Abstract
Human embryonic stem cells can differentiate into CD34+ hematopoietic progenitors by co-culture on murine feeders such as OP9 and S17. These CD34+ progenitors can be further differentiated into several cells of the hematopoietic lineage including macrophages. However, co-culture on murine feeders is time consuming and involves extensive manipulations. Furthermore, CD45 expression is low on hematopoietic cultures derived from stromal co-cultures. In this study we describe a novel and highly efficient system of generating differentiated macrophages from hematopoietic progenitors generated from embryoid body cultures of human embryonic stem cells. The hematopoietic progenitors generated from these embryoid bodies express higher numbers of CD45+ cells and are able to differentiate to macrophages when cultured in presence of cytokines. Using this system we were able to generate higher yields of CD14+ macrophages compared to traditional stromal cell culture methods. The embryoid body derived macrophages are phagocytic, respond to Toll-like receptor stimulation and express phenotypic markers of mature macrophages. Importantly, the embryoid body system generates hematopoietic progenitors suitable for clinical use by eliminating the need for murine feeder cells. Furthermore, this system is amenable to genetic manipulation and may thus be used to study important mechanisms of macrophage differentiation and function.
Introduction
Macrophages (Mφ’s) are a population of ubiquitously distributed mononuclear phagocytes that participate in both specific immunity via antigen presentation and cytokine production and in nonspecific immunity against bacterial, viral and fungal pathogens. They participate in tissue remodeling [1] and in clearing invading pathogens, cellular debris and apoptotic cellular waste [2]. Mφ’s also serve as presenters of antigens to T cells and secrete various immune-regulating cytokines [3]. Mφ’s have been characterized in various metabolic diseases such as atherosclerosis and type-2 diabetes mellitus as well as in autoimmune disorders such as multiple sclerosis [4, 5]. Despite being well understood in terms of their contribution to chronic inflammatory states, a thorough study of human Mφ biology has been restricted by the absence of a tractable genetic system. Current systems involve tumor-derived cell lines and also primary bone marrow-derived Mφ’s, peritoneal Mφ’s or peripheral blood derived monocytes that can be differentiated to Mφ’s. While these systems have generated significant data they are also limited. The cell lines may differ significantly from normal Mφ’s because of their origin while the existing sources of primary cells represent either a more quiescent state or more activated tissue Mφ state. Further, these adult cells are terminally differentiated and do not possess a great replicative potential nor are they very amenable to genetic manipulation.
Human embryonic stem cells (hESC) can be coaxed to differentiate into various tissue types of the body [6]. With respect to the hematopoietic system, hESC have been differentiated into both myeloid and erythroid lineages by co-culturing hESC on murine bone marrow stromal cells such as S17 and OP9 [7] and also by the formation of embryoid bodies (EB’s) that are cultured in the presence of a combination of cytokines [8] [9]. The EB’s spontaneously differentiate into cells of all three embryonic germ layers [10]. The differentiation toward a desired lineage can be potentiated by addition of appropriate cytokines [11]. Both culture methods result in the production of CD34+ hematopoietic progenitors that proceed through a sequential development and are capable of forming multilineage hematopoietic colonies in vitro [12]. hESC recapitulate aspects of embryonic hematopoiesis closely [13, 14]. The earliest progenitors arising from stromal co-cultures are erythroid in nature as determined by early expression of CD235 and CD34. Myeloid progenitors arise a little later and are accompanied by expression of CD45 on the CD34+ cells [15].
Mφ’s are derived from common myeloid progenitors following induction of myeloid differentiation by hematopoietic cytokines. In vivo, this process is regulated by the interaction of specific hematopoietic cytokines with their cognate receptors on the surface of progenitor cells present in the bone marrow resulting in the activation of various signal transduction pathways [16]. In vitro, Mφ’s can be differentiated from hESC by co-culture on S17 stroma for 15–17 days, followed by subsequent culture of the differentiated CD34+ cells in methylcellulose for 2 weeks followed by further suspension culture of the colonies obtained from methylcellulose for an additional 12–15 days [17]. Though this system is more pliable than existing adult sources of Mφ’s, the use of mouse stroma restricts its use in clinical contexts. Very recently, the differentiation of hESC into mature Mφ’s via EB cultures has been demonstrated [18]. However this system cannot be adapted for transplant purposes since these conditions do not identify the lineage progenitors that are transplantable and produce only the final product namely terminally differentiated Mφ’s. Hence we chose to determine whether the feeder-free EB system could be used to identify and obtain hematopoietic progenitors that can be specifically differentiated into mature Mφ’s. The hematopoietic progenitors can carry genetic modifications introduced into hESC and thus be utilized in gene therapy and transplantation studies to address Mφ specific disorders.
