Abstract
Myostatin, a secreted protein, is a negative regulator of skeletal muscle growth. Down-regulating its expression increases skeletal muscle mass that is accompanied by a marked change in the fibre composition from one reliant on mitochondrial oxidative metabolism to glycolysis. A comparative proteomic investigation of this altered metabolism was carried out on mitochondria from the gastrocnemius muscle of myostatin-null mice compared with wild-type. Most of the proteins identified showed no significant modulation between the 2 phenotypes, but give interesting insight into previous observations. Several proteins were modulated, of which only one was identified. This protein, having a sequence similar to that of aldehyde reductase, was up-regulated in myostatin-null mitochondria, but its importance was not established, although it might play a role in the detoxification of harmful products of lipid peroxidation.
Keywords: gastrocnemius muscle, mitochondria, myostatin, peptide mass fingerprinting, proteomics, 2-dimensional electrophoresis
1. Introduction
Myostatin (growth and differentiation factor-8) is a secreted protein predominantly expressed in skeletal muscle that negatively regulates muscle growth by altering transcription through the activin type II receptor (Lee, 2004). It is highly conserved, showing a high degree of homology across a broad spectrum of animals (McPherron and Lee, 1997). When myostatin is non-functional or absent, myoblast proliferation and differentiation increases with the fibre content of muscles being raised (hyperplasia) and/or the size of the individual muscle fibres (hypertrophy) (McCroskery et al., 2003), commonly referred to as ‘double-muscling’. This occurs naturally in Belgian Blue and Piedmontese cattle (Kambadur et al., 1997), and artificially by inhibition of the myostatin protein or deletion of the gene (McPherron et al., 1997; Lee and McPherron, 2001; Grobet et al., 2003; Whittemore et al., 2003).
The primary characteristic of the double-muscling phenotype is a decrease in fat deposition along with an increase in muscle mass (McPherron et al., 1997). The phenotype is also characterized by reduced connective tissue, a lower proportion of bone and a reduction in the size of organs. Due to a reduced respiratory capacity, less body surface per mass unit and poorer circulation, double-muscled cattle become more susceptible to stress and heat (Boccard, 1981). The animals have an altered fibre-type composition in that they have a larger proportion of type II fibres and a reduction in the type I fibres. There is also an increased amount of type IIB (glycolytic) compared with type IIA fibres (oxidative) (Girgenrath et al., 2005). This means double-muscled animals tire faster than normal animals because of increased metabolic acidosis due to reduced aerobic metabolism (Boccard, 1981) and blood circulation (Menissier, 1982) that normally removes lactic acid.
Type I and type IIB fibres generally differ in that the former have more mitochondria (Jackman and Willis, 1996; Schmidt and Herpin, 1997) and higher myoglobin content (Kim et al., 2004; Donoghue et al., 2005), related to this type of fibre relying on oxidative phosphorylation for ATP production. Type IIB fibres rely primarily on glycolysis for energy production, with greater expression of glycolytic enzymes to support it (Rivero et al., 1998; Hallauer and Hastings, 2002). Mitochondrial activity of oxidative compared with glycolytic muscle fibres is no different with respect to ATP synthase activity, but oxidative fibre mitochondria have a greater rate of ATP synthesis, probably due to the increased cytochrome c oxidase activity (Jackman and Willis, 1996; Schmidt and Herpin, 1997; Gueguen et al., 2005). Mitochondria from oxidative and glycolytic fibres both respire well (Schmidt and Herpin, 1997; Gueguen et al., 2005), but the former are better equipped to utilize fatty acids (Mogensen and Sahlin, 2005). While the role of mitochondria in glycolytic muscle fibres is poorly characterized, they could be utilized to support basal and recovery metabolism (Mogensen and Sahlin, 2005), although not during times of rapid contraction as the muscles would be operating anaerobically due to their reduced myoglobin content (Kim et al., 2004; Donoghue et al., 2005).
Comparative proteomics of the differences between oxidative and glycolytic muscle fibres shows modulation of the levels of contractile proteins, varying between fast twitch isoforms and slow twitch forms, and of some small heat shock proteins. Up-regulation of myoglobin levels was detected in the oxidative fibres, as well as several oxidative metabolic proteins. Glycolytic fibres had increased levels of enzymes associated with glycolysis (Donoghue et al., 2005; Okumura et al., 2005; Sayd et al., 2006). Likewise, proteomic investigations of double-muscled and normal animals have shown up-regulation of multiple glycolytic proteins (Bouley et al., 2005; Hamelin et al., 2006).
So far, little work has been done on the metabolism of myostatin KO (knockout) animals, and no one has assessed how the mitochondria are affected. This is despite the increased proportion of glycolytic fibres in double-muscled animals, which in turn provides a model to look at changes associated with differences in muscle fibre type metabolism. Our study used a comparative proteomic approach to determine if the mitochondria from myostatin KO skeletal muscle show modulated protein expression compared with those from WT (wild-type) animals.
