA [70]. It thus appears that Asn-325/409, an invariant Asn residue in the NAT-signature motif, is more important in defining substrate binding in purine transporters of the family, while its role in the uracil-transporting homolog UraA (Asn-291) is limited to a stabilizing hydrogen bonding interaction with His-24 (TM1) at the periphery of the binding site [31].
To corroborate their structural observations, Lu et al. generated and tested a number of UraA variants with Ala-replacements of selected residues and found that replacement of the substrate-binding relevant Glu-241 (TM8), His-245 (TM8) or Glu-290 (TM10) resulted in abrogation of [3H]uracil uptake and loss of uracil binding (as measured with scintillation proximity assay), while replacement of Phe-73 (TM3), Tyr-288 (TM10), Glu-291 (TM10) or Tyr-342 (TM12) resulted in retention of high uracil-uptake activity [31]. These experiments also established that wild-type UraA recognizes uracil with high affinity (KD 0.4μM, Km 0.5μΜ), comparable to the ones of XanQ for xanthine (Km 4μΜ) [55] and of UapA for xanthine or uric acid (Km or Ki 7-8μΜ) [47,70].
Cys-scanning analysis of the xanthine permease XanQ: lessons and rationales
Rationale of the Cys-scanning projects
On the background of a fully functional Cys-less version [75], residues of the XanQ sequence were subjected to Cys-replacement mutagenesis to create a library of approximately 220 single-Cys mutants [25,41,42,75-77,87]. After assaying the mutants for expression, activity and sensitivity to site-directed alkylation, positions of low or negligible mutant activity were examined further with site-directed mutagenesis and search for substrate profile variations. The compendium of these data was used to delineate a set of putatively important residues as targets for further analysis of structure-function relationships. Finally, homology modeling in conjunction with sequence phylogenetic analyses was applied to evaluate experimental conclusions on the role of individual residues in XanQ and other related transporters [25,26]. The above research strategy has been applied to a range of projects on different sets of XanQ positions, as follows:
Residues at the tips of predicted loops: To address the topology of XanQ permease, single-Cys replacements were introduced at the tips of predicted hydrophilic loops and analyzed with site-directed alkylation using the membrane-impermeant reagent MTSES– [41] to monitor the accessibility of each position to solvent from either side of the membrane. MTSES-accessibility data were also deduced for single-Cys mutants of the NAT-signature motif, a characteristic conserved sequence that contains at least 2 residues implicated in substrate binding [41]. The results showed that single-Cys mutants of predicted loops follow a pattern of alternating accessibility indicative of 12-14 transmembrane segments, while single-Cys mutants of the NAT-signature motif are highly accessible to solvent from the periplasmic side and one of them (N325C) is protected in the presence of substrate, implying that this region forms part of a substrate-accessible hydrophilic cavity [41]. Based on re-evaluation of the data after elucidation of the structure of the uracil-transporting homolog UraA [31], it is now clear that the NAT-signature motif corresponds to a short α-helical segment at the cytoplasmic half of TM10 and contributes to the substrate binding site [25]. In addition, the cysteine-based topology analysis indicated that XanQ may contain extended loop regions at the C-terminal third of the molecule, a contention supported by both circular dichroism spectra showing that the protein is less than 50% α-helical [89] and homology modeling showing extensive unwound regions at the C-terminal side [31].
Conserved short sequence motifs: Cys-scanning mutagenesis was applied in 15 conserved sequence motifs of XanQ (ranging in length from 2 to 11 amino acids) in conjunction with phylogenetic analysis of the 189 closest XanQ homologs (identity > 32%) predicted as putative transporters of xanthine and/or uric acid [87]. Positions of inactive single-Cys mutants in this set of residues included (a) invariant residues located at the core domain (TM1, TM3, TM8, TM10, TM11) or TM12 of the gate domain, (b) residues that conserve a polar side chain at the binding site (Asn-93, Asp-276) or at the periphery (Asp-304), and (c) residues that conserve a small side chain (Ser-68, Gly-71, Ala-129, Gly-152, Gly-217, Gly-436) or a hydrophobic one (Phe-94, Ile-432) and are distributed in several TMs. Further analysis of the conserved sequence motifs of the core domain revealed that Gln-324 and Asn-325 in the NATsignature motif are irreplaceable [75] and important for purine substrate binding [41], the conserved motif ExxGDxxAT of TM8 is crucial for the mechanism and contains the irreplaceable Glu-272 [77] and the essential carboxyl group of Asp-276 [42], the motif GxxxDG of TM9 contains the irreplaceable Asp-304 that is not directly involved in binding [77], while TM3 contains two conserved motifs, one of which contains Asn-93 and Phe-94 at the middle of the transmembrane segment that are important for substrate recognition and specificity [25].
Contiguous regions flanking the NAT-signature motif: Since several residues essential for function or specificity of the purine binding site were found to cluster in the 11-amino acid NAT-signature motif AQNxGxxxxTG [41], extensive sequence regions flanking this motif (58-amino acid upstream and 14-amino acid downstream) in XanQ were subjected to Cys-scanning and site-directed mutagenesis to search for other putative important sites [42]. The whole contiguous sequence (from His-257 to Pro-354) is now known to roughly correspond to the C-terminal repeat of the core domain of the permease, including TM8, TM9, TM10 and TM11 [31]. The results showed that this region, especially TM8 and TM10, is populated with an increased proportion of important residues relative to other segments of XanQ (Figure 2A). Such residues are the four irreplaceable ones (Glu-272, Asp-304, Gln-324, Asn-325), Asp-276 where a carboxyl group is essential for function, Thr-280 that appears to be crucial for packing interactions with other TMs [42] and Pro-318 that is needed for insertion/stability of the protein in the membrane [75]. In addition, there are several side chains of TM8 or TM10 where a single-Cys replacement is highly sensitive to inactivation by site-directed alkylation, implying that these residues play a role at the periphery of the binding site and their alkylation may hinder conformational flexibility or block the release of substrate from the binding pocket. Residues of this category include Gly-275, Ser-284, Ala-323, Asn-326, Thr-332 and Gly-333 [41,42].
Intramembrane polar residues: In a separate project, the full complement of highly polar (Gln or Asn) and putatively charged (Asp, Glu, His, Lys, or Arg) residues of XanQ that are predicted to lie in transmembrane segments were studied with Cys-scanning and site-directed mutagenesis [77]. The rationale was that such residues are expected to face other hydrophilic parts of the protein and/or the solvent-accessible environment of the binding pocket and often play crucial roles in substrate binding and the mechanism of energy coupling in active transport. The studies in XanQ revealed that polar/charged residues critical for function cluster in TM1 (His-31), TM3 (Asn-93), TM8 (Glu-272, Asp-276), TM9 (Asp-304), TM10 (Gln-324, Asn-325) and TM14 (Asn-430) [77]. In NAT/NCS2 family, these side chains are either absolutely conserved (His-31, Glu-272, Asn-325) or show distinct variation patterns among relatives (Asn/Ser/Thr/Ala-93, Asp/Asn/Thr/His-276, Asp/Asn/Glu-304, Gln/Glu/Pro-324, Asn/Ser/Thr-430) [42,77]. Apart from the five essential residues of the C-terminal repeat of the core domain mentioned above [42], these scanning studies delineated three polar residues that are associated with defining the binding affinity or specificity. Of them, the invariably conserved His-31 at the middle of TM1 appears to be involved in a crucial hydrogen bonding interaction required for high-affinity binding of xanthine [77]. Based on homology modeling on the structure of UraA [31], such an interaction is probably partnered by Asn-325 at the periphery of the binding site. An important hydrogen bonding role is also apparent for this His residue of TM1 in other NAT/NCS2 transporters, which is crucial for either the overall structural folding (as proposed in UapA) [47,71] or the constitution of a proper affinity and specificity binding site (as proposed in hSVCT1 and hSVCT2) [65,66] or for both. The other two affinity-and/or specificity-linked polar residues seem to play distinct roles, the one associated directly with purine substrate selectivity (Asn-93 in TM3) and the other located at a strategic and conformationally dynamic position that affects binding indirectly (Asn-430 in TM14). Overall, 6 of the 8 polar side chains of XanQ delineated as mechanistically important fall in the substrate binding shelter formed by TM1, TM3, TM8 and TM10 of the core domain (Figure 2B).