Our studies show for the first time a highly efficient method of obtaining high yields of Mφ’s from hematopoietic progenitors derived from EB differentiation of hESC. Cells derived from EB’s display multiple features characteristic of Mφ’s including expression of definitive markers of Mφ’s such as CD14, CD68 and LOX1 (lectin-like Ox-LDL receptor 1) [19, 20]. We also find that these EB derived cells are competent to phagocytose opsonized and non-opsonized bacterial proteins and show normal responses to Toll-like receptor (TLR) ligands. Further, this process is not inhibited by genetic modification of the hESC cells themselves. Compared to the murine stromal co-culture system, the EB system produces higher percentages of CD34+ hematopoietic progenitors that in turn yield higher numbers of functional Mφ’s.
Materials and Methods
Growth and propagation of human embryonic stem cells
The H1 line (NIH code WA01) was obtained from WiCell Wisconsin. hESC colonies were propagated on irradiated mouse embryonic fibroblast feeders in DMEM F12 (Invitrogen, Carlsbad, CA) containing 20% Serum Replacer (Invitrogen), 2mM L-glutamine, 100μM non-essential amino acids and 10ng/ml basic fibroblast growth factor (Invitrogen). The cells were passaged on a weekly basis with Collagenase IV (Invitrogen). The UCLA IRB and ESCRO committees approved all work with hESC.
Formation and Differentiation of Embryoid Bodies
Two days prior to passage, undifferentiated H1 cells were treated with 0.5mg/ml Dispase (Invitrogen) for 20 minutes at 37°C. The dissociated colonies were lifted off the plates by gentle pipetting, washed in DMEM F12 medium and plated overnight in ultra low attachment plates (Corning Incorporated, Corning, NY) in Iscove’s Modified Dulbecco Medium (IMDM) (Invitrogen) supplemented with 15% non-heat-inactivated defined serum (HyClone, Logan, UT), 1mM L-glutamine, 1% non-essential amino acids and 0.1 mM β2-mercaptoethanol. Medium was changed every alternate day. The medium was supplemented with different combinations of the following cytokines for the subsequent media changes starting at day 4. The cytokines used were SCF at 50 ng/ml (Amgen, CA), IL-3 at 20ng/ml (Invitrogen), IL-6 at 10ng/ml (Peprotech, RockyHill, NJ), M-CSF at 20ng/ml (Peprotech), BMP-4 at 10ng/ml (R andD Biosystems, Minneapolis, MN), FLT-3L at 10ng/ml (R andD Biosystems), G-CSF at 20ng/ml (Peprotech) and GM-CSF at 20ng/ml (Peprotech). BMP-4 was removed from the medium at day 12. The EB’s were harvested on the day of experiments, washed 2X in IMDM and digested with 0.25% Trypsin –EDTA (Invitrogen) supplemented with 2% chick serum (Sigma, St. Louis, MO) for 30–45 minutes at 37°C. The resulting cell suspension was washed twice and filtered through a 40μ filter to obtain a single cell suspension.
Differentiation of Human Embryonic Stem Cells on OP9
H1 cells were differentiated on OP9 as described [21], [22]. Briefly, gelatinized 6 well plates were seeded with 2 × 104 OP9 cells /well and cultured for 5 days with medium change on day 4. Undifferentiated H1 cells were harvested with Collagenase IV (1mg/ml). The resulting clumps were plated onto OP9 cells and maintained for 16 days in α-Modified Minimal Essential Medium supplemented with 10% defined serum (Invitrogen) and 100μM monothioglycerol (Sigma) with half medium changes every alternate day. At the termination of the culture, single cell suspensions were obtained by treatment with collagenase IV (Invitrogen), for 20 minutes at 37° C followed by treatment with Trypsin-EDTA (0.05%) for 15 minutes at 37° C. Cell clumps were disrupted by further pipetting, washed twice and filtered through a 40μ filter to obtain a single cell suspension.
