Abstract
Alfalfa (Medicago sativa L.) roots contain large quantities of β-amylase, but little is known about its role in vivo. We studied this by isolating a β-amylase cDNA and by examining signals that affect its expression. The β-amylase cDNA encoded a 55.95-kD polypeptide with a deduced amino acid sequence showing high similarity to other plant β-amylases. Starch concentrations, β-amylase activities, and β-amylase mRNA levels were measured in roots of alfalfa after defoliation, in suspension-cultured cells incubated in sucrose-rich or -deprived media, and in roots of cold-acclimated germ plasms. Starch levels, β-amylase activities, and β-amylase transcripts were reduced significantly in roots of defoliated plants and in sucrose-deprived cell cultures. β-Amylase transcript was high in roots of intact plants but could not be detected 2 to 8 d after defoliation. β-Amylase transcript levels increased in roots between September and October and then declined 10-fold in November and December after shoots were killed by frost. Alfalfa roots contain greater β-amylase transcript levels compared with roots of sweetclover (Melilotus officinalis L.), red clover (Trifolium pratense L.), and birdsfoot trefoil (Lotus corniculatus L.). Southern analysis indicated that β-amylase is present as a multigene family in alfalfa. Our results show no clear association between β-amylase activity or transcript abundance and starch hydrolysis in alfalfa roots. The great abundance of β-amylase and its unexpected patterns of gene expression and protein accumulation support our current belief that this protein serves a storage function in roots of this perennial species.
β-Amylase catalyzes the hydrolysis of α-1,4-glucosidic linkages from the nonreducing ends of starch molecules releasing maltose and producing β-limit dextrin (Thomas et al., 1971). It is abundant in seeds and roots of certain species (Doehlert et al., 1982; Yoshida and Nakamura, 1991; Boyce and Volenec, 1992a) and is also present in other vegetative tissues (Beck and Ziegler, 1989). Because of its high abundance and its perceived role in starch metabolism, β-amylase has been the focus of several physiological and molecular studies. Molecular analyses of plant β-amylase have been conducted using cDNAs or genomic sequences from both dicots (Monroe et al., 1991; Yoshida and Nakamura, 1991; Totsuka and Fukazawa, 1993) and monocots (Sadowski et al., 1993; Yoshigi et al., 1994; Wagner et al., 1996; Wang et al., 1997). Encoded amino acid sequences for these plant β-amylases are highly conserved, with amino acid similarity ranging from 60% to 96%. An endosperm-specific β-amylase has been described for rye (Rorat et al., 1991) and barley (Yoshigi et al., 1994) that contains Gly-rich repetitive sequences in the carboxyl terminus of the protein.
The mode of regulation of plant β-amylase genes appears complex and at times contradictory. In many plant systems β-amylase transcript accumulation is regulated by sugars. Arabidopsis β-amylase mRNA levels increased in rosette leaves when plants or excised, fully expanded leaves were supplied with Suc, Glc, and Fru but were not affected by mannitol or sorbitol (Mita et al., 1995). Exposure of Arabidopsis plants to light was essential for the accumulation of the β-amylase transcript. Light also induced accumulation of the β-amylase transcript in mustard cotyledons (Sharma and Shopfer, 1987). Sweet potato β-amylase gene expression occurs in darkness if leaf-petiole cuttings are supplied with Suc (Nakamura et al., 1991). Dipping sweet potato leaf-petiole cuttings in polygalacturonic acid or chitosan also induced β-amylase mRNA accumulation, whereas mechanical wounding of leaves only occasionally induced β-amylase gene expression (Ohto et al., 1992). ABA induced the expression of sweet potato β-amylase in leaf-petiole cuttings within 12 h of treatment (Ohto et al., 1992). However, in rice aleurone cells, ABA inhibited de novo synthesis of β-amylase and reduced β-amylase transcript levels (Wang et al., 1996). In most systems studied to date increases in β-amylase activity and accumulation of β-amylase transcripts were associated with starch deposition in tissue. This raises questions regarding the in vivo role of plant β-amylase as a starch hydrolase.
Alfalfa (Medicago sativa L.) is an excellent system in which to study mechanisms of starch utilization and accumulation. In agricultural ecosystems alfalfa is completely defoliated at approximately 30-d intervals. Rapid herbage regrowth after defoliation has been positively associated with quantities of carbon and nitrogen reserves in taproots, including starch (Graber et al., 1927; Smith, 1962; Hendershot and Volenec, 1993b). Previous reports showed that defoliation results in a decline in both root amylase activity (>99% β-amylase) and starch concentration (Volenec and Brown, 1988; Volenec et al., 1991). Roots of other forage legumes such as sweetclover (Melilotus officinalis L.), red clover (Trifolium pratense L.), and birdsfoot trefoil (Lotus corniculatus L.) contain about one-half (red clover) or less of the β-amylase activity levels found in alfalfa roots (Li et al., 1996). Like alfalfa, roots of sweetclover and red clover exhibit a decline in β-amylase activity that parallels a decline in root starch concentration after defoliation (Li et al., 1996). Very low total amylase specific activity was observed in roots of birdsfoot trefoil, even though root starch was depleted in a manner similar to that of alfalfa after defoliation (Boyce et al., 1992). Clearly, the pattern of decreased β-amylase activity coincident with large declines in root starch concentration are inconsistent with the perceived role of β-amylase as a starch hydrolase in roots of alfalfa and the other forage legumes.
Nitrogen-containing compounds in alfalfa roots (such as amino acids and proteins) have been shown to be positively associated with the rate of herbage regrowth after defoliation (Kim et al., 1991; Hendershot and Volenec, 1993b; Ourry et al., 1994; Barber et al., 1996; Volenec et al., 1996) and in the spring when shoot growth resumes (Volenec et al., 1991; Hendershot and Volenec, 1993a; Li et al., 1996). Three polypeptides constituting approximately 40% of the root's soluble protein pool have been isolated and characterized (Cunningham and Volenec, 1996). We believe that these polypeptides are VSPs because they accumulate in alfalfa taproots in early autumn and disappear in the spring and after plants are defoliated, in a manner that is consistent with functions assigned to VSPs (Cyr and Bewley, 1990). Boyce and Volenec (1992b) purified a 57.5-kD β-amylase protein from alfalfa taproots and found that it constituted 8% of root-soluble protein. The seasonal pattern of β-amylase activity followed the trends in concentration of root VSPs, increasing in autumn and declining markedly in spring (Volenec et al., 1991; Hendershot and Volenec, 1993a; Li et al., 1996). Because defoliation reduces nitrogenase activity (Vance et al., 1979) and uptake of nitrogen from the soil (Kim et al., 1993), we speculate that β-amylase, like VSPs, may be hydrolyzed to its constituent amino acids, which are then transported to regrowing shoots to provide some of the nitrogen needed for herbage growth in spring and regrowth after defoliation in the summer (Boyce and Volenec, 1992b).