Transmembrane segment TM3: The entire TM3 and flanking hydrophilic sequences were subjected to mutagenesis to elucidate the role of residues upstream and downstream of Asn-93 [25]. In conjunction with homology modeling and sequence phylogenetic analysis these studies showed that Asn-93 is at the junction between a short β-sheet and an α-helix at the middle of TM3 and confers to substrate recognition and specificity together with the neighboring Phe-94. Phe-94 appears to contribute to the proper recognition of xanthine analogs that differ in the pyrimidine moiety (2-thio and 6-thioxanthine, 3-methylxanthine) while Asn-93 is essential to prevent aberrant recognition of substrates or analogs that differ in the imidazole moiety (8-methyl and 7-methylxanthine, uric acid). In addition, replacement of Asn-93 with a smaller polar (Ser) or hydrophobic residue (Ala) has the unique property of allowing uptake of uric acid, which is strictly excluded as a substrate from the binding site of wild-type XanQ [25]. The most probable explanation is that the side chain replacing Asn-93 is oriented away from the substrate-binding relevant Glu-272 and relaxes the specificity constraint by leaving more space for interactions in the binding pocket [25]. In addition to the substrate-interacting residues, TM3 contains important residues clustered in a conserved GxGLx motif at its cytoplasmic unwound-sequence region. This motif may permit conformational flexibility needed to dictate the unique architectural fold of TM3 (mainly through Gly-83, that is irreplaceable for the permease integrity/stability in the membrane) or establish appropriate interactions with other TMs needed in the alternating access mechanism (mainly through Leu-86, that represents a highly alkylation-sensitive position at the binding site periphery) [25].
Transmembrane segment TM14: The majority of important residues of XanQ identified through Cys-scanning and site-directed mutagenesis fall in transmembrane segments of the core domain. On the other hand, TM14 of the gate domain had been indicated to interact functionally with the NAT-signature motif (corresponding to TM10 of the core domain), based on second-site suppressor analysis of mutants replacing Gln-408 in the fungal UapA [69]. These studies on UapA led to the proposition that an aromatic residue at the middle of TM14 (Phe-528) acts as a purine selectivity filter that is distinct from the binding site [72] and a similar role was later proposed for Thr-526 [74]. To understand the role of TM14 in detail, a systematic study of this segment was undertaken using both Cys-scanning mutagenesis of XanQ and chimeric XanQ/UapA transporter analysis [76]. Strikingly, two residues at the middle of TM14, Asn-430 and Ile-432 that correspond to the non-conserved important side chains of Thr-526 and Phe-528 in UapA, were found to contribute to optimal activity and specificity of XanQ and allow rescue of xanthine uptake activity in an inactive XanQ/UapA chimera [76]. The remaining residues of TM14 were shown to have minor role [76] but contain molecular determinants that are important to optimize activity or specificity in a chimeric XanQ/UapA background [90]. Since the single-Cys N430C was found to be alkylation-sensitive and protected from alkylation in the presence of 2-thioxanthine, it was originally thought that Asn-430 may be at the vicinity of the binding site and interfere with the substrate analogue directly [76]. In view of the structural model of UraA [31], it is now clear that TM14 is distal from the binding site (Figure 2C) and the ligand effect on N430C is possibly due to a conformational interaction of TM14 with TM10 or other TMs of the core domain. Such an interaction has been investigated using paired-Cys replacements and cross-linking assays (see next section).
Delineation of important residues
To date, the combined Cys-scanning and sitedirected mutagenesis of the XanQ permease has yielded a total of about 385 single-replacement mutants that provided information on the role of about 220 native amino acid residues to various extents. Among these residues, 29 have been annotated as important based on criteria deduced from the properties of the single-replacement mutants at these positions. In brief, residues annotated as important are either (1) irreplaceable with respect to function (Glu-272, Asp-304, Gln-324, Asn-325) or expression in the membrane (Gly-83, Pro-318), (2) replaceable with a limited number of other side chains (His-31, Asn-93, Phe-94, Asp-276, Ala-279, Thr-280, Gly-305, Gly-351, Pro-354, Ile-432), or (3) highly sensitive to inactivation of a substituted Cys mutant by N-ethylmaleimide (Leu-86, Val-261, Gly-275, Ser-284, Ala-323, Asn-326, Gly-327, Val-328, Ile-329, Thr-332, Gly-333, Ser-336, Asn-430). Most of these 29 residues are highly conserved in NAT/NCS2 family which facilitates analysis of their role not only in XanQ but also in homologous transporters [26]. To address the putative role of these residues in XanQ (for example, whether they are involved in binding, specificity, conformation cycling or crucial intramolecular interactions) one can use more specific criteria, as follows:
Binding site: To interrogate whether a residue is at the binding site or at the periphery, the effects of substrate on both the activity and the side chain reactivity of mutants should be taken into account. Residues in the binding site that interact directly with substrate (xanthine) are expected to be irreplaceable either for detectable transport activity or for binding xanthine and analogues with decent affinity. In addition, residues at the periphery of the binding site that are not irreplaceable for substrate affinity are expected to display steric protection of their single-Cys mutants in the presence of saturating concentrations of xanthine. Therefore, a strong biochemical indication that a residue is at the binding site or at the periphery can be based on two criteria: (a) abrogation of affinity for substrate and analogues with any of the site-replacement mutants; (b) very low or negligible uptake with any of the mutants and substrate-caused protection of the single-Cys mutant against site-specific alkylation independently of the reagent used. In XanQ, two sequential residues of the NAT-signature motif (Gln-324, Asn-325) meet either one of these criteria. Mutants replacing Gln-324 display very low or negligible activity and the marginally active ones (Q324E, Q324N) are grossly impaired with respect to affinity for xanthine (as measured with transport kinetics assay) or xanthine analogues (as measured with ligand competition assays) [75], and the single-Cys Q324C also fails to bind substrate as implied from the results of site-directed alkylation assays [41]. Mutants replacing Asn-325 are essentially inactive but site-directed alkylation analysis of N325C shows that high-affinity binding of xanthine is retained and protects the Cys at this position from alkylation with either N-ethylmaleimide (NEM) or 2-sulfonatoethyl methanethiosulfonate (MTSES-) most probably due to steric hindrance [41]. Thus, the data support that Gln-324 interacts directly with substrate in the binding site (criterion a) while Asn-325 is at the periphery and interferes with substrate binding sterically (criterion b). Both the functional properties of relevant UapA mutants [70] and homology modeling on the template of UraA structure [31] are in agreement with the above conclusions.