2. Materials and methods
2.1. Sample collection
Gastrocnemius muscle was dissected from the hind limbs of 14-week-old WT and myostatin KO mice. The mice were asphyxiated with carbon dioxide before the muscle was removed and placed directly in ice-cold mitochondrial isolation buffer (Rustin et al., 1994). Mitochondria were isolated on the same day. Muscles tissues were acquired from the Growth and Development Group at AgResearch Ruakura. The method of myostatin KO in these mice was via genetic deletion (McPherron et al., 1997). All procedures involving the use of animals had the ethical approval of the University of Waikato Animal Ethics Committee.
2.2. Proteomic analysis of KO mitochondrial protein levels
Only the gastrocnemius muscle is used since it is easily isolated and of reasonable size. It also has a mixed fibre composition and therefore can display any changes in the mitochondria that occur due to a switch in muscle metabolism.
2.2.1. Mitochondrial isolation
A mitochondria-enriched fraction was isolated from the gastrocnemius muscle using the method of Rustin et al. (1994) with slight modifications. The dissected muscle was placed in ice-cold mitochondrial isolation buffer, finely diced and homogenized at 11500 rev./min for 15 s (IKA T10 basic ULTRA-TURRAX). The homogenate was filtered through a 100 μm nylon net, centrifuged at 2000 rcf for 8 min (Eppendorf 5415R) to remove cell debris and nuclei. The supernatant was centrifuged at 10 000 rcf for 10 min to pellet the mitochondria. The pellet was washed once more by resuspending in mitochondrial isolation buffer supplemented with 5% (v/v) Percoll and centrifuged at 10 000 rcf for 10 min. The mitochondria-enriched pellet was retained and frozen at −70°C until required.
2.2.2. Protein solubilization
An IPG strips (immobilized pH gradient strips) rehydration/solubilization solution was prepared fresh daily (8 M urea, 2 mM TBP (tributylphosphine), 2% CHAPS and 0.2% pH 3–10 ampholytes). The 8.5 M urea stock was treated with mixed-bed ion-exchange resin for 10 min to remove charged species. It was vacuum filtered through no.1 filter paper to remove the ion-exchange beads and frozen at −20°C until required.
The IPG strips rehydration/solubilization solution was added directly to the mitochondrial pellet, vortex-mixed (Grant-Bio) and left for 1 h at 20°C. The solution was centrifuged at 14 000 rcf (Eppendorf miniSpin plus) for 4 min to pellet the insoluble material. The supernatant was kept at 20°C until required. Protein concentration of this solution was measured with a BCA (bicinchoninic acid) protein assay kit (Pierce) according to the manufacturer's instructions, but with minor modifications. Because the IPG rehydration/solubilization solution contains 8 M urea, which interferes with the assay, it was used as a diluent, a blank and to make the BSA standard.
2.2.3. 2DE (2-dimensional electrophoresis)
Protein solutions were diluted with the IPG strips rehydration/solubilization solution prior to loading on to IPG strips. To track the progress of the IEF (isoelectric focusing), 0.0007% Bromophenol Blue was added to the diluted solution. The sample was laid along the length of a 17 cm IEF focusing tray (Bio-Rad) and 17 cm pH 3–10 NL (non-linear) IPG strips (Bio-Rad) were laid on top. This was left for 1 h before being overlaid with mineral oil. The focusing tray lid was replaced and the IPG strips were left to rehydrate passively for another 16 h at 20°C in a Protean IEF Cell (Bio-Rad).
Before running the IEF, the IPG strips were lifted at each end and an electrode wick (Bio-Rad) wetted with MilliQ water was placed between the electrode and the gel. The IEF was run at 20°C where the gels were held at 250 V for 15 min and then subjected to a rapid ramp over 3 h at 10000 V, where the voltage was maintained at 10 000 V for 60 000 V h. After completion of the IEF, the gels were held at 500 V until removed from the IEF cell to limit diffusion of the proteins. After removing excess mineral oil, the IPG strips were placed gel side up in a rehydration/equilibration tray (Bio-Rad), wrapped in cling film and frozen at −70°C until the second dimension was run.
The IPG strips were prepared for the second dimension by equilibrating them in SDS/PAGE equilibration buffer (6 M urea, 0.375 M Tris/HCl, pH 8.8, 2% SDS and 20% glycerol) supplemented with 2% dithiothreitol for 15 min to reduce disulfide bonds. This was decanted off and replaced with SDS/PAGE equilibration buffer supplemented with 2.5% iodoacetamide for another 15 min to acetylate the free thiol groups (Herbert et al., 2001).
After removing excess liquid, the IPG strips were laid on top of an 18.5×20 cm2, 1.5 mm thick, SDS/12% PAGE gel. A 1% agarose gel plug containing 10 kDa protein markers (GibcoBRL) was placed next to the acidic end of the IPG strips and both were covered in 2DE agar overlay (0.5% agarose and 0.003% Bromophenol Blue). The gels were run at 35 mA per gel (Power Supply 1000/500; Bio-Rad) for 5 h and 45 min, during which they were maintained at 15°C by circulating chilled water (RB-12A/TE-8D; Techne) through the cooling chamber.