In view of the great abundance of β-amylase in alfalfa roots and its unexpected pattern of activity during root starch loss, it is important to understand how β-amylase gene expression is regulated by environmental cues such as defoliation and cold temperatures. Our objectives were: (a) to isolate a cDNA for alfalfa root β-amylase; (b) to determine tissue-specific expression of the β-amylase gene; (c) to examine the extent to which roots of other perennial forage legumes accumulate β-amylase mRNA; (d) to determine genomic organization of the β-amylase gene; and (e) to determine how defoliation, Suc deprivation in cell-suspension cultures, and winter hardening influence β-amylase transcript levels, β-amylase activity, and root starch concentrations.
MATERIALS AND METHODS
Plant Material Used for Isolation of β-Amylase cDNA
An alfalfa (Medicago sativa L.) germ plasm (Norseman-H) that had undergone three cycles of selection from cv Norseman for decreased fall dormancy was used for β-amylase cDNA isolation. Seedlings were established at the Agronomy Research Center of Purdue University in April. Randomization of field plots and management practices were as described previously (Cunningham et al., 1998). Plants were defoliated in mid-August, and roots were sampled on October 15. Roots were washed free of soil under a stream of cold water. Nodules were removed and discarded to ensure root tissue specificity. The top 5 cm of the roots were immersed in liquid nitrogen, packed in solid CO2, and transported to the laboratory, where tissues were stored at −80°C.
Total RNA Extraction
Total RNA was isolated using hot phenol and the procedure of Ougham and Davis (1990) with minor modifications. Polyvinylpolypyrrolidone (0.4 g) was added to tubes (Nalgene) containing 2 g of frozen root tissue that had been ground in liquid nitrogen. Water-saturated phenol (8 mL, 65°C) was added to each tube and the tubes were placed in a 65°C water bath. When samples thawed, 8 mL of RNA-extraction buffer (0.2 m sodium acetate, pH 5.2, 0.01 m EDTA, 1% SDS) was added to each tube, and the samples were incubated at 65°C for 15 min. The tubes were removed from the 65°C water bath and cooled to 25°C, and 8 mL of chloroform was added. Agitation of tubes, centrifugation, and recovery of the RNA pellet was as described by Ougham and Davis (1990).
Isolation and Identification of Alfalfa β-Amylase cDNA
For the isolation of the β-amylase cDNA, the first-strand cDNA was synthesized using 0.2 μm of a modified oligo(dT)-primer, 5′-GAGAAGCT12GC-3′, and 200 units of reverse transcriptase (Superscript II, BRL/Life Technologies) according to the manufacturer's protocol. The total volume for the reaction was 20 μL. White clover (Trifolium repens L.) RNA was used as an internal control because the upstream primer used for reverse transcriptase-PCR amplification of putative alfalfa β-amylase cDNA clones had been made to sequences of the 5′ region of white clover β-amylase cDNAs (accession no. AF049098). PCR amplification was in a 50-μL reaction volume containing 2 μL of the first-strand cDNA mixture, 1× PCR buffer, 0.2 mm final concentration of the deoxyribonucleotide triphosphates, 0.2 μm final concentration of the upstream primer (5′-CAA, GGC, CAC, TTC, TAA, CAA, CAT, G-3′), 0.2 μm final concentration of the modified oligo(dT)-primer (5′-GAGAAGT12GC-3′), and 5 units of Taq DNA polymerase. The temperature profile for the thermocycler was 94°C for 5 min, 40 cycles at 94°C for 30 s, 50°C for 30 s, 72°C for 2 min, and then 75°C for 15 min after PCR. The PCR products were analyzed on a 1.5% agarose gel. Four bands with approximate sizes of 635, 969, 1345, and 1701 bp were eluted from the gel and subcloned into the pGEM-T vector (Promega). The 1.7-kb band from alfalfa roots also corresponded to a band of similar size from white clover and was further analyzed.
DNA Sequencing and Analysis
A plasmid containing the 1.7-kb putative β-amylase cDNA designated pMSBA1 (M. sativa β-amylase 1) was digested with HindIII and liberated an approximately 800-bp 3′ fragment. This fragment was subcloned into pBluescript SK(−) plasmid (Stratagene). The two subclones (pMSBA1 minus the HindIII fragment and pBluescript plus the HindIII fragment) were sequenced on both strands using an automated DNA sequencer (Pharmacia) with both universal and specific primers. Homology searches were obtained with the Basic Local Alignment Search Tool of the National Center for Biotechnology Information (Altschul et al., 1990). Predicted amino acid sequences were analyzed using software from the Genetics Computer Group (Madison, WI).
Tissue- and Species-Specific Expression of β-Amylase
Seeds of five forage legumes, alfalfa Pioneer Brand cv 5454, white clover cv Prestige, red clover (Trifolium pratense L. cv Arlington), sweetclover (Melilotus officinalis L.), and birdsfoot trefoil (Lotus corniculatus L. cv Fergus) were inoculated with the appropriate Rhizobium species (Li et al., 1996) and planted in 1-L plastic pots containing a 1:1 mixture of silt loam soil:sand. Pots were placed in a greenhouse in which air temperature was 25°C ± 5°C and the photoperiod was extended to 15 h with fluorescent and incandescent lights (160 μmol m−2 s−1). Plants were watered as needed with water purified by reverse osmosis and were also provided with 50 mL of nitrogen-free Hoagland solution twice weekly. Plants were grown to the flowering stage. To determine the tissue-specific expression of β-amylase in alfalfa, the leaves, stems, and roots (without nodules) were sampled in three replicates. To determine species-specific expression of β-amylase, three replicates of root tissues of each of the five forage legume species were sampled at the time of flowering. Tissues were processed and stored for RNA analysis as described above.
Effects of Defoliation on Starch Accumulation and β-Amylase Expression
Plants of alfalfa cv 5454 were established in 1-L plastic pots in the greenhouse as previously described. Plants were grown to flowering (approximately 75 d). To determine how defoliation influenced β-amylase expression, one-half of the alfalfa plants were defoliated, leaving a 5-cm stubble. Roots were sampled immediately (0 h), at 3, 6, and 12 h, and at 1, 2, 4, 8, 12, 16, 20, 24, and 28 d after defoliation. Herbage of the remaining alfalfa plants was left intact to serve as undefoliated controls, and roots of these plants were sampled at the same time as the defoliated plants. Roots were washed free of soil under a stream of cold water. Nodules were removed and discarded. The top 5 cm of taproots was selected for analysis to avoid errors due to plant-to-plant variations in root length. Previous studies have shown that results obtained using the uppermost 5 cm of taproot were closely associated with results obtained using the reminder of the root system (J.J. Volenec, unpublished data). Root tissues were immersed in liquid nitrogen, packed in solid CO2, and taken to the laboratory, where tissues were stored at −80°C. The remaining root tissue was lyophilized, ground to pass a 1-mm screen, and stored at −20°C until analyzed for starch concentration and β-amylase activity. In this experiment pots were arranged in a randomized, complete block design with three replications. For statistical analysis, variation was partitioned into replicate, defoliation treatment, sampling time after defoliation, and corresponding interactions.