Substrate accessible cavity: Residues that are at a distance from the binding site or at the vicinity but not interacting with binding might define a substrate-accessible hydrophilic space within the protein which roughly delineates the translocation pathway of substrate. Two criteria can be used to identify such residues from the Cys-scanning analysis data: (a) sensitivity of the single-Cys mutant at this position to inactivation by N-ethylmaleimide (NEM) [5]; (b) reactivity of the single-Cys mutant at this position with MTSES- or another membrane impermeant hydrophilic reagent. Sensitivity to inactivation by NEM indicates a position where the bulky maleimidyl adduct prevents the release of substrate from the binding pocket by blocking the conformational changes needed in the alternating access mechanism [5). Reactivity of a transmembrane-segment residue with MTSES- indicates that this position is accessible to solvent from outside of the membrane which is expected of a residue contributing to the internal substrate- and water-accessible cavity [21]. In XanQ permease, both criteria (sensitivity to NEM and reactivity with MTSES-) are met by several residues of the NAT-signature motif in TM10 (Ala-323, Asn-326, Gly-327, Val-328, Ile-329, Thr-332, Gly-333, Ser-336) [41]. In addition, the criterion of sensitivity to NEM is displayed by residues of TM8 (Val-261, Gly-275, Ser-284) [42], TM3 (Leu-86) [25] and TM14 (Asn-430) [76]. These data support that the substrate translocation pathway is lined in part by a short α-segment of TM10, a long α-helical face of TM8 and residues at strategic positions of TM3 and TM14. Homology modeling of the permease structure in an inward-open substrate-bound conformation [25,31] substantiates further the above conclusions with regard to the cytoplasmic parts of TM10, TM8 and TM3.
Specificity: Residues that contribute importantly to substrate specificity may be revealed by the properties of the relevant site-directed replacement mutants. A specificity-related residue is expected to fulfil at least one of the following criteria: (a) different activity or specificity profile with different mutants indicating a pattern of constraints in site chain replacement at this position; (b) radically altered profile for recognition of analogues with one or more sitedirected replacements; and (c) clear deviation in substrate selectivity (such as capacity of transporting a new, non-wild-type substrate) displayed by at least one of the site-replacement mutants. In XanQ, all the above criteria are fulfilled with Asn-93 (TM3), which appears to be crucial for specificity with respect to the imidazole moiety of xanthine and for the counter-selectivity against uric acid [25]. In addition, the first two criteria are fulfilled with several residues, most of which appear to contribute directly or indirectly to the specificity of the binding site with respect to the imidazole moiety, i.e. the exclusion of analogues substituted at positions 7 or 8 of xanthine. Site-directed XanQ mutants that display a radically altered profile include H31Q (TM1), allowing recognition of 8-methylxanthine and, to a marginal extent, of 7-methylxanthine and uric acid [77], N93A or N93S (TM3), allowing high-affinity recognition of 8-methylxanthine and low but detectable transport of uric acid [25], F94Y (TM3), disrupting affinity for 2-thio and 6-thioxanthine [25], D276E (TM8), allowing high-affinity recognition of 8-methylxanthine but disrupting recognition of any analogue at the pyrimidine moiety [42], T332N (TM10), allowing recognition of 8-methylxanthine and compromising affinity of 1-methylxanthine [75], G333R (TM10), allowing recognition of 7-methyl and 8-methylxanthine and, to a marginal extent, of uric acid [41], S336N (TM11), disrupting recognition of 6-thioxanthine [41], N430C and I432T (TM14), enhancing recognition of 2-thioxanthine but disrupting affinity for other analogues [76], and I432Q, I432N or I432V (TM14), disrupting affinity for analogues at the pyrimidine moiety except 2-thioxanthine [76]. Three of the above mutants (N93A, N93S, D276E) enable binding of 8-methylxanthine with high affinity (Ki values 32μM, 48μM, 79μM, respectively). Two of them (N93A and N93S) can transport uric acid (8-oxy-xanthine) in addition to the wildtype substrate (xanthine), a property that is unique among all XanQ mutants tested [25]. However, the capacity of these mutants for uric acid uptake is very low [25].
Conformation sensors: Residues of which the single-Cys replacement mutant is reactive with N-ethylmaleimide (NEM) and its reactivity or sensitivity to inactivation is altered significantly in the presence of substrate are probably at key positions in the tertiary structure of the permease that move to a different environment with the conformational change elicited by substrate binding. Such residues are often found at sites that are distant or oriented away from the binding site, with most dynamic changes usually reported at the cytoplasmic or the periplasmic entrance of the internal hydrophilic cavity [5,6]. In XanQ, increased sensitivity to inactivation by NEM at saturating concentrations of substrate (xanthine) has been reported with G275C (TM8) (IC50 change from 14.5μM to 7.9μM) [42] and A323C (TM10) (IC50 change from 34μM to 14μM) [41], implying diverging movements of the cytoplasmic parts of TM8 and TM10 during the mechanism of transport that expose the relevant side chains to the reagent. This interpretation is consistent with structural modeling which predicts that the binding site is accessible from the cytoplasm at the substrate-bound conformation [31] (Figure 2B). In any event, it should be noted that the substrate effects on side-directed alkylation of the single-Cys XanQ mutants have not been studied extensively, except in the sequence region of the NAT-signature motif (residues 321-339) [41]. In the NAT-signature sequence, all single-Cys mutants were found highly reactive with both NEM and the membrane-impermeant MTSES- and, with the exception of A323C and the substrate-relevant N325C (see above), their accessibility or reactivity is practically unaltered by substrate [41]. These results confirm that the motif is close to the binding site and not at distal regions of the internal cavity that would be subject to more dynamic conformational changes [41].
Intramolecular interactions: Although the majority of important XanQ residues have been found at the core domain which is implicated directly in defining binding affinity and specificity, more dynamic structural elements that contribute to the alternating access mechanism by controlling access to and release from the binding site are expected to be located at the gate domain [31]. In this respect, it is important to identify the interactions between helices of the core and the gate domains of XanQ and study their putative mechanistic roles. One site of such interactions involves the junction between the unwound region and the short α-helix of the last transmembrane segment (TM14), which includes the specificity-related Asn-430 and Ile-432 and has been proposed to “cross-talk” functionally with the NAT-signature motif (TM10) based on the properties of corresponding double-replacement mutants [76]. Homology modeling indicates that this site of TM14 is relatively close to the periplasmic ends of TM3 and TM8 at the interface between the core and the gate domains (Figure 2C). Paired-Cys replacements have been used to investigate such a putative interaction via cadmiumsensitivity assays and cross linking [91]. The double mutant E429C/H257C was found to be sensitive to inhibition by cadmium chloride, indicating that positions Glu-429 (TM14) and His-257 (TM8) are sufficiently close to allow interaction of the two substituted Cys residues through coordination of a cadmium ion (Cd2+) resulting in severe limitations in the conformational movements and inhibition of active transport. Experiments are underway to analyze the extent and significance of such interactions between TM3/TM8/TM14 by Cys-Cys crosslinking assays [91].