On completion of the electrophoresis, the gel was removed and stained using a Silver Stain Plus kit (Bio-Rad). Silver Stain Plus does not cross-link lysine residues like conventional silver stain, and therefore allows for downstream MS analysis. All glassware and gel containers used for silver staining were washed with 2 M nitric acid overnight and rinsed in MilliQ water.
2.2.4. Image analysis
The gel image was captured on a luminescent image analyser (LAS-1000 plus; FujiFilm) controlled by Image Reader LAS-1000 Lite (FujiFilm) with an exposure time of 1/60 s. The resulting spot intensities were analysed using Image Gauge Version 4.0 (FujiFilm). Spots were selected manually and the pixel intensities [expressed in AU (arbitrary units)] were divided by the size of the area selected (in mm2) to take into account the different spot sizes present. The background, also divided by its spot size, was subtracted from the AU/mm2. Statistical significance was determined using a 2-tailed Student's t test comparing the replicate spot intensities from the WT gels with those of the KO gels.
2.2.5. In-gel tryptic digest
Spots of interest were excised using 200 μl pipette tips (Greiner Bio-one) cut off at the first mark, 12 mm from the tip. These were stored at −20°C until required. The gel plugs were destained in 1% H2O2, added directly to the tube in which the plugs were stored and shaken (MS1 mini shaker; IKA) overnight at 600 rev./min. The destain solution was removed and the plugs were washed with 25 mM NH4HCO3 for 15 min and equilibrated in 1:1 25 mM NH4HCO3/acetonitrile for 1 h before dehydrating with acetonitrile. Acetonitrile was removed after 15 min and the gel plugs were left to air-dry in a hood for 2 h.
Trypsin Gold (0.005 μg/μl; Promega), made up in 25 mM NH4HCO3, was added to each dehydrated gel plug and left at 4°C to rehydrate the gel for 1 h. Excess trypsin solution was removed and the closed tubes were placed in a sealed container with a wet paper towel to maintain humidity, which was incubated at 37°C for 6 h (Contherm Digital Series). The resulting peptides were extracted in 20% acetonitrile/0.1% TFA (trifluoroacetic acid) overnight at 4°C.
2.2.6. MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS) of tryptic digest products
Tryptic digest samples for PMF (peptide mass fingerprinting) were prepared for MALDI–TOF-MS by applying 0.5 μl of the tryptic digest to a thin layer of α-cyano-4-hydroxycinnamate on a 600 μm anchorchip (Bruker Daltonics). Once the sample had air-dried, it was washed with 0.1% TFA in 10 mM NH4H2PO4 for ∼5 s before removing and allowing to air-dry again.
An AutoFlex II MALDI–TOF mass spectrometer (Bruker Daltonics) operated using FlexControl (Bruker Daltonics) was used to assess the mass of the peptides produced in the tryptic digest. The instrument was calibrated against a peptide calibration standard (Bruker Daltonics) that was loaded in the same manner as the samples on to calibanchors, located at the centre of every 4 sample anchors. Sample spectra were acquired over a 500–4000 m/z range by summing 500 shots, with an acceleration voltage of 19 kV and a reflector voltage of 20 kV. Pulsed-ion extraction of 80 ns was used to build up the concentration of ions in the ion source, and ions below 500 m/z were suppressed to avoid detector saturation from matrix ions.
Spectra were automatically annotated using FlexAnalysis (Bruker Daltonics) to peak the mono-isotopic peaks within 800–4000 m/z (SNAP algorithm). Mascot database searches were performed via BioTools 3.0 (Bruker Daltonics), searching the NCBInr database for sequences from either Mammalia (mammals) or Mus musculus (house mouse) with a fixed modification of ‘Carbidomethyl’, 0 or 1 missed cleavages and a mass accuracy of 75–150 p.p.m.