Influence of Suc Deprivation on Starch Accumulation and β-Amylase Expression in Cell Suspensions
Because sugar influenced β-amylase transcript abundance in other plant systems, we were interested in using cell-suspension cultures to alter the sugar availability to alfalfa cells and study its impact on β-amylase gene expression. Alfalfa cv Pioneer Brand 5929 seeds were surface-sterilized in 20% (v/v) commercial bleach for 10 min and washed three washes with sterile water. Seeds were germinated on filter-paper bridges at 25°C under a 16-h photoperiod of fluorescent lights (25 μmol m−2 s−1) for 7 d. Callus was initiated from 5-cm-long root sections on Murashige-Skoog medium (Murashige and Skoog, 1962) supplemented with 1 mg L−1 2,4-D. Gamborg et al. (1968) B5g liquid medium supplemented with 1 mg L−1 2,4-D was used for induction of suspension cultures. Control cells were grown in B5g medium containing 20 g L−1 Suc. For the Suc-depleted B5g media, Suc was replaced with an equimolar concentration of mannitol. Cells were sampled 24, 48, and 72 h after transfer to Suc-deprived or normal B5g media. Cells were recovered from suspensions and immediately frozen in liquid nitrogen and stored at −80°C for analysis. The cell-culture experiment was replicated three times, and flasks were arranged in a randomized, completed block design. For statistical analysis, experimental variation was partitioned into replicate effects, Suc treatment effects, sampling time effects, and corresponding interactions using analysis of variance.
Effects of Fall Hardening on Starch Utilization and β-Amylase Expression in Contrasting Alfalfa Germ Plasms
Winter hardening alters root carbohydrate metabolism in alfalfa. We wanted to determine how winter hardening influenced starch levels and β-amylase expression in closely related alfalfa germ plasms that possess contrasting fall dormancy and winter hardiness. The six diverse alfalfa cultivars used for this study included fall nondormant, nonhardy cv CUF 101-O, fall dormant, winter hardy cv Norseman-O (O = original cultivar), and populations from the third cycle of selection for contrasting fall dormancy from within these two cultivars (Cunningham et al., 1998). The contrasting populations of cv Norseman-O and cv CUF-101-O, cv Norseman-L and cv CUF 101-L for germ plasms selected for increasing fall dormancy, and cv Norseman-H and cv CUF 101-H were designated for the germ plasms selected for decreasing fall dormancy. Seedlings were established in April at the Agronomy Research Center. The top 5-cm root sections were sampled on September 10, October 15, November 18, and December 17, and tissues were handled as described above for RNA isolation. In addition, the remaining root tissue was cut into 5-cm segments, packed in solid CO2, transported to the laboratory, lyophilized, ground to pass a 1-mm screen, and stored at −20°C until analysis. This study was replicated four times. Data were analyzed as a split-plot design with repeated sampling of plants from within rows over time. Data from starch concentrations and β-amylase activity were analyzed using statistical analysis software (SAS Institute, 1989).
Northern Hybridization Analysis
Total RNA (20 μg) was separated on 1.5% agarose formaldehyde gels (Lehrach et al., 1977) and transferred to Zeta-probe membranes (Bio-Rad). The membranes were prehybridized for 4 h at 42°C (Stewart and Walker, 1989) with slow shaking. The insert (β-amylase cDNA) from pMSBA1 was labeled with [32P]dCTP using random priming (Feinberg and Vogelstein, 1983). Hybridization and washing of membranes were as described by Gana et al. (1997). To determine the amount of total RNA loaded in each lane, the 32P-labeled β-amylase probe was stripped from the membranes using the manufacturer's protocol (Bio-Rad) and rehybridized with a 32P-labeled alfalfa 18S ribosomal probe to correct for RNA loading differences. Membranes were exposed to radiographic film (Fuji, Tokyo, Japan) at −80°C. Signal intensities were quantified using an imager (Packard Instruments, Downers Grove, IL).
Genomic Southern Analysis
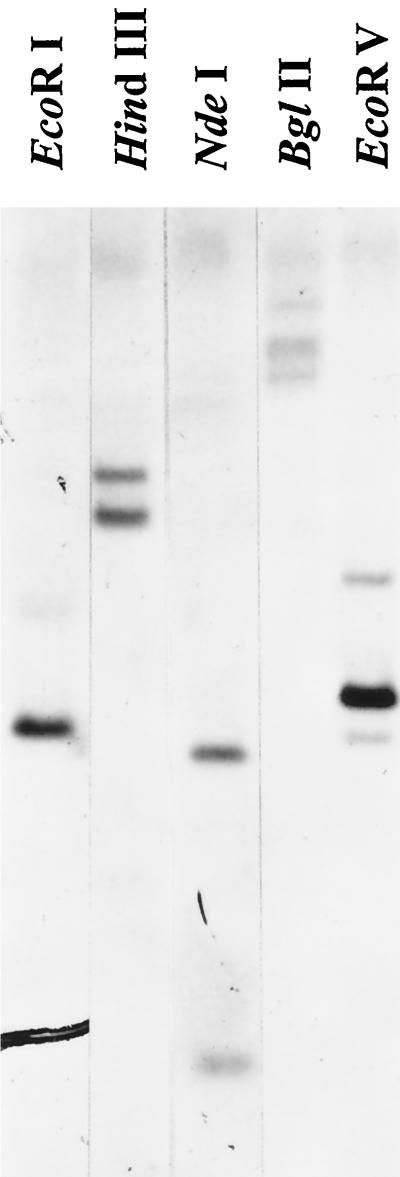
Genomic DNA was extracted from roots of field-grown alfalfa using a urea-based DNA miniprep procedure (Shure et al., 1983). The DNA (10 μg) was digested separately with HindIII, NdeI, EcoRI, EcoRV, and BglII. Fragments were separated on a 1% agarose gel and transferred to Zeta-probe membranes (Bio-Rad). Membranes were prehybridized and hybridized as described by the manufacturer (Bio-Rad) using the 32P-labeled β-amylase probe described above.
Starch Analysis
For starch analysis, 30 mg of ground, freeze-dried tissue or 100 mg of fresh root tissue ground in liquid nitrogen was used. Sugars were initially extracted using 80% (v/v) ethanol. Starch in the ethanol-extracted residue was analyzed as previously described (Li et al., 1996), except that the α-amylase product A-2643 (Sigma) was substituted for the α-amylase product A-0273 (Sigma).