Study of homologous transporters
NAT/NCS2 family encompasses at least 2,000 structurally homologous members with diverse substrate preferences or specificities but very few of the members are characterized experimentally. The information derived from the Cys-scanning analysis of XanQ permease, in conjunction with the structure model of UraA and the existing genetic evidence from the fungal UapA, can serve as a basis to study structure-function relationships in other related members of the NAT/NCS2 family which are not yet characterized. In this respect, it is challenging that most of the functionally known homologs including the well studied XanQ and UapA are selective for xanthine or uric acid (8-oxy-xanthine) or for both and display distinct variations in their specificity profiles [55-58].
Inspection of conserved sequence motifs and sequence alignment analysis of the different xanthine and/or uric acid-transporting homologs indicated interesting patterns of correlation with changes between xanthine-selective and xanthine/uric acid dual-selectivity transporters, especially at the characteristic NAT-signature motif [41]. However, such sequence differences did not correlate with clear-cut changes in substrate selectivity of corresponding mutants which were made in either XanQ [41,75] or UapA [70,73]. The most pronounced change in XanQ was accomplished with replacement of Gly-333 to Arg (at the carboxyl-terminal end of the motif sequence) yielding aberrant recognition of 7-methyl and 8-methylxanthine (which are not wild-type ligands) but without affecting the selectivity preference for xanthine [41]. Recognition of 8-methylxanthine has also been observed with a number of other single-replacement XanQ mutants and even with UapA/XanQ chimeras [76], implying that several changes at different sites in this xanthinespecific transporter can confer a degree of promiscuity for the recognition of analogues at the imidazole moiety of xanthine. However, none of these changes resulted in a clear selectivity change (most notably, none allowed recognition or uptake of uric acid except N93A/S which transport uric acid to a marginal extent) [25]. Thus, it is evident that the strict preference of XanQ for xanthine is not easily modifiable and a more systematic approach is needed to address the basis of xanthine/8-oxy-xanthine selectivity differences. Such an approach is offered through the exploitation of evidence from the Cys-scanning analysis of XanQ and the elucidation of the function of new, closely related but distinct-selectivity homologs. An application example of this approach has been described recently for elucidation of the molecular basis of selectivity of UacT, a newly characterized uric acid transporter from E. coli [26].
In practice, UacT was identified functionally in a screen for characterization of the complement of putative purine transporters of E. coli K-12 [26]. UacT is related in sequence and genomic locus with XanQ and is found along with it in a cluster of putative purine catabolic genes. Although the pathway of catabolic oxidation of uric acid to allantoin in E. coli is unclear, UacT was identified as a proton-gradient dependent, low-affinity (Km 0.5mM) and high-capacity transporter for uric acid [26]. UacT can also transport xanthine, but with disproportionately low capacity [26]. The clear selectivity difference between UacT (uric acid) and XanQ (xanthine) which share an approximately 30% identity in sequence prompted a rationally designed site-directed mutagenesis study of UacT based on the data available from the Cys-scanning analysis of XanQ [26]. Mutagenesis targets were selected from UacT residues corresponding to the set of the important residues of XanQ (Supplemental Figure S1). Residues of this set that had been found to be irreplaceable for function or essential for substrate affinity/specificity in XanQ were subjected to extensive mutagenesis in UacT. Of the remaining residues of this set, the ones that correspond to non-conserved side chains were replaced with the corresponding amino acid found in XanQ and vice versa [26].
The most prominent results from this comprehensive first-time mutagenesis study of UacT permease were that His-37 (TM1), Glu-270 (TM8), Asp-298 (TM9), Gln-318 and Asn-319 (TM10) are functionally irreplaceable, and Thr-100 (TM3) is essential for the uric acid selectivity. The five irreplaceable residues of UacT are conserved in all purine transporters of the NAT/NCS2 family and four out of five are also irreplaceable for function in XanQ [75,77] and in UapA [70,73]. Structural modeling indicates that two of them (Glu-270 and Gln-318, corresponding to Glu-272 and Gln-324 of XanQ) are in the substrate binding pocket and may form direct hydrogen bonds with substrate [31]. On the other hand, the selectivity-associated Thr-100 (Asn-93 in XanQ) which is also at the vicinity of the binding site displays poor conservation in NAT/NCS2 family but retains invariably a polar character in the known nucleobase-transporting members (Asn, Thr or Ser) while the ascorbate-transporting members have an Ala [25]. The conclusion that the side chain of Thr-100 is linked with the transporter ability to differentiate between uric acid (8-oxy-xanthine) and xanthine was based on the finding that replacement of this residue with Ala changes the uric acid-selective UacT to a dual-selectivity variant transporting both uric acid and xanthine with comparable efficiencies [26]. A similar but less pronounced effect is evident with the corresponding amino acid replacement in XanQ (N93A) which converts the xanthine-selective XanQ to a dual-selectivity variant albeit with very low capacity for the non-wild type substrate (Figure 3).
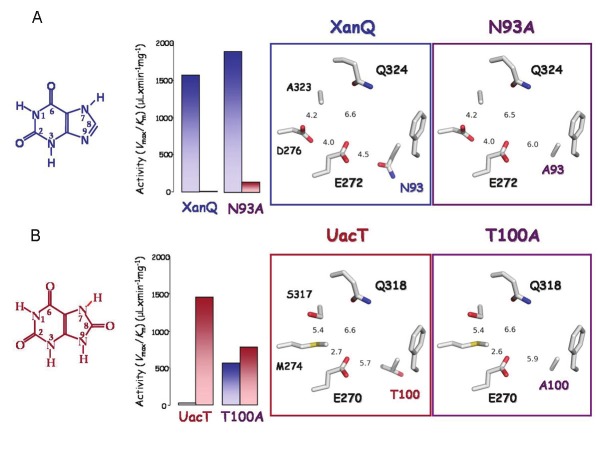
Figure 3.
Determinants of the xanthine/8-oxy-xanthine distinction between permeases XanQ and UacT. The major substrate, relative efficiencies for the uptake of xanthine (blue bars) and uric acid (red bars) and the arrangement of side chains in the presumed binding site are shown for XanQ and its mutant N93A (A) or UacT and its mutant T100A (B). The two mutants shown were the only ones leading to clear-cut changes in selectivity in a mutagenesis screen of 16 important residue positions [for a complete account of this set of mutagenesis targets, see Supplemental Figure S1]. Among the other binding-site residues, E270/272 and Q318/324 are functionally irreplaceable in either transporter, S317 contributes to the uric acid selectivity to a lesser extent and M274 has a stabilization role on the binding site integrity [26]. Transport efficiencies (Vmax/Km) for uric acid were measured at 25°C [26]. Transport efficiencies (Vmax/Km) for xanthine were measured at 25°C (A) or 37°C (B) [25,26]. The sequence of XanQ or UacT was threaded on the known x-ray structure of UraA [31] using the SWISSPROT modeling server and displayed with PyMOL v1.4 (Schrödinger, LLC). Numbers represent minimal distances in Ǻ between the indicated side chains.