3. Results
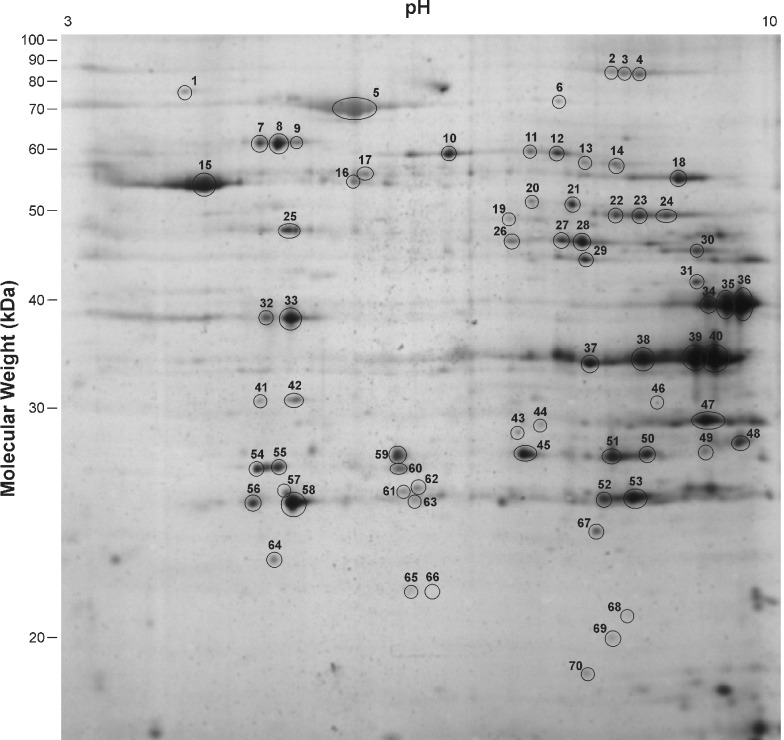
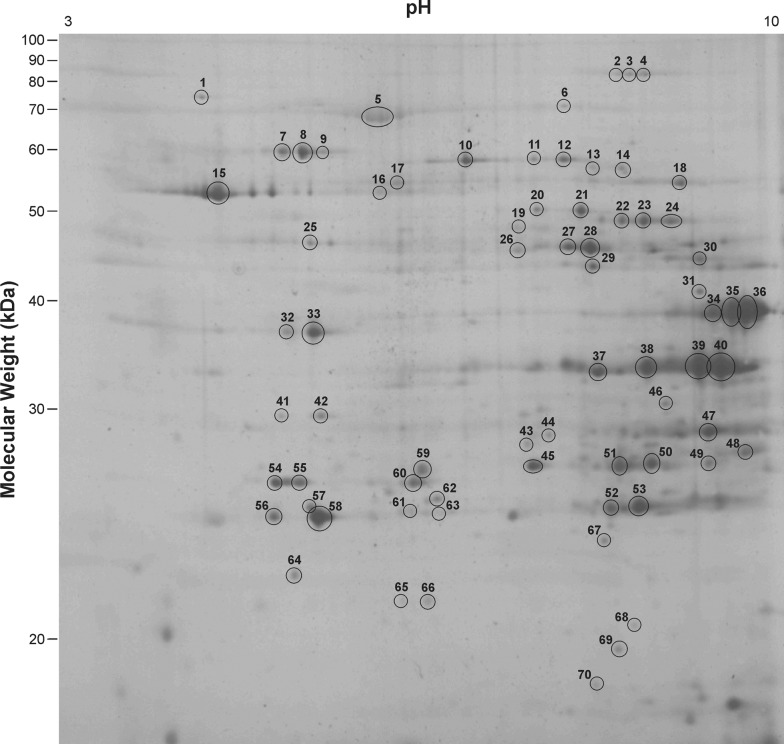
Mitochondrial proteins from the gastrocnemius muscle of WT and myostatin KO mice were separated by their pI and their molecular mass. This 2DE gel set consisted of 3 WT gels and 3 KO gels. Figures 1 and 2 show 2DE gels from the WT and KO samples respectively, showing the numbers assigned to the spots that had their intensities assessed. These spot numbers were assigned to spots that already had proteins identified and others that showed visible differences in intensity. The pixel intensity of only these spots was measured. In these gels, ∼160 individual protein spots were detected by silver staining. Proteins were concentrated in the alkaline region of the gel and in the higher molecular mass acidic region. A few proteins were resolved in the neutral pH region. No proteins of Mr>90 kDa were observed and proteins of a lower mass were not well resolved, showing large amounts of diffusion.
Figure 1. 2DE gel of mitochondrial protein from WT gastrocnemius muscle showing the assigned spot numbers.
The ellipses are not an indication of the spot circumference, but serve to highlight the location of the spot.
Figure 2. 2DE gel of mitochondrial protein from myostatin KO gastrocnemius muscle showing the assigned spot numbers.
The ellipses are not an indication of the spot circumference, but serve to highlight the location of the spot.
Seventy of these spots had their intensities measured and the majority quantified showed no difference in expression between the WT and KO mitochondria (Table 1). However, several proteins did show changes in their levels of expression. Spot numbers 49 and 68 were up-regulated in the KO sample and spots 63 and 67 were down-regulated, but these differences were not statistically significant (P<0.1). Spot number 59 was expressed 1.2 times lower in the KO mitochondria (P<0.05). Unfortunately, all these proteins were expressed at levels too low to be identified during the present study. Spot number 46, which was expressed 2.4 times higher (P<0.005) in the KO mitochondria, was identified as an unnamed protein from M. musculus (accession number BAB23853).