β-Amylase Assay
Total soluble protein extraction and the β-amylase assay procedures were conducted at 4°C or on ice unless otherwise indicated. Soluble protein was extracted from 30 mg of ground, freeze-dried tissue or 100 mg of fresh root tissue ground in liquid nitrogen using 1 mL of 100 mm imidazole buffer, pH 6.5, containing 10 mm 2-mercaptoethanol and 1 mm PMSF. Samples were vortexed three times and centrifuged at 14,000g, and the soluble protein concentration of the supernatant was estimated using protein dye-binding (Bradford, 1976). β-Amylase activity was determined by diluting the buffer-soluble proteins 1:10 with 100 mm imidazole buffer, pH 7.5, and incubating 25 μL of the diluted sample with 50 μL of p-nitrophenol-α-d-maltopentaose, a specific β-amylase substrate (Betamyl kit, Megazyme International, Bray, County Wicklow, Ireland). Samples were incubated for 5 min at 40°C and the reactions were terminated by the addition of 2 mL of 1.65 m Tris, and the A410 was determined. One unit of activity was defined as the quantity of enzyme that released 1 μm of p-nitrophenol min−1.
RESULTS
Isolation and Characterization of Alfalfa β-Amylase
Reverse transcriptase-PCR amplification of RNA from roots of field-grown alfalfa produced four bands ranging in size from approximately 600 to 1700 bp, as observed in ethidium-bromide-stained agarose gels (data not shown). Partial DNA sequence analyses revealed that three of the four cloned inserts corresponded to β-amylase sequences. The 1.7-kb band was analyzed further because it was judged to be close to a full-length cDNA, based on our previous molecular characterization of the β-amylase polypeptide (Boyce and Volenec, 1992b).
The complete nucleotide sequence was 1671 bp long, and the longest open reading frame was 1553 bp. We designated this cDNA pMSBA1 and the sequence was deposited in GenBank (accession no. AF026217). The open reading frame encodes a protein of 496 amino acid residues, with a calculated molecular mass of 55,950 D and a predicted pI of 5.31. The first ATG codon is found in position 4 of the nucleotide sequence and is presumed to be the translation initiation codon after comparison with the soybean and white clover β-amylases. Three potential polyadenylation signals (Joshi, 1987) were found in the 141-bp 3′-untranslated region.
The MSBA1-encoded protein contains the two β-amylase signature motifs predicted by the PROSITE program (Bairoch, 1994). Alignment of the amino acid sequences of β-amylase from several plant species (Fig. 1A) shows that the encoded alfalfa β-amylase contains the eight highly conserved motifs identified by Pujadas et al. (1996). These authors indicated that these motifs define structural and/or functional elements of β-amylases. The predicted amino acid sequence of MSAB1 is nearly identical with those of white clover (96%) and soybean (92%). Phylogenetic dendrogram analysis reveals that MSBA1 is more closely related to white clover and soybean β-amylases than with other plant β-amylases (Fig. 1B). The dendrogram shows that MSBA1 is clustered with β-amylases from dicots and is clearly separated from monocots. The MSBA1 lacks the Gly-rich repeated motif found in the COOH terminus of rye and barley endosperm β-amylases (Fig. 1A).
Figure 1.
A, Deduced amino acid sequence of MSBA1 aligned with sequences of some selected plant β-amylases obtained from GenBank. The multiple alignment was done with the PILEUP program of the DNA sequence analysis package from Genetics Computer Group. The eight highly conserved regions of all β-amylases identified by Pujadas et al. (1997) are boxed. B, Phylogenetic tree of plant β-amylases. β-Amylase protein sequences of different species were obtained from GenBank. The protein sequences were aligned using PILEUP. DISTANCE and GROWTREE programs (Genetics Computer Group) were used to analyze the evolutionary distances and create the phylogenetic dendrogram.
Genome Analysis of the β-Amylase Gene
To determine distribution of the β-amylase gene in the tetraploid alfalfa genome, genomic DNA was digested with various restriction enzymes (Fig. 2). Two strong hybridizing bands were seen in the lanes where DNA was digested with HindIII, EcoRV, and NdeI. Three hybridizing bands were obtained for BglII and one strong band with several weak bands for EcoRI. Because pMSBA1 does not have NdeI or BglII sites, we expected to observe one hybridizing band if β-amylase were present as a single gene (except for interruptions by an intron), but instead these enzymes gave more than one band. We expected two bands from the HindIII and EcoRV digests, since both have a single restriction site in the pMSBA1 cDNA. Because of the hybridization patterns we obtained, it was difficult to tell whether β-amylase was present as a single gene. We then made a probe to only one of the HindIII fragments from pMSBA1 and reprobed the Southern blots. We obtained the same hybridization pattern (Fig. 2) as when the entire cDNA was used as a probe. Our conclusion was that β-amylase was encoded by more than one gene.
Tissue- and Species-Specific Expression of β-Amylase
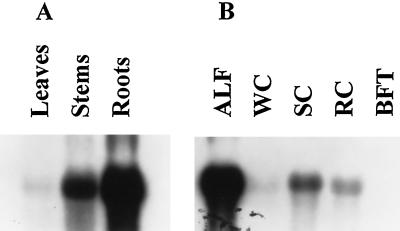
Expression of the β-amylase gene was examined in leaves, stems, and roots of alfalfa and in the roots of sweetclover, white clover, red clover, and birdsfoot trefoil. Transcripts for β-amylase were 100- and 50-fold more abundant in alfalfa roots than in leaves and stems, respectively (Fig. 3A). The β-amylase transcript was 20-fold more abundant in roots of alfalfa compared with roots of sweetclover and 50-fold more abundant compared with roots of white clover and red clover (Fig. 3B). The β-amylase transcript was not detected in roots of birdsfoot trefoil.
Figure 3.
Tissue- and species-specific expression of the β-amylase gene. A, Total RNA from leaves, stems, and roots of alfalfa. B, Total RNA from roots of alfalfa (ALF), white clover (WC), sweetclover (SC), red clover (RC), and birdsfoot trefoil (BFT) at the time of flowering. Northern blots were hybridized with the 32P-labeled pMSBA1 β-amylase insert.