Homology modeling of the purine-transporting members of the NAT/NCS2 family indicates that the relevant position of TM3 (Thr-100/Asn-93) is vicinal to the substrate binding site but the Thr or Ser replacing Asn-93 in UacT and all dual-selectivity homologs is distal from the conserved, substrate-relevant glutamate of TM8 (minimal distance between oxygen atoms, 6.0 Ǻ) while Asn-93 in XanQ is significantly closer (distance between oxygen atoms, 4.5 Ǻ). In the dually selective UapA, occupation of the Asn-93 position by Ser orients this residue away from the substrate-relevant Glu of TM8, leading to relaxation of a constraint for the recognition of analogues at position 8 and allowing binding and transport of uric acid (8-oxy-xanthine). In XanQ, the mutants replacing Asn-93 with Ser or Ala also orient this residue away from the substrate-relevant Glu (Glu-272) and yield efficient recognition of 8-methylxanthine and low but significant uptake of uric acid [25], mimicking in part the dual-selectivity homolog (Figure 3A). In UacT, Thr-100 is also oriented away from the carboxyl group of Glu-270 (TM8) leaving more space in the substrate binding pocket but, at the same time, the pKa of Glu-270 may be distorted significantly relative to the corresponding carboxylic acid in XanQ due to its vicinity with hydrophobic groups from Thr-100 and Met-274 (Figure 3B). These changes on the substrate binding Glu-272/Glu-270 might account for the selectivity difference between UacT (uric acid) and XanQ (xanthine). Replacement of Thr-100 with Ala leads to a dual-selectivity profile possibly due to partial alleviation of the hydrophobicity and space constraints with respect to Glu-270 in UacT (Figure 3B). On the other hand, replacement of Asn-93 with Thr in XanQ cannot imitate the UacT profile but leads to low affinity for all xanthine analogues [25], possibly due to interference of the methyl group of Thr-93 in the vicinity of the essential Glu-272 which is not counterbalanced by other changes. Based on these observations, it is evident that further combinatorial replacements are needed to promptly convert XanQ to the UacT selectivity profile or vice versa. Targets for such replacements can be selected from residues of the important set taking into account their orientation relative to the binding site in the modeled structures and functional properties of the relevant single-replacement mutants [26].
The above comparative studies of xanthine and/or uric acid-selective transporter homologs open the way for more systematic insight on the molecular basis of the evolution of substrate profile preferences in NAT/NCS2 family, by means of characterizing and concisely analyzing selected new homologs for structurefunction relationships.
Concluding remarks
The systematic Cys-scanning analysis of the xanthine permease XanQ from E. coli K-12 has allowed delineation of a set of important residues in NAT/NCS2 family and usage of the properties of the corresponding mutants as a basis to study new homologs with novel substrate profiles. The analysis of XanQ as a study prototype for the evolutionarily ubiquitous NAT/NCS2 family was launched after initial functional indications on the role of one characteristic sequence motif of the family (the NAT-signature motif) about seven years ago and was completed to a major extent before elucidation of the first x-ray structure of a related homolog (UraA) one year ago. In retrospect, structural modeling has supported many of the conclusions deduced from analysis of the Cys-scanning data while the combination of the structure predictions with functional evidence has permitted more focused further mutagenesis designs. As is clear from this review, the work on XanQ permease has set the stage for more rationalized structure-function-specificity analyses to address ab initio studied homologs and understand the spectrum of specificity variations in purine transporters of the family. From a broader methodological point of view, the data show that comprehensive analysis of structure-function relationships in newly characterized transporters can be accomplished with relatively few site-directed replacements, based on the knowledge available from Cys-scanning mutagenesis of one prototypic homolog.
Acknowledgements
Work in the author’s laboratory has been supported in part by the European Union, European Social Fund (ESF) and National Sources, within the framework of programs NONEU (Collaborations with Research and Technology Organizations outside Europe; Greece-USA), PENED (Reinforcement Programme of Human Research Manpower) and Heraklitos II (Education and Lifelong Learning, Hellenic Ministry of Education). I wish to thank Ekaterini Georgopoulou, Ekaterini Karena, George Mermelekas and Konstantinos Papakostas for key research contributions in the context of their participation in the above programs.
Supporting Information
References
- 1.Boudker Ο, Verdon G. Structural perspectives on secondary active transporters. Trends Pharmacol Sci. 2010;31:418–426. doi: 10.1016/j.tips.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forrest LR, Krämer R, Ziegler C. The structural basis of secondary active transport mechanisms. BBA Bioenergetics. 2011;1807:167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Kaback HR, Smirnova I, Kasho V, Nie Y, Zhu Y. The alternating access transport mechanism in LacY. J Membr Biol. 2011;239:85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weyand S, Shimamura T, Beckstein O, Sansom MS, Iwata S, Henderson PJ, Cameron AD. The alternating access mechanism of transport as observed in the sodium-hydantoin transporter Mhp1. J Synchrotron Radiat. 2011;18:20–23. doi: 10.1107/S0909049510032449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frillingos S, Sahin-Tóth M, Wu J, Kaback HR. Cys-scanning mutagenesis: a novel approach to structure-function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 6.Kaback HR, Dunten R, Frillingos S, Venkatesan P, Kwaw I, Zhang W, Ermolova N. Site-directed alkylation and the alternating access model for LacY. Proc Natl Acad Sci USA. 2007;104:491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorgen PL, Hu Y, Guan L, Kaback HR, Girvin ME. An approach to membrane protein structure without crystals. Proc Natl Acad Sci USA. 2002;99:14037–14040. doi: 10.1073/pnas.182552199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura N, Konishi S, Yamaguchi A. Mechanisms of drug/H+ antiport: complete cysteine-scanning mutagenesis and the protein engineering approach. Curr Opin Chem Biol. 2003;7:570–579. doi: 10.1016/j.cbpa.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 9.van Iwaarden PR, Pastore JC, Konings WN, Kaback HR. Construction of a functional lactose permease devoid of cysteine residues. Biochemistry. 1999;30:9595–9600. doi: 10.1021/bi00104a005. [DOI] [PubMed] [Google Scholar]
- 10.Weissborn AC, Botfield MC, Kuroda M, Tsuchiya T, Wilson TH. The construction of a cysteine-less melibiose carrier from E. coli . BBA Biomembranes. 1997;1329:237–244. doi: 10.1016/s0005-2736(97)00116-8. [DOI] [PubMed] [Google Scholar]