Lentiviral Transduction of hESC
The virus was produced as described earlier by co-transfection of three plasmids: (i) pSIN18.cPPT.hEF1α.EGFP.WPRE, (ii) the vesicular stomatitis virus G (VSVG) expression plasmid pHCMVG and (iii) the packaging plasmid pCMVΔR8.2DVPR in 293T cells as previously described [22]. Undifferentiated H1 colonies were mechanically disrupted and mixed with virus, incubated for 2 h at 37°C, washed and plated on fresh mouse fibroblast feeders. Newly grown colonies were analyzed under UV microscopy and GFP-positive regions were selectively excised and passaged, eventually enriching for GFP expression to near complete homogeneity.
Cell Sorting and Flow Cytometry
hESC differentiated as either OP9 co-cultures or as EB’s were stained with monoclonal antibodies to CD34, CD45, CD14, HLADR, CD11c, CD4, CD68, CD86, (Beckman Coulter, Fullerton, CA) conjugated to FITC (fluorescein isothiocyanate), PE (phycoerythrin), APC (allophcocyanin), ECD (electron-coupled dye) or PE-Cy7. LOX-1 antibody was purchased from Hycult Biotechnology, Canton, MA and CD209 from BD Biosciences, Bedford, MA. Cells were analyzed for fluorochrome expression with a Coulter FC500 flow cytometer and FloJo software (Tree Star Inc, Ashland, OR). At indicated times post differentiation, CD34+ cells were sorted by magnetic sorting. Cells were labeled with anti-CD34 microbeads (Miltenyi Biotech, Auburn, CA) and the positive fraction was obtained on an AutoMACS cell sorter (Miltenyi). Greater than 85% enrichment was obtained by this process. CD14 positive cells were sorted using CD14- PE (Beckman Coulter) monoclonal antibody followed by staining with anti-PE microbeads (Miltenyi Biotech). The cells were sorted using the AutoMACS.
Macrophage Differentiation
Sorted CD34+ cells were plated at 300,000/well in 6 well plates and expanded in IMDM + 20% heat-inactivated fetal bovine serum (FBS) supplemented with IL-3 at 20ng/ml, M-CSF at 20ng/ml and SCF at 50ng/ml. Half medium change was performed at day 4 post plating and the floating cells were transferred to new wells.
Following additional 4–5 days of culture, the adherent cells were collected using PBS + 2mM EDTA and washed. The cells were counted and replated in new 6 well plates at 500,000 cells/well in IMDM + 20% FBS with 20ng/ml M-CSF for additional 5–6 days. Mφ’s from human peripheral blood were obtained by the method of adherence. Peripheral blood mononuclear cells were obtained from the Virology Core, UCLA.
Hematopoietic Colony Forming Assays
H1 cells differentiated by co-culture on OP9 or as EB’s were dissociated to single cell suspensions. H1 cells co-cultured with OP9 were plated in complete methylcellulose medium, Methocult GF+ H4435 (Stem Cell Technologies, Vancouver, BC, Canada) at a concentration of 105 cells/ml. Cells cultured as EB’s were plated at a concentration of 25000/ml. All assays were performed in quadruplicates. Colonies were counted at 2 weeks post-plating. P values were determined using the Student’s t-test. Cytospin preparation from colonies were made using a cytospin centrifuge (Shandon, Thermo Electron Corporation, Pittsburgh, PA) and stained with Wright-Giemsa stain (Sigma-Aldrich). Images were captured using an Olympus IX51 inverted microscope.
Immunoblotting
H1 derived macrophages at 1×10e6/ml were stimulated with 100ng/ml of LPS for 6 hours. Cells were lysed in lysis buffer (1% Triton-X-100, 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1.0 mM EDTA, 1.0 mM inhibitor mixture). Na3VO4, and a protease Protein samples were separated on 10% SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with antibodies against anti-phospho-STAT-1 (BD Pharmingen) antibody to determine the activation of STAT-1. The blots were also re-probed with antibodies against β-actin protein as loading control.
PCR
H1 derived macrophages were stimulated with 100ng/ml of LPS for 6 hours. Total RNA was isolated with Trizol reagent (Invitrogen) and was reverse-transcribed with SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. PCR was performed for TNFα and GAPDH. The primers used were as follows. TNF α: 5′CAGAGGGAAGAGTCCCCCAGGGACC 3′ and 3′CCTTGGTCTGGTAGGAGACGGCGATG 5′. GAPDH primers were: 5′CTGGGCTACAC TGAGCACCAG 3′ and 3′CCAGCGTCAAAGGT GGAG 5′.