Defoliation-Induced Changes in Starch Concentrations, β-Amylase Activity, and β-Amylase Transcript Levels
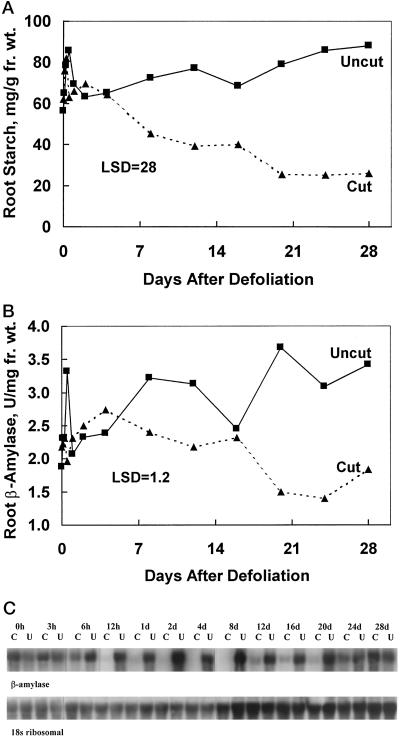
We investigated the effect of defoliation on root starch concentrations, β-amylase activity, and β-amylase gene expression because of the potential role of β-amylase in starch degradation. Defoliation reduced root starch concentrations within 7 d after herbage removal, and starch concentrations in roots of these plants remained low for the remainder of the study (Fig. 4A). Because root protein levels also decline after defoliation, β-amylase activity expressed on a protein basis was similar in roots of defoliated and intact alfalfa plants (data not shown). However, when expressed on a fresh-weight basis, β-amylase activity declined from d 4 to 24 (Fig. 4B).
Figure 4.
Changes in starch concentration, β-amylase activity, and β-amylase transcript levels as influenced by defoliation. Greenhouse-grown plants were defoliated at flowering. Roots were sampled immediately (0 h), at 3, 6, and 12 h, and at 1, 2, 4, 8, 12, 16, 20, 24, and 28 d after defoliation and analyzed for starch (A) and for β-amylase activity (B). In B, U is units. The lsd is shown at the 5% probability level. C, Total RNA (20 μg) was analyzed by northern analysis using radiolabeled β-amylase cDNA, and the membrane was stripped and reprobed with a 32P-labeled alfalfa 18S ribosomal cDNA. C, Cut (defoliated) plants; U, uncut, intact control plants. Fr. wt., Fresh weight.
Northern analysis was used to determine how defoliation affected β-amylase transcript abundance and its relationship to defoliation-induced declines in β-amylase activity. β-Amylase mRNA abundance declined within 12 h after defoliation, attaining very low levels on d 2, 4, and 8 (Fig. 4C). Reaccumulation of β-amylase transcript began on d 12 and reached levels similar to that observed in roots of intact plants by d 28. The β-amylase transcript levels did not change in roots of intact plants during this period. The decline in β-amylase transcript levels was more rapid than the decline in β-amylase activity (Fig. 4, B and C). This lag in the loss of β-amylase activity may be due to the relative slow turnover and stability of the β-amylase polypeptide. β-Amylase from mustard was also shown to be relatively stable, with a slow turnover rate (Subbaramaiah and Sharma, 1989).
Levels of Starch, β-Amylase Activity, and mRNA in Cells Grown in Suc-Rich and Suc-Deprived Medium
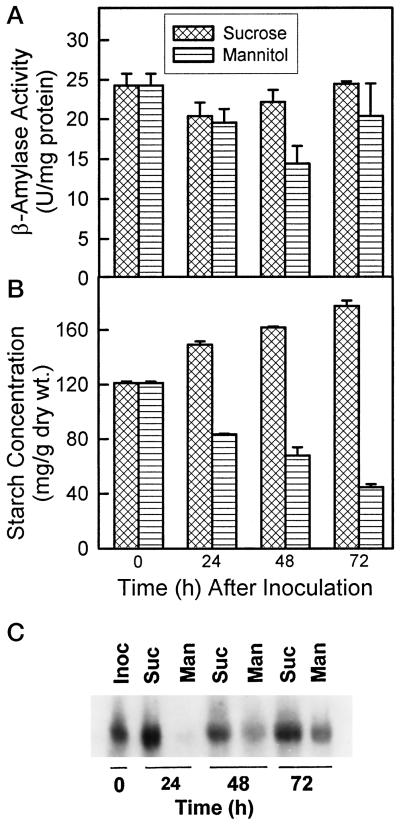
Sugar-inducible expression of β-amylase has been reported in other species (Nakamura et al., 1991; Ohto et al., 1992; Mita et al., 1995, 1997). We determined how starch concentrations, β-amylase activity, and the β-amylase transcript levels were influenced by alfalfa cell cultures grown in B5g medium with or without Suc. Starch levels declined between 0 and 72 h after inoculation when cells were grown in B5g medium containing mannitol (Fig. 5B). Starch concentrations increased during this same time when cells were grown in B5g medium containing Suc. β-Amylase activity did not change significantly in Suc-grown cells, whereas β-amylase activity declined significantly in mannitol-grown cells at 24 and 48 h (Fig. 5A). The decline in starch levels in mannitol-grown cells is not associated with elevated β-amylase activity. High levels of β-amylase mRNA were maintained in cells grown in Suc, but β-amylase mRNA declined substantially within 24 h for cells grown in mannitol (Fig. 5C). As observed for the plant defoliation experiment above, the decline in β-amylase transcript levels was more rapid and substantial than the decline in β-amylase activity, suggesting high stability of the polypeptide.
Figure 5.
Levels of starch (A), β-amylase activity (B), and β-amylase transcript (C) during incubation of alfalfa cell cultures in Suc-containing and Suc-deprived B5g medium. Cell cultures were sampled at the time of inoculation (Inoc; 0 h) and 24, 48, and 72 h after cultures were transferred to Suc-free medium, and then starch, β-amylase, and β-amylase mRNA were measured. U, Units.
Effect of Fall Hardening on Starch Concentrations, β-Amylase Activity, and β-Amylase Transcript Levels
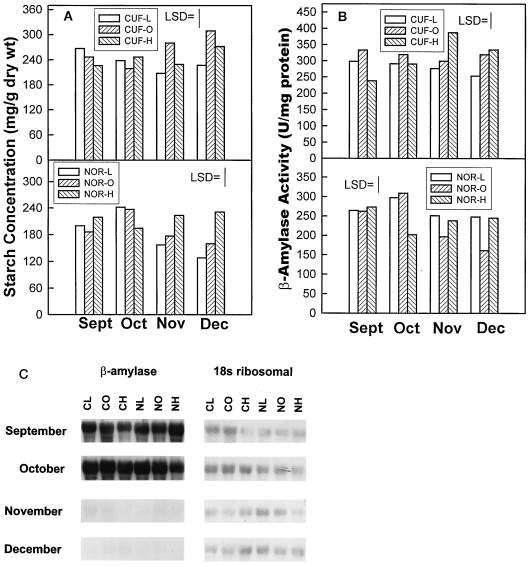
Alfalfa root starch levels decline and sugar concentrations increase in roots of dormant, winter-hardy cultivars. Our objective in this study was to determine the relationship between root starch concentration, β-amylase activity, and β-amylase transcript abundance. In general, trends in β-amylase activity paralleled changes in starch concentrations. Starch concentrations increased significantly from September to October in roots of cv Norseman-L and cv Norseman-O, the two most fall-dormant, winter-hardy germ plasms, and then, as expected, declined between October and December (Fig. 6A). Starch concentrations also declined in roots of cv CUF 101-L, the most fall-dormant, winter-hardy cv CUF 101 germ plasm, between September and November (Fig. 6A). In the remaining populations starch concentrations either remained unchanged or increased between September and December. β-Amylase activity was measured to determine whether the decline in starch concentration was associated with high β-amylase activity. When expressed on a dry-weight basis, β-amylase activity did not interact with germ plasm but did increase from 8.9 units/mg in September to 10.3 units/mg in November and December.