Table 1. Modulation of proteins in myostatin KO mitochondria, as shown by changes in 2DE spot intensities.
| Spot(s)a | Protein | Species | Mrb | pIc | Accession no. | Changed | Significancee |
|---|---|---|---|---|---|---|---|
| Chaperone and regulator proteins | |||||||
| 1 | Glucose-regulated protein, 78 kDa | M. musculus | 72.5 | 4.9 | NP_071705 | N/C | |
| 7, 8, 9 | Heat-shock protein 60 | M. musculus | 61.1 | 5.8 | NP_034607 | N/C | |
| 6 | Inner membrane protein | M. musculus | 84.2 | 6.2 | NP_083949 | N/C | |
| 41, 42 | Prohibitin | M. musculus | 29.9 | 5.5 | NP_032857 | N/C | |
| 52, 53 | Superoxide dismutase 2 | M. musculus | 24.8 | 9.5 | NP_038699 | N/C | |
| 60 | Thioredoxin-dependent peroxide reductase | M. musculus | 28.3 | 8.0 | NP_031478 | N/C | |
| 38, 39, 40 | Voltage-dependent anion channel 1 | M. musculus | 30.9 | 9.2 | NP_035824 | N/C | |
| 45 | Voltage-dependent anion channel, chain A | Homo sapiens | 32.2 | 9.5 | 2JK4_A | N/C | |
| Oxidative phosphorylation | |||||||
| 56, 58 | ATP synthase, subunit d | M. musculus | 18.8 | 5.4 | NP_082138 | N/C | |
| 18 | ATP synthase, F1 complex, α subunit | M. musculus | 54.7 | 8.9 | EDL09448 | N/C | |
| 15 | ATP synthase, F1 complex, β subunit | H. sapiens | 48.1 | 4.8 | ABD77240 | N/C | |
| 47 | Electron transferring flavoprotein, α subunit | M. musculus | 35.4 | 9.5 | AAH03432 | N/C | |
| 37 | Electron transferring plavoprotein, β subunit | M. musculus | 21.9 | 9.6 | EDL22659 | N/C | |
| 55 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | M. musculus | 27.6 | 7.8 | NP_082664 | N/C | |
| Tricarboxylic acid cycle enzymes | |||||||
| 2, 3, 4 | Aconitase hydratase | M. musculus | 86.2 | 8.9 | NP_542364 | N/C | |
| 22, 23, 24 | Fumarate hydratase 1 | M. musculus | 54.6 | 9.7 | NP_034339 | N/C | |
| 29 | Isocitrate dehydrogenase 3, β subunit | M. musculus | 42.5 | 9.4 | NP_570954 | N/C | |
| 34, 35, 36 | Malate dehydrogenase | M. musculus | 36.0 | 9.8 | NP_032643 | N/C | |
| 69 | Nucleoside-diphosphate kinase B | H. sapiens | 9.6 | 10.1 | NP_001185611 | N/C | |
| 25 | Succinate-CoA ligase, β subunit | M. musculus | 48.2 | 5.7 | EDL35853 | N/C | |
| Other metabolic enzymes | |||||||
| 13 | Aldehyde dehydrogenase 6a1 | M. musculus | 50.0 | 7.7 | AAH31148 | N/C | |
| 30 | Aldolase A, fructose-bisphosphate | M. musculus | 39.8 | 9.2 | NP_031464 | N/C | |
| 10, 11, 12 | Dihydrolipoamide dehydrogenase | M. musculus | 54.8 | 9.0 | NP_031887 | N/C | |
| 21 | Enolase 3, β subunit | M. musculus | 49.0 | 6.9 | EDL12600 | N/C | |
| 48 | Hydroxyacyl-CoA dehydrogenase 2 | M. musculus | 27.4 | 9.6 | NP_058043 | N/C | |
| 26 | Isovaleryl CoA dehydrogenase | M. musculus | 46.7 | 9.3 | NP_062800 | N/C | |
| 32, 33 | Pyruvate dehydrogenase, β subunit | M. musculus | 35.2 | 5.6 | AAH02188 | N/C | |
| 46 | Unnamed protein | M. musculus | 29.1 | 6.6 | BAB23853 | ↑ 2.4 | P<0.005 |
| Miscellaneous | |||||||
| 57 | Adenylate kinase 1 | M. musculus | 21.6 | 5.6 | NP_001185719 | N/C | |
| 54 | Apolipoprotein A-1 | Bos taurus | 30.3 | 5.6 | NP_776667 | N/C | |
| 5 | BSA | B. taurus | 71.3 | 5.8 | NP_851335 | N/C | |
| 27, 28 | Creatine kinase, M-type | M. musculus | 43.2 | 6.6 | NP_031736 | N/C | |
| 50 | ES1 | M. musculus | 28.4 | 9.9 | NP_613067 | N/C | |
| 63 | Unidentified | ↓ 2.1 | P<0.1 | ||||
| 67 | Unidentified | ↓ 1.3 | P<0.1 | ||||
| 59 | Unidentified | ↓ 1.2 | P<0.05 | ||||
| 49 | Unidentified | ↑ 1.3 | P<0.1 | ||||
| 68 | Unidentified | ↑ 1.4 | P<0.1 |
Spot numbers correspond to those shown in Figure 1; those not listed were not able to be identified by PMF and did not show statistically significant changes in expression between KO and WT.
Theoretical molecular mass (kDa).
Theoretical pI.
Change in protein expression is expressed as a fold increase (↑) or decrease (↓) in the KO spot intensity compared to WT spot intensity; N/C signifies no statistically significant change in protein expression.