Phagocytosis Assays
Phagocytosis assays with E. coli expressing green fluorescence protein (GFP) was performed as previously described [23]. Briefly, the green fluorescence protein (GFP) expressing E. coli (a kind gift from Dr. Genhong Cheng, UCLA) was grown from a single colony in Luria- Bertani broth containing 100ng/ml ampicillin. A subculture was started to bring the bacteria to the logarithmic phase of growth. The bacteria were washed twice in phosphate buffered saline (PBS) and the optical density was measured using a spectrophotometer. hESC derived Mφ’s and peripheral blood derived Mφ’s were stimulated with and without LPS for 24hrs. The cells were mixed with bacteria at an MOI of 50 and 100 respectively. Optimal time for phagocytosis was determined to be approximately 90 minutes. The cells were then washed in cold PBS and pictures were taken using an Olympus IX51 inverted camera at 200× magnification. The cells were then collected from the plates and fixed in 1% paraformaldehyde and analysed on a Coulter FC500 flow cytometer. Polyinosinic acid (Sigma) was used at a concentration of 0.5mg/ml. For phagoctyosis assays to determine Fcγ receptor specificity, fluorescently labeled staphylococcus aureus bioparticles were purchased from Molecular Probes along with the opsonizing reagent. The protocol provided by the company was followed to determine phagocytosis. Human AB serum (Sigma) was used as a blocking agent.
TNFα ELISA
H1 and PBMC derived Mφ’s were stimulated for 6hrs with LPS and lysed. The lysates were tested for TNFα expression following instructions provided by the manufacturer (BD Biosciences, San Jose, CA).
Results
Human Embryonic Stem Cell Derived Embryoid Bodies Give Rise to CD14 Expressing Cells in the Presence of Cytokines
To determine if we could differentiate Mφ’s from hESC derived EB’s, we began by generating hematopoietic progenitors from EB cultures [8, 13, 24] and determining the expression of CD14 on the differentiated progenitors, as expression of CD14 is a defining characteristic of monocytes/Mφ’s. In order to generate EB’s, intact hESC colonies are detached from supporting stroma by enzymatic treatment and transferred to low attachment, feeder-free plates where they form 3-dimensional aggregates in suspension. For comparison, we co-cultured H1 cells on OP9 stromal feeder layers, a method that is known to result in myeloid differentiation [21]. The hESC line was also cultured as EB’s in the presence of Interleukin 3 (IL-3), Interleukin 6 (IL-6), Stem Cell Factor (SCF), Flt-3 ligand (FLT-3L) and bone morphogenetic protein 4 (BMP-4) [8, 25] (BASAL medium, Table 1). In order to compare the efficiency of the two methods in obtaining CD34 expressing hematopoietic progenitors, we first dissociated the cultures after 14–15 days on OP9 or as EB cultures and plated the cells in methylcellulose supplemented with various growth factors to determine multi-lineage hematopoietic colony forming ability. The 2 week culture time point was deemed to be optimal for both methods based on hematopoietic progenitor formation assayed over 21 days of culture (data not shown). As seen in Figure 1A, the H1 cells cultured as EB’s had significantly higher potential to form hematopoietic colonies as compared to cells cultured on OP9. Further, the EB cultures yielded almost 2 fold higher numbers of myeloid colonies as compared to the progenitors derived from the OP9 co-cultures. Interestingly, the erythroid potential of cells cultured on OP9 as well as in EB cultures was relatively low with the EB culture having the lesser potential. To evaluate the hematopoietic progenitors obtained, we analyzed expression of CD34 and CD45 on cells from both systems. As seen in Figure 1B, at day 15, the EB culture system yields higher numbers of CD34+ cells.
Table I.
Composition of three different cytokine cocktails tested on embryoid body cultures
| Basal | Basal +G/GM | Basal+G/M |
|---|---|---|
|
| ||
| IL-3–20 ng/ml | IL-3–20 ng/ml | IL-3–20 ng/ml |
| IL-6–10 ng/ml | lL-6–10ng/ml | IL-6–10 ng/ml |
| SCF-50 ng/ml | SCF-50 ng/ml | SCF-50 ng/ml |
| FIt-3–10 ng/ml | FIt-3–10 ng/ml | FIt-3–10 ng/ml |
| BMP4–10 ng/ml | BMP4–10 ng/ml | BMP4–10 ng/ml |
| G-CSF-20ng/ml | G-CSF-20ng/ml | |
| GM-CSF-20ng/ml | M-CSF-20ng/ml | |
Figure 1.