Figure 6.
Changes in starch concentration, β-amylase activity, and β-amylase transcript abundance in roots of alfalfa germ plasms exhibiting contrasting fall dormancy during winter hardening in autumn. Roots were sampled from field plots in September, October, November, and December. Starch levels (A) and β-amylase activity (B) were measured. The lsd is shown at the 5% probability level. U, Units. C, Total RNA (20 μg) was analyzed by northern analysis using radiolabeled β-amylase cDNA and the membrane was stripped and reprobed with a 32-P-labeled alfalfa 18S ribosomal cDNA.
When expressed on a protein basis, the harvest by germ plasm interaction was significant for β-amylase (Fig. 6B). β-Amylase activity declined from September to December in roots of cv CUF 101-L but did not change in cv CUF 101-O, whereas β-amylase activity increased in cv CUF 101-H roots from September to November and then declined in December. β-Amylase activity increased from September to October in roots of cv Norseman-L and cv Norseman-O and then, like root starch concentrations, decreased after October. In contrast, β-amylase activity decreased from September to October and then increased in November in roots of cv Norseman-H. This consistent association of high β-amylase activity with high root starch concentrations suggests that declines in root starch concentrations were not caused by higher β-amylase activity. Northern analysis showed that β-amylase transcript levels increased from September to October in all germ plasms except Norseman-H (Fig. 6C). The high transcript levels for β-amylase in September and October declined markedly by November and December for all germ plasms. Although large declines in β-amylase mRNA occurred between October and December, β-amylase activity remained high in roots of all populations, suggesting high stability in planta for the β-amylase polypeptide.
DISCUSSION
We have isolated and sequenced a β-amylase cDNA that we believe encodes the β-amylase polypeptide previously described (Boyce and Volenec, 1992b) for several reasons. First, the molecular mass of the encoded β-amylase polypeptide (approximately 55,950 D) is similar to the 57-kD estimate for the β-amylase polypeptide obtained previously using SDS-PAGE (Boyce and Volenec, 1992b). Second, tissue distribution of the β-amylase transcript and β-amylase activities agree with both being much more abundant in roots than in leaves or stems. Third, β-amylase transcript levels are very abundant in roots of alfalfa. Lesser amounts were detected in roots of red clover, sweet- clover, and white clover, and amounts were below our detection limit in roots of birdsfoot trefoil. β-Amylase activity was also found to be more prevalent in roots of alfalfa than red clover, with virtually no activity in roots of birdsfoot trefoil (Boyce et al., 1992; Li et al., 1996). The β-amylase polypeptide constitutes 8% of the total soluble protein fraction in roots (Boyce and Volenec, 1992b). Similarly, we found large quantities of β-amylase mRNA in alfalfa roots, as indicated by the short exposure time (less than 5 min) needed for obtaining signals on radiographic films.
The physiological role of β-amylase in alfalfa roots remains obscure. In this study we examined how treatments known to reduce tissue starch concentrations (defoliation, Suc-deprivation of suspension-cultured cells, and cold acclimation) influence β-amylase activity and transcript abundance. Taproots of uncut alfalfa plants showed high starch concentrations, high β-amylase activities, and abundance of the β-amylase transcript (Fig. 4). Defoliation reduced root starch levels but, contrary to our expectations for starch hydrolase, also resulted in lower β-amylase activity and transcript levels. We have also shown that suspension-cultured cells grown in Suc-rich medium maintain high starch levels, elevated β-amylase activity, and higher quantities of the β-amylase transcript compared with cells deprived of Suc, which are forced to use starch as a carbon source (Fig. 5).
In agreement with previous studies (Volenec et al., 1991; Li et al., 1996), we found that cold acclimation of alfalfa stimulated starch degradation in autumn in winter-hardy germ plasms but without a concomitant increase in β-amylase activity or transcript level (Fig. 6). Roots of birdsfoot trefoil contain very low β-amylase activity (Li et al., 1996), and the β-amylase transcript was not detectable; yet starch degradation occurs in a similar manner in roots of birdsfoot trefoil and alfalfa after defoliation (Boyce and Volenec, 1992b) and during cold acclimation (Li et al., 1996). The amount of β-amylase mRNA in alfalfa, red clover, and birdsfoot trefoil correlates to the specific activity of total amylase activities observed previously for these forage legumes (Li et al., 1996). Our data suggest that β-amylase is not a key enzyme in starch degradation in roots of these forage legumes, because accumulation of the transcript and enzymatic activity did not coincide with the loss of root starch.
Large quantities of β-amylase are found in starch-storing organs (Doehlert et al., 1982; Yoshida and Nakamura, 1991; Boyce and Volenec, 1992a). We found high β-amylase mRNA levels in stems of alfalfa even though this tissue contains little starch. Wang et al. (1995) isolated a phloem-specific β-amylase from Streptanthus tortuosus (Brassicaceae). Because monoclonal antibodies made to this β-amylase cross-reacted with β-amylase from Arabidopsis, it was suggested that the major form of Arabidopsis β-amylase could be a phloem-specific enzyme. These authors speculated that the phloem-specific β-amylase may function to hydrolyze maltodextrins in sieve elements to prevent the accumulation of large polysaccharides that could impede carbon transport (Wang et al., 1995). Pea epicotyl β-amylase was shown to hydrolyze maltodextrins with few Glc molecules (Lizotte et al., 1990). Whether alfalfa β-amylase has a similar role in the hydrolysis of maltodextrin in phloem tissue of alfalfa stems is not known.
Since the function of alfalfa β-amylase is not clear, perhaps signals that enhance or repress expression of the β-amylase gene may lend some clue as to its function. Sugar-inducible expression of the β-amylase gene is well documented in Arabidopsis and sweet potato (Nakamura et al., 1991; Ohto et al., 1992; Mita et al., 1995, 1997). In agreement, cell cultures grown in Suc-free medium reduced β-amylase mRNA, activity, and starch concentrations (Fig. 5). Ironically, roots of field-grown alfalfa accumulate a significant quantity of soluble sugars in November and December (Cunningham et al., 1998), and yet during this period levels of β-amylase mRNA are very low compared with the high mRNA levels in September and October (Fig. 6), when sugar concentrations are low. Mita et al. (1997) proposed that the gene for Arabidopsis β-amylase is regulated by a combination of both positive and negative signals that are dependent on the level of sugars. Southern analysis shows that β-amylase is encoded by a small gene family (Fig. 2), members of which may be regulated by different sugars or sugar concentrations. Three β-amylase isozymes reported previously from alfalfa taproots (Doehlert et al., 1982; Habben and Volenec, 1991) may each be encoded by a different member of the β-amylase gene family. In contrast, β-amylase from Arabidopsis and maize is encoded by a single gene (Mita et al., 1995; Wang et al., 1997).