Statistical significance.
4. Discussion
4.1. Proteins identified from 2DE gels
While most of the proteins identified in this study did not show modulation in the myostatin KO mitochondria, spot number 46 showed increased expression in the KO mitochondria and was identified as indicated above. Its amino acid sequence (Figure 3) is extremely similar to ALR (aldehyde reductase; accession number AAH46762), which is up-regulated in response to lipid peroxidation (Rittner et al., 1999; Oberschall et al., 2000) and is found in the mitochondrial inner membrane (Udovikova and Wojtczak, 1998). Its increased level of expression in the myostatin KO mitochondria could reflect a higher level of oxidative stress; however, other proteins commonly up-regulated in oxidative stress were not significantly affected. These include isocitrate dehydrogenase, superoxide dismutase 2 and thioredoxin-dependent peroxide reductase, which are all involved in detoxifying reactive oxygen species produced by aerobic respiration (Halliwell and Chirico, 1993; Chang et al., 2004; Kim et al., 2005). Since several other proteins normally associated with oxidative stress were not modulated in the myostatin KO mitochondria, it is probable that the increase in expression of the unidentified protein is unrelated to increased lipid peroxidation.
Figure 3. Sequence alignment of ALR (accession number AAH46762) and the unnamed mouse protein (UNP; accession number BAB23853) identified in spot 46.
The remainder of the proteins identified in the 2DE gels (Table 1) were high-abundance mitochondrial proteins that were not modulated in the myostatin KO muscle. They include those responsible for the import of nuclear-encoded proteins into the mitochondria and links among glycolysis, the tricarboxylic acid cycle, the electron transport system and oxidative phosphorylation. This suggests that muscle mitochondria from myostatin KO mice have the potential to operate in a similar capacity as those from WT mice.
The presence of several proteins, considered to be cytoplasmic proteins, suggests a low level of cytoplasmic contamination of the mitochondrial preparation. However, aldolase and enolase have been shown to be present in or interact with the mitochondria (Giegé et al., 2003; Brandina et al., 2006). Apolipoprotein A-1 interacts with the β subunit of ATP synthase (Martinez et al., 2003), and prohibitin is a protein that acts as a chaperone to stabilize newly synthesized proteins in the mitochondria (Nijtmans et al., 2000). Creatine kinase is used in the mitochondria for ATP homoeostasis, as it transfers a phosphate group from phosphocreatine stores (Bruton et al., 2003). The BSA identified in spot 5 was most likely carry over from the mitochondrial isolation buffer.
4.2 Identifications relevant to previous studies
Two of the mitochondrial enzymes identified in the present study are known to have decreased activity in glycolytic muscle mitochondria compared with mitochondria from oxidative fibres. Mogensen and Sahlin (2005) reported a 2-fold decrease in 3-hydroxyacyl-CoA dehydrogenase activity in extensor digitorum longus muscle mitochondria (type IIB fibres) compared with soleus muscle mitochondria (type I fibres). Jackman and Willis (1996) found that malate dehydrogenase was decreased in glycolytic fibre mitochondria. Our present study showed no significant change in the expression levels of these enzymes between WT and myostatin KO mitochondria. As the myostatin KO mice serve as a model for glycolytic muscle metabolism due to their increased proportion of type IIB fibres (Girgenrath et al., 2005), it suggests that the increased enzymatic activity observed by these researchers could be due to either enzyme activation in the oxidative fibre mitochondria or inhibition in the glycolytic fibre mitochondria.
Proteomic studies analysing skeletal muscle samples from oxidative and glycolytic fibres have reported modulation of mitochondrial metabolic proteins between the fibre types. Aconitase, ATP synthase subunits, malate dehydrogenase, mitochondrial creatine kinase and NADH dehydrogenase are down-regulated in glycolytic fibres (Donoghue et al., 2005; Okumura et al., 2005; Sayd et al., 2006). A similar trend was observed in proteomic studies using double-muscled animals, with an increase in the presence of glycolytic enzymes and reduction in those involved in oxidative phosphorylation (Bouley et al., 2005; Hamelin et al., 2006). Our present study, using only mitochondrial samples isolated from the muscle, shows no significant change in expression of these proteins, suggesting that the observations made by others could be due to a decrease in the density of mitochondria in these fibre types. This reiterates observations made by other investigators that there is a decrease in the number of mitochondria present in glycolytic muscle fibres (Jackman and Willis, 1996; Schmidt and Herpin, 1997).
4.3 Future directions
Estimates of the total number of proteins present in the mitochondria vary between 1000 and 2000; these numbers are gauged from genomic analysis, with only 685 of them being identified (Gibson, 2005). In the final 2DE gel set, approx. 160 spots were visualized using Silver Stain Plus (Bio-Rad). This corresponds to approximately one tenth of the proteins present in the mitochondrial proteome. Granted that many of the proteins present in the mitochondria are hydrophobic proteins and difficult to resolve using IPG strips, there are still many more hydrophilic proteins present in the mitochondria that cannot be seen in this gel set.