- 11.Jung H, Rübenhagen R, Tebbe S, Leifker K, Tholema N, Quick M, Schmid R. Topology of the Na+/proline transporter of Escherichia coli . J Biol Chem. 1998;273:26400–26407. doi: 10.1074/jbc.273.41.26400. [DOI] [PubMed] [Google Scholar]
- 12.Sahin-Tóth M, Frillingos S, Lawrence MC, Kaback HR. The sucrose permease of Escherichia coli: functional significance of cysteine residues and properties of a cysteine-less transporter. Biochemistry. 2000;39:6164–6169. doi: 10.1021/bi000124o. [DOI] [PubMed] [Google Scholar]
- 13.Slotboom DJ, Konings WN, Lolkema JS. Cysteine-scanning mutagenesis reveals a highly amphipathic, pore-lining membrane-spanning helix in the glutamate transporter GltT. J Biol Chem. 2001;276:10775–10781. doi: 10.1074/jbc.M011064200. [DOI] [PubMed] [Google Scholar]
- 14.Culham DE, Hillar A, Henderson J, Ly A, Vernikovska YI, Racher KI, Boggs JM, Wood JM. Creation of a fully-functional cysteine-less variant of osmosensor and proton-osmoprotectant symporter ProP from Escherichia coli and its application to assess the transporter’s membrane orientation. Biochemistry. 2003;42:11815–11823. doi: 10.1021/bi034939j. [DOI] [PubMed] [Google Scholar]
- 15.Akabas MH, Stauffer DA, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 16.Yan RT, Maloney PC. Identification of a residue in the translocation pathway of a membrane carrier. Cell. 1993;75:37–44. [PubMed] [Google Scholar]
- 17.Wu J, Kaback HR. A general method for determining helix packing in membrane proteins in situ: helices I and II are close to helix VII in the lactose permease of Escherichia coli . Proc Natl Acad Sci USA. 1996;93:14498–14502. doi: 10.1073/pnas.93.25.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung K, Jung H, Wu J, Privé GG, Kaback HR. Use of site-directed fluorescence labeling to study proximity relationships in the lactose permease of Escherichia coli. Biochemistry. 1993;32:12273–12278. doi: 10.1021/bi00097a001. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Frillingos S, Kaback HR. Dynamics of lactose permease of Escherichia coli determined by site-directed chemical labeling and fluorescence spectroscopy. Biochemistry. 1995;34:8257–8263. doi: 10.1021/bi00026a007. [DOI] [PubMed] [Google Scholar]
- 20.Voss J, Hubbell WL, Kaback HR. Distance determination in proteins using designed metal ion binding sites and site-directed spin labeling: application to the lactose permease of Escherichia coli . Proc Natl Acad Sci USA. 1995;92:12300–12303. doi: 10.1073/pnas.92.26.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frillingos S, Kaback HR. Probing the conformation of the lactose permease of Escherichia coli by in situ site-directed sulfhydryl modification. Biochemistry. 1996;35:3950–3956. doi: 10.1021/bi952601m. [DOI] [PubMed] [Google Scholar]
- 22.Guan L, Kaback HR. Site-directed alkylation of cysteine to test solvent accessibility of membrane proteins. Nature Protocols. 2007;2:2012–2017. doi: 10.1038/nprot.2007.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang X, Nie Y, Kaback HR. Site-directed alkylation studies with LacY provide evidence for the alternating access model of transport. Biochemistry. 2011;50:1634–1640. doi: 10.1021/bi101988s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavoulari S, Frillingos S. Substrate selectivity of the melibiose permease (MelY) from Enterobacter cloacae . J Mol Biol. 2008;376:681–693. doi: 10.1016/j.jmb.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Karena E, Frillingos S. The role of transmembrane segment TM3 in the xanthine permease XanQ of Escherichia coli . J Biol Chem. 2011;286:39595–39605. doi: 10.1074/jbc.M111.299164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papakostas K, Frillingos S. Substrate selectivity of YgfU, a uric acid transporter from Escherichia coli . J Biol Chem. 2012;287:15684–15695. doi: 10.1074/jbc.M112.355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Möhlmann T, Bernard C, Hach S, Neuhaus HE. Nucleoside transport and associated metabolism. Plant Biol. 2010;(suppl 1):26–34. doi: 10.1111/j.1438-8677.2010.00351.x. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki S, Henderson PJ. The hydantoin transport protein from Microbacterium liquefaciens . J Bacteriol. 2006;188:3329–3336. doi: 10.1128/JB.188.9.3329-3336.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O’Reilly J, Ma P, Saidijam M, Patching SG, Hope RJ, Norbertczak HT, Roach PC, Iwata S, Henderson PJ, Cameron AD. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimamura T, Weyand S, Beckstein O, Rutherford NG, Hadden JM, Sharples D, Iwata S, Henderson PJ, Cameron AD. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328:470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu F, Li S, Jiang Y, Jiang J, Fan H, Lu G, Deng D, Dang S, Zhang X, Wang J, Yan N. Structure and mechanism of the uracil transporter UraA. Nature. 2011;472:243–246. doi: 10.1038/nature09885. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto S, Inoue K, Murata T, Kamigaso S, Yasujima T, Maeda J, Yoshida Y, Ohta K, Yuasa H. Identification and functional characterization of the first nucleobase transporter in mammals: implication in the species difference in the intestinal absorption mechanism of nucleobases and their analogs between higher primates and other mammals. J Biol Chem. 2010;285:6522–6531. doi: 10.1074/jbc.M109.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao SY, Ng AM, Cass CE, Baldwin SA, Young JD. Nucleobase transport by human equilibrative nucleoside transporter 1 (hENT1) J Biol Chem. 2011;286:32552–32562. doi: 10.1074/jbc.M111.236117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 35.King AE, Ackley MA, Cass CE, Young JD, Baldwin SA. Nucleoside transporters: from scavengers to novel therapeutic drugs. Trends Pharmacol Sci. 2006;27:416–425. doi: 10.1016/j.tips.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Lanfear SM. Transporters for drug delivery and as drug targets in parasitic protozoa. Clin Pharmacol Ther. 2010;87:122–125. doi: 10.1038/clpt.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdés R, Arastu-Kapur S, Landfear SM, Shinde U. An ab initio structural model of a nucleoside permease predicts functionally important residues. J Biol Chem. 2009;284:19067–19076. doi: 10.1074/jbc.M109.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riegelhaupt PM, Frame IJ, Akabas MH. Transmembrane segment 11 appears to line the purine permeation pathway of the Plasmodium falciparum equilibrative nucleoside transporter 1 (PfENT1) J Biol Chem. 2010;285:17001–17010. doi: 10.1074/jbc.M110.115758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallace LJ, Candlish D, de Koning HP. Different substrate recognition motifs of human and trypanosome nucleobase transporters: Selective uptake of purine antimetabolites. J Biol Chem. 2002;277:26149–26156. doi: 10.1074/jbc.M202835200. [DOI] [PubMed] [Google Scholar]