Figure 2.
Southern analysis of the β-amylase gene. Genomic DNA (10 μg) was digested separately with restriction enzymes, as indicated in each lane, and analyzed by hybridization with the radiolabeled β-amylase probe.
Although the in vivo starch hydrolase function of alfalfa β-amylase is not clear, it has been proposed that β-amylase may function as a VSP, providing the nitrogen needed for shoot regrowth (Hendershot and Volenec, 1993b; Li et al., 1996). This is an attractive hypothesis because β-amylase conforms to the definition of VSPs based on the perceived physiological function described by Cyr and Bewley (1990). VSPs are preferentially synthesized in storage organs and exhibit depletion during reactivation of meristems, and their abundance greatly exceeds that of other proteins in perenniating organs. β-Amylase conforms to these definitions of a VSP because it is more abundant in alfalfa roots than in stems and leaves. Defoliation reactivates meristems on crowns for shoot regrowth and substantially reduces β-amylase activity and β-amylase transcripts.
Eight percent of the soluble protein in alfalfa taproots is β-amylase (Boyce and Volenec, 1992a). Defoliation reduces nitrogenase activity and nitrogen fixation (Vance et al., 1979). Alfalfa root β-amylase shows patterns of activity similar to three abundant VSPs we have characterized from alfalfa roots (Cunningham and Volenec, 1996). These four polypeptides (including β-amylase) accumulate in alfalfa taproots in early autumn and disappear in spring, when shoot growth resumes, and during shoot regrowth when plants are defoliated. As shown by 15N labeling, significant movement of nitrogen occurs from this VSP-enriched protein pool to shoots after defoliation (Avice et al., 1996; Barber et al., 1996). Our data support the view that β-amylase from alfalfa roots functions as a VSP.
We have presented new information concerning the mode of regulation of the β-amylase gene by winter hardening and defoliation stress in alfalfa roots. Further work will concentrate on enhancing our understanding of the function of β-amylase using antisense DNA transformation studies. Work is also under way to clone and characterize the cDNAs for the three alfalfa taproot VSPs that we described previously (Cunningham and Volenec, 1996). We hope that understanding the structural and biological features of these VSPs will reveal the reasons that β-amylase serves a similar role in the roots of this species.
Abbreviation:
- VSP
vegetative storage protein
Footnotes
The work was supported by U.S. Department of Agriculture grant no. 96-35100-3141.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Avice J-C, Ourry A, Volenec JJ, Lemaire G, Boucard J. Defoliation-induced changes in abundance and immuno-localization of vegetative storage proteins in taproots of Medicago sativa. Plant Physiol Biochem. 1996;34:1–11. [Google Scholar]
- Bairoch A. PROSITE: recent developments. Nucleic Acids Res. 1994;22:3578–3580. [PMC free article] [PubMed] [Google Scholar]
- Barber LD, Joern BC, Volenec JJ, Cunningham SM. Supplemental nitrogen effects on alfalfa regrowth and taproot nitrogen mobilization. Crop Sci. 1996;36:1217–1223. [Google Scholar]
- Beck E, Ziegler P. Biosynthesis and degradation of starch in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:95–117. [Google Scholar]
- Boyce PJ, Penaloza E, Volenec JJ. Amylase activities in roots of Medicago sativa L. and Lotus corniculatusL. following defoliation. J Exp Bot. 1992;43:1053–1059. [Google Scholar]
- Boyce PJ, Volenec JJ. Taproot carbohydrate concentrations and stress tolerance of contrasting alfalfa genotypes. Crop Sci. 1992a;32:757–761. [Google Scholar]
- Boyce PJ, Volenec JJ. β-Amylase from taproots of alfalfa. Phytochemistry. 1992b;31:427–431. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cunningham SM, Volenec JJ. Purification and characterization of vegetative storage proteins from alfalfa (Medicago sativaL.) taproots. J Plant Physiol. 1996;147:625–632. [Google Scholar]
- Cunningham SM, Volenec JJ, Teuber LR. Plant survival and root and bud composition of alfalfa populations selected for contrasting fall dormancy. Crop Sci. 1998;38:962–969. [Google Scholar]
- Cyr DR, Bewley JD. Proteins in roots of the perennial weeds chicory (Cichorium intylus L.) and dandelion (Taraxacum officinaleWeber) are associated with overwintering. Planta. 1990;182:370–374. doi: 10.1007/BF02411387. [DOI] [PubMed] [Google Scholar]
- Doehlert DC, Duke SH, Anderson L. Beta-amylases from alfalfa (Medicago sativaL.) roots. Plant Physiol. 1982;69:1096–1102. doi: 10.1104/pp.69.5.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller NA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Gana JA, Sutton F, Kenefick DG. cDNA structure and expression patterns of a low-temperature-specific wheat gene tacr7. Plant Mol Biol. 1997;34:643–650. doi: 10.1023/a:1005852703506. [DOI] [PubMed] [Google Scholar]
- Graber LH, Nelson NT, Luekel WA, Albert WB (1927) Organic food reserves in relation to the growth of alfalfa and other perennial herbaceous plants. Wis Agric Exp Stn Bull 80
- Habben JE, Volenec JJ. Amylolytic activity in taproots of diploid and tetraploid Medicago sativaL. Ann Bot. 1991;68:393–400. [Google Scholar]
- Hendershot KL, Volenec JJ. Taproot nitrogen accumulation and use in overwinter alfalfa (Medicago sativaL.) J Plant Physiol. 1993a;141:68–74. [Google Scholar]
- Hendershot KL, Volenec JJ. Nitrogen pools in roots of Medicago sativaL. after defoliation. J Plant Physiol. 1993b;141:129–135. [Google Scholar]
- Joshi CP. Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res. 1987;15:9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Ourry A, Boucaud J, Lemaire G. Changes in source sink relationship for nitrogen during regrowth of lucerne (Medicago sativaL.) following removal of shoots. Aust J Plant Physiol. 1991;18:593–602. [Google Scholar]
- Kim TH, Ourry A, Boucard J, Lemaire G. Partitioning of nitrogen derived from N2 fixation and reserves in nodulated Medicago sativaL. during regrowth. J Exp Bot. 1993;44:555–562. [Google Scholar]