Several strategies could have been adopted to detect more of the mitochondrial proteome. The amount of protein loaded on to the gel could have been increased, and the inherent loss in resolution compensated for using a smaller pH gradient during the IEF. However, sample availability did not allow for this during the present study. Also, a more sensitive detection method, such as fluorescent staining, could have been used. This was not undertaken because increasing the staining sensitivity would still not allow for the newly detected proteins to be identified.
Of the 160 protein spots detected in the 2DE gels, 50 were identified as known proteins. Since silver stain was used during this study as opposed to Coomassie Blue stain, many of the spots observed were not present at high enough levels to be identified using PMF by MALDI MS. To identify more of the proteins seen in the 2DE gels (via PMF by MALDI MS), more protein would have to be loaded on to the gel. Preparative gels could have been run, but was not applied during this study due to sample restriction. An attempt was made to identify the protein in spot 59, by combining gel plugs from 3 WT 2DE gels and performing a tryptic digest as usual, but this yielded only a low-intensity spectrum.
In conclusion, 2DE gels of mitochondria from WT and myostatin KO skeletal muscle showed 160 proteins, of which 50 were identified as known proteins. Most of these proteins were high-abundance mitochondrial proteins that showed no significant modulation between the WT and KO samples. This suggests that the results observed in previous proteomic studies have been due to a lower number of mitochondria in the glycolytic muscle samples used, as opposed to a lower expression in the mitochondria themselves. A protein showing sequence similarity to ALR was up-regulated in the myostatin KO mitochondria, but its significance was not established.
Acknowledgments
We thank Dr Ravi Kambadur (AgResearch Ruakura) for access to the samples we used in this study. Alex Henebry (AgResearch Ruakura), Julie Goldsbury, Kerry Allen, Raewyn Towers and Wendy Jackson (University of Waikato) are acknowledged for their valuable technical assistance.
Footnotes
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Author contribution
Jonathan Puddick carried out the experiments described in the study as part of his MSc research programme at the University of Waikato under the supervision of Ryan Martinus.
References
- Boccard R. Facts and reflections on muscular hypertrophy in cattle: double muscling or culard. In: Lawrie R, editor. Developments in meat science. London: Applied Science Publishers; 1981. pp. 1–28. [Google Scholar]
- Bouley J, Meunier B, Chambon C, De Smet S, Hocquette JF, Picard B. Proteomic analysis of bovine skeletal muscle hypertrophy. Proteomics. 2005;5:490–500. doi: 10.1002/pmic.200400925. [DOI] [PubMed] [Google Scholar]
- Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. BBA Bioenergetics. 2006;1757:1217–28. doi: 10.1016/j.bbabio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Bruton JD, Dahlstedt AJ, Abbate F, Westerblad H. Mitochondrial function in intact skeletal muscle fibres of creatine kinase deficient mice. J Physiol. 2003;552:393–402. doi: 10.1113/jphysiol.2003.050732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TS, Cho CS, Park S, Yu S, Kang SW, Rhee SG. Peroxiredoxin III, a mitochondrion-specific peroxidase, regulates apoptotic signaling by mitochondria. J Biol Chem. 2004;279:41975–84. doi: 10.1074/jbc.M407707200. [DOI] [PubMed] [Google Scholar]
- Donoghue P, Doran P, Dowling P, Ohlendieck K. Differential expression of the fast skeletal muscle proteome following chronic low-frequency stimulation. BBA Proteins Proteom. 2005;1752:166–76. doi: 10.1016/j.bbapap.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Gibson BW. The human mitochondrial proteome: oxidative stress, protein modifications and oxidative phosphorylation. Int J Biochem Cell B. 2005;37:927–34. doi: 10.1016/j.biocel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Giegé P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell. 2003;15:2140–51. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenrath S, Song K, Whittemore LA. Loss of myostatin expression alters fiber-type distribution and expression of myosin heavy chain isoforms in slow- and fast-type skeletal muscle. Muscle Nerve. 2005;31:34–40. doi: 10.1002/mus.20175. [DOI] [PubMed] [Google Scholar]
- Grobet L, Pirottin D, Farnir F, Poncelet D, Royo LJ, Brouwers B. Modulating skeletal muscle mass by postnatal, muscle-specific inactivation of the myostatin gene. Genesis. 2003;35:227–38. doi: 10.1002/gene.10188. [DOI] [PubMed] [Google Scholar]