- 40.Gournas C, Papageorgiou I, Diallinas G. The nucleobase-ascorbate transporter (NAT) family: genomics, evolution, structure-function relationships and physiological role. Mol Biosyst. 2008;4:404–416. doi: 10.1039/b719777b. [DOI] [PubMed] [Google Scholar]
- 41.Georgopoulou E, Mermelekas G, Karena E, Frillingos S. Purine substrate recognition by the nucleobase-ascorbate transporter motif in the YgfO xanthine permease: Asn-325 binds and Ala-323 senses substrate. J Biol Chem. 2010;285:19422–19433. doi: 10.1074/jbc.M110.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mermelekas G, Georgopoulou E, Kallis A, Botou M, Vlantos V, Frillingos S. Cysteinescanning analysis of helices TM8, TM9a, TM9b and intervening loops in the YgfO xanthine permease: a carboxyl group is essential at position Asp-276. J Biol Chem. 2010;285:35011–35020. doi: 10.1074/jbc.M110.170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cecchetto G, Amillis S, Diallinas G, Scazzocchio C, Drevet C. The AzgA purine transporter of Aspergillus nidulans: Characterization of a protein belonging to a new phylogenetic cluster. J Biol Chem. 2004;279:3132–3141. doi: 10.1074/jbc.M308826200. [DOI] [PubMed] [Google Scholar]
- 44.Goudela S, Tsilivi H, Diallinas G. Comparative kinetic analysis of AzgA and Fcy21p, prototypes of the two major fungal hypoxanthine-adenine-guanine transporter families. Mol Membr Biol. 2006;23:291–303. doi: 10.1080/09687860600685109. [DOI] [PubMed] [Google Scholar]
- 45.Mansfield TA, Schultes NP, Mourad GS. AtAzgA1 and AtAzgA2 comprise a novel family of purine transporters in Arabidopsis . FEBS Lett. 2009;583:481–486. doi: 10.1016/j.febslet.2008.12.048. [DOI] [PubMed] [Google Scholar]
- 46.Diallinas G, Gournas C. Structure-function relationships in the nucleobase-ascorbate transporter (NAT) family: Lessons from model microbial genetic systems. Channels. 2008;2:363–372. doi: 10.4161/chan.2.5.6902. [DOI] [PubMed] [Google Scholar]
- 47.Amillis S, Kosti V, Pantazopoulou A, Mikros E, Diallinas G. Mutational analysis and modeling reveal functionally critical residues in transmembrane segments 1 and 3 of the UapA transporter. J Mol Biol. 2011;411:567–580. doi: 10.1016/j.jmb.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 48.Diallinas G, Scazzocchio C. A gene coding for the uric acid-xanthine permease of Aspergillus nidulans: inactivational cloning, characterisation and sequence of a cis-acting mutation. Genetics. 1989;122:341–350. doi: 10.1093/genetics/122.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorfinkiel L, Diallinas G, Scazzocchio C. Sequence and regulation of the uapA gene encoding a uric acid-xanthine permease in the fungus Aspergillus nidulans . J Biol Chem. 1993;268:23376–23381. [PubMed] [Google Scholar]
- 50.Diallinas G, Gonrfinkiel L, Arst HN Jr, Cecchetto G, Scazzocchio C. Genetic and molecular characterization of a gene encoding a wide specificity purine permease of Aspergillus nidulans reveals a novel family of transporters conserved in prokaryotes and eukaryotes. J Biol Chem. 1995;270:8610–8622. doi: 10.1074/jbc.270.15.8610. [DOI] [PubMed] [Google Scholar]
- 51.Andersen PS, Frees D, Fast R, Mygind B. Uracil uptake in Escherichia coli K-12: isolation of uraA mutants and cloning of the gene. J Bacteriol. 1995;177:2008–2013. doi: 10.1128/jb.177.8.2008-2013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner RJ, Lu Y, Switzer RI. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J Bacteriol. 1994;176:3708–3722. doi: 10.1128/jb.176.12.3708-3722.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christiansen LC, Schou S, Nygaard P, Saxild HH. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz AC, Nygaard P, Saxild HH. Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator. J Bacteriol. 2001;183:3293–3302. doi: 10.1128/JB.183.11.3293-3302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karatza P, Frillingos S. Cloning and functional characterization of two bacterial members of the NAT/NCS2 family in Escherichia coli . Mol Membr Biol. 2005;22:251–261. doi: 10.1080/09687860500092927. [DOI] [PubMed] [Google Scholar]
- 56.Goudela S, Karatza P, Koukaki M, Frillingos S, Diallinas G. Comparative substrate recognition by bacterial and fungal purine transporters of the NAT/NCS2 family. Mol Membr Biol. 2005;22:263–275. doi: 10.1080/09687860500093016. [DOI] [PubMed] [Google Scholar]
- 57.Goudela S, Reichard U, Amillis S, Diallinas G. Characterization and kinetics of the major purine transporters in Aspergillus fumigatus . Fungal Genet Biol. 2008;45:459–472. doi: 10.1016/j.fgb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Argyrou E, Sophianopoulou V, Schultes N, Diallinas G. Functional characterization of a maize purine transporter by expression in Aspergillus nidulans . Plant Cell. 2001;13:953–964. doi: 10.1105/tpc.13.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maurino VG, Zielinski J, Schild A, Fischer K, Flügge UI. Identification and expression analysis of twelve members of the nucleobaseascorbate transporter (NAT) gene family in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:1381–1393. doi: 10.1093/pcp/pcl011. [DOI] [PubMed] [Google Scholar]
- 60.Faaland CA, Race JE, Ricken G, Warner FJ, Williams WJ, Holtzman EJ. Molecular characterization of two novel transporters from human and mouse kidney and from LLC-PK1 cells reveals a novel conserved family that is homologous to bacterial and Aspergillus nucleobase transporters. Biochim Biophys Acta. 1998;1442:353–360. doi: 10.1016/s0167-4781(98)00151-1. [DOI] [PubMed] [Google Scholar]
- 61.Tsukaguchi H, Tokui T, Mackenzie B, Berger UV, Chen XZ, Wang Y, Brubaker RF, Hediger MA. A family of mammalian Na+-dependent Lascorbic acid transporters. Nature. 1999;399:70–75. doi: 10.1038/19986. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, Dutta B, Huang W, Devoe LD, Leibach FH, Ganapathy V, Prasad PD. Human Na+-dependent vitamin transporter 1 (hSVCT1): primary structure, functional characteristics and evidence for a non-functional splice variant. Biochim Biophys Acta. 1999;1461:1–9. doi: 10.1016/s0005-2736(99)00182-0. [DOI] [PubMed] [Google Scholar]
- 63.Rajan DP, Huang W, Dutta B, Devoe LD, Leibach FH, Ganapathy V, Prasad PD. Human placental sodium-dependent vitamin transporter 2 (hSVCT2): molecular cloning and transport function. Biochem Biophys Res Commun. 1999;262:762–768. doi: 10.1006/bbrc.1999.1272. [DOI] [PubMed] [Google Scholar]
- 64.Guimarães MJ, Bazan JF, Zlotnik A, Wiles MV, Grimaldi JC, Lee F, McClanahan T. A new approach to the study of haematopoietic development in the yolk sac and embryonic bodies. Development. 1995;121:3335–3346. doi: 10.1242/dev.121.10.3335. [DOI] [PubMed] [Google Scholar]
- 65.Varma S, Campbell CE, Kuo SM. Functional role of conserved transmembrane segment 1 residues in human sodium-dependent vitamin C transporters. Biochemistry. 2008;47:2952–2960. doi: 10.1021/bi701666q. [DOI] [PubMed] [Google Scholar]