- Lehrach H, Diamond D, Wozney JM, Booedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Li R, Volenec JJ, Joern BC, Cunningham SM. Seasonal changes in nonstructural carbohydrates, proteins, and macronutrients in roots of alfalfa, red clover, sweetclover, and birdsfoot trefoil. Crop Sci. 1996;36:617–623. [Google Scholar]
- Lizotte PA, Henson CA, Duke SH. Purification and characterization of pea epicotyl β-amylase. Plant Physiol. 1990;94:1033–1039. doi: 10.1104/pp.92.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Hirano H, Nakamura K. Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana. Plant Physiol. 1997;114:575–582. doi: 10.1104/pp.114.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Suzuki-ujii K, Nakamura K. Sugar-inducible expression of a gene for beta-amylase in Arabidopsis thaliana. Plant Physiol. 1995;107:895–904. doi: 10.1104/pp.107.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe JD, Salminen MD, Preiss J. Nucleotide sequence of a cDNA clone encoding a beta-amylase from Arabidopsis thaliana. Plant Physiol. 1991;97:1599–1601. doi: 10.1104/pp.97.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Nakamura K, Ohto M, Yoshida N, Nakamura K. Sucrose-induced accumulation of β-amylase occurs concomitant with the accumulation of starch and sporamin in leaf-petiole cuttings of sweet potato. Plant Physiol. 1991;96:902–909. doi: 10.1104/pp.96.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M-A, Nakamura-Kito K, Nakamura K. Induction of expression of genes coding for sporamin and β-amylase by polygalacturonic acid in leaves of sweet potato. Plant Physiol. 1992;99:422–427. doi: 10.1104/pp.99.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ougham HJ, Davis TGE. Leaf development in Lolium temulentum. Gradients of RNA complement and plastid and non-plastid transcripts. Physiol Plant. 1990;79:331–338. [Google Scholar]
- Ourry A, Kim TH, Boucard J. Nitrogen reserves mobilization during regrowth of Medicago sativaL. Plant Physiol. 1994;105:831–837. doi: 10.1104/pp.105.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas G, Ramirex FM, Valero R, Palau J. Evolution of β-amylase: patterns of variation and conservation in subfamily sequences in relation to parsimony mechanisms. Proteins Structure Function Genet. 1996;25:456–472. doi: 10.1002/prot.6. [DOI] [PubMed] [Google Scholar]
- Rorat T, Sadowski J, Grellet F, Daussant J, Delsny M. Characterization of cDNA clones for rye endosperm-specific β-amylase and analysis of β-amylase deficiency in rye mutant lines. Theor Appl Genet. 1991;83:257–263. doi: 10.1007/BF00226260. [DOI] [PubMed] [Google Scholar]
- Sadowski J, Rorat T, Cooke R, Delseny M. Nucleotide sequence of a cDNA clone encoding ubiquitous beta-amylase in rye (Secale cerealeL.) Plant Physiol. 1993;102:315–316. doi: 10.1104/pp.102.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (1989) SAS/STAT Guide for Personal Computer, version 6. SAS Institute, Cary, NC
- Sharma R, Schopfer P. Phytochrome-mediated regulation of β-amylase mRNA in mustard (Sinapsis albaL.) cotyledons. Planta. 1987;171:313–320. doi: 10.1007/BF00398676. [DOI] [PubMed] [Google Scholar]
- Shure M, Wessler S, Fedoroff N. Molecular identification and isolation of the waxy locus in maize. Cell. 1983;35:235–242. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Smith D. Carbohydrate root reserves in alfalfa, red clover and birdsfoot trefoil under several management schedules. Crop Sci. 1962;2:75–78. [Google Scholar]
- Stewart W, Walker C. Comparison of nylon membranes. Methods Mol Cell Biol. 1989;1:73–76. [Google Scholar]
- Subbaramaiah K, Sharma R. β-Amylase from mustard (Sinapsis albaL) cotyledons. Plant Physiol. 1989;89:860–866. doi: 10.1104/pp.89.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Spradlin JE, Dygert S (1971) Plant and animal amylases. In PD Boyer, ed, The Enzymes, Ed 3, Vol 5. Academic Press, New York, pp 115–189
- Totsuka A, Fukazawa C. Expression and mutation of soybean β-amylase in Escherichia coli. J Biochem. 1993;214:787–794. doi: 10.1111/j.1432-1033.1993.tb17981.x. [DOI] [PubMed] [Google Scholar]
- Vance CP, Heichel GH, Barnes DK, Bryan JW, Johnson LE. Nitrogen fixation, nodule development, and vegetative regrowth of alfalfa (Medicago sativaL.) following harvest. Plant Physiol. 1979;64:1–8. doi: 10.1104/pp.64.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volenec JJ, Boyce PJ, Hendershot KL. Carbohydrate metabolism in taproots of Medicago sativaL. during winter adaptation and spring regrowth. Plant Physiol. 1991;96:786–793. doi: 10.1104/pp.96.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volenec JJ, Brown GA (1988) Characterization of starch degrading enzymes in alfalfa roots. In Agronomy Abstracts. American Society of Agronomy, Madison, WI, p 199
- Volenec JJ, Ourry A, Joern BC. A role for nitrogen reserves in forage regrowth and stress tolerance. Physiol Plant. 1996;97:185–193. [Google Scholar]
- Wagner G, Haeger K-P, Ziegler P. Nucleotide sequence of a cDNA from wheat leaves encoding ubiquitous β-amylase (accession no. X98504) (PGR 96-123) Plant Physiol. 1996;112:1735–1736. [Google Scholar]
- Wang S-H, Lue W-L, Eimert K, Chen J. Phytohormone-regulated β-amylase gene expression in rice. Plant Mol Biol. 1996;31:975–982. doi: 10.1007/BF00040716. [DOI] [PubMed] [Google Scholar]
- Wang S-H, Lue W-L, Wu S-Y, Huang H-W, Chen J. Characterization of a maize β-amylase cDNA clone and its expression during seed germination. Plant Physiol. 1997;113:403–409. doi: 10.1104/pp.113.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Monroe J, Sjolund RD. Identification and characterization of a phloem-specific β-amylase. Plant Physiol. 1995;109:743–750. doi: 10.1104/pp.109.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Nakamura K. Molecular cloning and expression in Escherichia coliof cDNA encoding the subunit of sweet potato β-amylase. J Biochem. 1991;110:196–201. doi: 10.1093/oxfordjournals.jbchem.a123556. [DOI] [PubMed] [Google Scholar]
- Yoshigi N, Okada Y, Sahara H, Koshino S. PCR cloning and sequencing of the beta-amylase cDNA from barley. J Biochem. 1994;115:47–51. doi: 10.1093/oxfordjournals.jbchem.a124303. [DOI] [PubMed] [Google Scholar]