- Gueguen N, Lefaucheur L, Fillaut M, Vincent A, Herpin P. Control of skeletal muscle mitochondria respiration by adenine nucleotides: differential effect of ADP and ATP according to muscle contractile type in pigs. Comp Biochem Physiol B: Biochem Mol Biol. 2005;140:287–97. doi: 10.1016/j.cbpc.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Hallauer PL, Hastings KEM. Coregulation of fast contractile protein transgene and glycolytic enzyme expression in mouse skeletal muscle. Am J Physiol Cell Physiol. 2002;282:C113–C24. doi: 10.1152/ajpcell.00294.2001. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Chirico S. Lipid peroxidation: its mechanism, measurement, and significance. Am J Clin Nutr. 1993;57:715S–24S. doi: 10.1093/ajcn/57.5.715S. [DOI] [PubMed] [Google Scholar]
- Hamelin M, Sayd T, Chambon C, Bouix J, Bibé B, Milenkovic D. Proteomic analysis of ovine muscle hypertrophy. J Anim Sci. 2006;84:3266–76. doi: 10.2527/jas.2006-162. [DOI] [PubMed] [Google Scholar]
- Herbert BJ, Schulenberg B, Patton WF, Capaldi RA. A novel subfractionation approach for mitochondrial proteins: a three-dimensional mitochondrial proteome map. Electrophoresis. 2001;22:950–9. doi: 10.1002/1522-2683()22:5<950::AID-ELPS950>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Jackman MR, Willis WT. Characteristics of mitochondria isolated from type I and type IIb skeletal muscle. Am J Physiol Cell Physiol. 1996;270:C673–C78. doi: 10.1152/ajpcell.1996.270.2.C673. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Sharma M, Smith TPL, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–5. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kang BS, Park J-W. Cellular defense against heat shock-induced oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. Free Radical Res. 2005;39:441–8. doi: 10.1080/10715760500066265. [DOI] [PubMed] [Google Scholar]
- Kim N-K, Joh J-H, Park H-R, Kim O-H, Park B-Y, Lee C-S. Differential expression profiling of the proteomes and their mRNAs in porcine white and red skeletal muscles. Proteomics. 2004;4:3422–8. doi: 10.1002/pmic.200400976. [DOI] [PubMed] [Google Scholar]
- Lee SJ. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- Lee SJ, McPherron AC. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci USA. 2001;98:9306–11. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LO, Jacquet S, Esteve JP, Rolland C, Cabezon E, Champagne E. Ectopic β-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nature. 2003;421:75–9. doi: 10.1038/nature01250. [DOI] [PubMed] [Google Scholar]
- McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–47. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–61. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGFβ superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Menissier F. General survey of the effect of double muscling on cattle performance. In: King JWB, Menissier F, editors. Muscle Hypertrophy of Genetic Origin and its Use to Improve Beef Production. The Hague: Martinus Nijhoff Publishers; 1982. pp. 23–53. [Google Scholar]
- Mogensen M, Sahlin K. Mitochondrial efficiency in rat skeletal muscle: influence of respiration rate, substrate and muscle type. Acta Physiol Scand. 2005;185:229–36. doi: 10.1111/j.1365-201X.2005.01488.x. [DOI] [PubMed] [Google Scholar]
- Nijtmans LGJ, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–51. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberschall A, Deák M, Török K, Sass L, Vass I, Kovács I. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J. 2000;24:437–46. doi: 10.1046/j.1365-313x.2000.00885.x. [DOI] [PubMed] [Google Scholar]
- Okumura N, Hashida-Okumura A, Kita K, Matsubae M, Matsubara T, Takao T. Proteomic analysis of slow- and fast-twitch skeletal muscles. Proteomics. 2005;5:2896–906. doi: 10.1002/pmic.200401181. [DOI] [PubMed] [Google Scholar]
- Rittner HL, Hafner V, Klimiuk PA, Szweda LI, Goronzy JJ, Weyand CM. Aldose reductase functions as a detoxification system for lipid peroxidation products in vasculitis. J Clin Invest. 1999;103:1007–13. doi: 10.1172/JCI4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero JLL, Talmadge RJ, Edgerton VR. Fibre size and metabolic properties of myosin heavy chain-based fibre types in rat skeletal muscle. J Muscle Res Cell Motil. 1998;19:733–42. doi: 10.1023/a:1005482816442. [DOI] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, Gérard B, Rötig A, Saudubray JM. Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Sayd T, Morzel M, Chambon C, Franck M, Figwer P, Larzul C. Proteome analysis of the sarcoplasmic fraction of pig semimembranosus muscle: implications on meat color development. J Agric Food Chem. 2006;54:2732–7. doi: 10.1021/jf052569v. [DOI] [PubMed] [Google Scholar]
- Schmidt I, Herpin P. Postnatal changes in mitochondrial protein mass and respiration in skeletal muscle from the newborn pig. Comp Biochem Physiol B: Biochem Mol Biol. 1997;118:639–47. doi: 10.1016/s0305-0491(97)00268-x. [DOI] [PubMed] [Google Scholar]
- Udovikova EA, Wojtczak L. Mitochondrial aldehyde reductase: Identification and characterization in rat liver and kidney cortex. Int J Biochem Cell Biol. 1998;30:597–608. doi: 10.1016/s1357-2725(97)00143-x. [DOI] [PubMed] [Google Scholar]
- Whittemore LA, Song K, Li X, Aghajanian J, Davies M, Girgenrath S. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun. 2003;300:965–71. doi: 10.1016/s0006-291x(02)02953-4. [DOI] [PubMed] [Google Scholar]