- 66.Ormazabal V, Zuñiga FA, Escobar E, Aylwin C, Salas-Burgos A, Godoy A, Reyes AM, Vera JC, Rivas CI. Histidine residues in the Na+-coupled ascorbic acid transporter-2 (SVCT2) are central regulators of SVCT2 function, modulating pH sensitivity, transporter kinetics, Na+ cooperativity, conformational stability, and subcellular localization. J Biol Chem. 2010;285:36471–36485. doi: 10.1074/jbc.M110.155630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diallinas G, Valdez J, Sophianopoulou V, Rosa A, Scazzocchio C. Chimeric purine transporters of Aspergillus nidulans define a domain critical for function and specificity conserved in bacterial, plant and metazoan homologues. EMBO J. 1998;17:3827–3837. doi: 10.1093/emboj/17.14.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meintanis C, Karagouni AD, Diallinas G. Amino acid residues N450 and Q449 are critical for the uptake capacity and specificity of UapA, a prototype of a nucleobase-ascorbate transporter family. Mol Membr Biol. 2000;17:47–57. doi: 10.1080/096876800294489. [DOI] [PubMed] [Google Scholar]
- 69.Amillis S, Koukaki M, Diallinas G. Substitution F569S converts UapA, a specific uric acidxanthine transporter, into a broad specificity transporter for purine-related solutes. J Mol Biol. 2001;313:765–774. doi: 10.1006/jmbi.2001.5087. [DOI] [PubMed] [Google Scholar]
- 70.Koukaki M, Vlanti A, Goudela S, Pantazopoulou A, Gioule H, Tournaviti S, Diallinas G. The nucleobase-ascorbate transporter (NAT) signature motif in UapA defines the function of the purine translocation pathway. J Mol Biol. 2005;350:499–513. doi: 10.1016/j.jmb.2005.04.076. [DOI] [PubMed] [Google Scholar]
- 71.Pantazopoulou A, Diallinas G. The first transmembrane segment (TMS1) of UapA contains determinants necessary for expression in the plasma membrane and purine transport. Mol Membr Biol. 2006;23:337–348. doi: 10.1080/09687860600738239. [DOI] [PubMed] [Google Scholar]
- 72.Vlanti A, Amillis S, Koukaki M, Diallinas G. A novel-type substrate-selectivity filter and ER-exit determinants in the UapA purine transporter. J Mol Biol. 2006;357:808–819. doi: 10.1016/j.jmb.2005.12.070. [DOI] [PubMed] [Google Scholar]
- 73.Papageorgiou I, Gournas C, Vlanti A, Amillis S, Pantazopoulou A, Diallinas G. Specific interdomain synergy in the UapA transporter determines its unique specificity for uric acid among NAT carriers. J Mol Biol. 2008;382:1121–1135. doi: 10.1016/j.jmb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 74.Kosti V, Papageorgiou I, Diallinas G. Dynamic elements at both cytoplasmically and extracellularly facing sides of the UapA transporter selectively control the accessibility of substrates to their translocation pathway. J Mol Biol. 2010;397:1132–1143. doi: 10.1016/j.jmb.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 75.Karatza P, Panos P, Georgopoulou E, Frillingos S. Cysteine-scanning analysis of the nucleobase-ascorbate transporter signature motif in YgfO permease of Escherichia coli: Gln-324 and Asn-325 are essential and Ile-329-Val-339 form an alpha-helix. J Biol Chem. 2006;281:39881–39890. doi: 10.1074/jbc.M605748200. [DOI] [PubMed] [Google Scholar]
- 76.Papakostas K, Georgopoulou E, Frillingos S. Cysteine-scanning analysis of putative helix XII in the YgfO xanthine permease: Ile-432 and Asn-430 are important. J Biol Chem. 2008;283:13666–13678. doi: 10.1074/jbc.M800261200. [DOI] [PubMed] [Google Scholar]
- 77.Karena E, Frillingos S. Role of intramembrane polar residues in the YgfO xanthine permease: His-31 and Asn-93 are crucial for affinity and specificity, and Asp-304 and Glu-272 are irreplaceable. J Biol Chem. 2009;284:24257–24268. doi: 10.1074/jbc.M109.030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varma S, Sobey K, Campbell CE, Kuo SM. Hierarchical contribution of N- and C-terminal sequences to the differential localization of homologous sodium-dependent vitamin C transporters, SVCT1 and SVCT2, in epithelial cells. Biochemistry. 2009;48:2969–2980. doi: 10.1021/bi802294v. [DOI] [PubMed] [Google Scholar]
- 79.Subramanian VS, Marchant JS, Said HM. Molecular determinants dictating cell surface expression of the human sodium-dependent vitamin C transporter-2 in human liver cells. Am J Physiol Gastrointest Liver Physiol. 2010;298:G267–G274. doi: 10.1152/ajpgi.00435.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Velho AM, Jarvis SM. Topological studies of hSVCT1, the human sodium-dependent vitamin C transporter and the influence of N-glycosylation on its intracellular targeting. Exp Cell Res. 2009;315:2312–2321. doi: 10.1016/j.yexcr.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 81.Pantazopoulou A, Diallinas G. Fungal nucleobase transporters. FEMS Microbiol Rev. 2007;31:657–675. doi: 10.1111/j.1574-6976.2007.00083.x. [DOI] [PubMed] [Google Scholar]
- 82.Gournas C, Amillis S, Vlanti A, Diallinas G. Transport-dependent endocytosis and turnover of a uric acid-xanthine permease. Mol Microbiol. 2010;75:246–260. doi: 10.1111/j.1365-2958.2009.06997.x. [DOI] [PubMed] [Google Scholar]
- 83.Gournas C, Oestreicher N, Amillis S, Diallinas G, Scazzocchio C. Completing the purine utilisation pathway of Aspergillus nidulans . Fungal Genet Biol. 2011;48:840–848. doi: 10.1016/j.fgb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 84.Lemuh ND, Diallinas G, Frillingos S, Mermelekas G, Karagouni AD, Hatzinikolaou DG. Purification and partial characterization of the xanthine-uric acid transporter (UapA) of Aspergillus nidulans . Protein Expr Purif. 2009;63:33–39. doi: 10.1016/j.pep.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 85.Leung J, Karachaliou M, Alves C, Diallinas G, Byrne B. Expression and purification of a functional uric acid-xanthine transporter (UapA) Protein Expr Purif. 2010;72:139–146. doi: 10.1016/j.pep.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 86.Leung J, Cameron A, Diallinas G, Byrne B. Stabilizing the heterologously expressed uric acid-xanthine transporter UapA from the lower eukaryote Aspergillus nidulans . Mol Membr Biol. 2012;29 doi: 10.3109/09687688.2012.690572. in press. [DOI] [PubMed] [Google Scholar]
- 87.Karena E, Tatsaki E, Frillingos S. The threedimensional arrangement of important and conserved residues in the xanthine transporter XanQ of E. coli . Hell Soc Biol Sci. 2012;34:94–95. [Google Scholar]
- 88.Stroud RM, Choe S, Holton J, Kaback HR, Kwiatkowski W, Minor DL, Riek R, Sali A, Stahlberg H, Harries W. 2007 Annual progress report synopsis of the Center for Structures of Membrane Proteins. J Struct Funct Genomics. 2009;10:193–208. doi: 10.1007/s10969-008-9058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mermelekas G, Vourvou E, Frillingos S. Purification and biophysical study of xanthine permease XanQ. Hell Soc Biol Sci. 2011;33:196–197. [Google Scholar]
- 90.Georgopoulou E, Frillingos S. Analysis of cross-homolog chimeras reveals the evolvability dynamics of xanthine permease YgfO (XanQ) Hell Soc Biochem Mol Biol. 2010;61:52–53. [Google Scholar]
- 91.Karena E, Papakostas K, Frillingos S. Interaction between the core and the gate domain of the xanthine permease XanQ of E. coli as determined with cadmium-sensitivity assays. Hell Soc Biochem Mol Biol. 2011;62:222–223. [Google Scholar]
- 92.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.