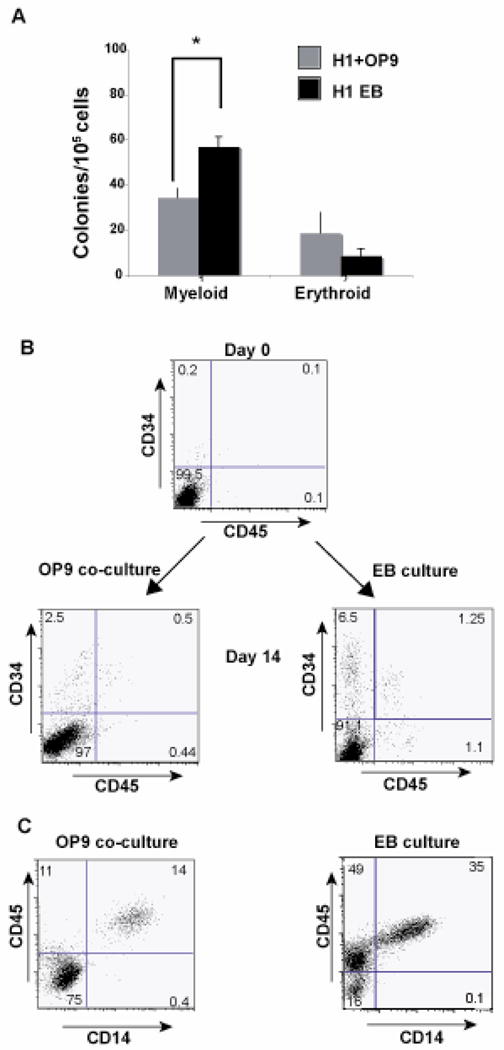
Embryoid body cultures yield more myeloid colonies as compared to OP9 cocultures : A) H1 cells co-cultured with OP9 feeders or as EB’s (in BASAL medium as shown in Table1) were dissociated into single cells following 14 days of culture. The single cells were plated in complete methylcellulose medium and analyzed for colony formation 2 weeks post plating. The figure is representative of the average of 3 independent experiments with OP9 feeders and 3 independent experiments as embryoid body cultures. Values of colonies that are significantly different between OP9 cultures and EB cultures are indicated by asterisks (*, P <0.0015). Probability values were calculated using Students t- test. (B) H1 cells co-cultured on OP9 feeders or as EB’s (in BASAL medium, Table1) were analyzed for expression of CD34 and CD14 at day 0 of culture. Following 14 days of culture in both conditions, cells were dissociated and analyzed for CD34 and CD45 expression by flow cytometry. (C) The CD34+ cells were sorted and differentiated further in presence of cytokines IL3, SCF and M-CSF and assayed for expression of CD14 and CD45. The figure is representative of 3 independent experiments.
In addition, numbers of CD34+ cells expressing CD45 are higher in the EB culture system when compared to the OP9 system that has previously been shown to have lower levels of CD45 expression [8], [21, 22]. This observation coincides with earlier studies suggesting that CD34+ cells are precursors of CD45+ cells [24]. Subsequently, CD34+ progenitors were isolated from both systems and cultured in the presence of IL-3, SCF and M-CSF for 8–9 days followed by further differentiation in the presence of M-CSF for an additional 6–7 days. We then determined the level of CD14 expression on these cells. As shown in Figure 1C, the numbers of CD14 expressing cells were dramatically higher from the EB derived CD34+ hematopoietic progenitors as compared to the OP9 co-cultures. From equivalent numbers of CD34+ cells (1 × 106) from either culture system, the EB derived CD34+ cells consistently yield 400,000–450,000 CD14 expressing cells when subjected to our cytokine differentiation approach, while the OP9 co-cultures yield 125,000–150,000 CD14 expressing cells (an approximate 4 fold increase in efficiency) (Figure 1B). Thus the EB system promotes the development of cells expressing the monocyte/Mφ marker CD14 in significantly higher numbers compared to traditional co-culture on murine stromal lines.
Optimization of Embryoid Body Culture Conditions for Hematopoietic Progenitor Differentiation
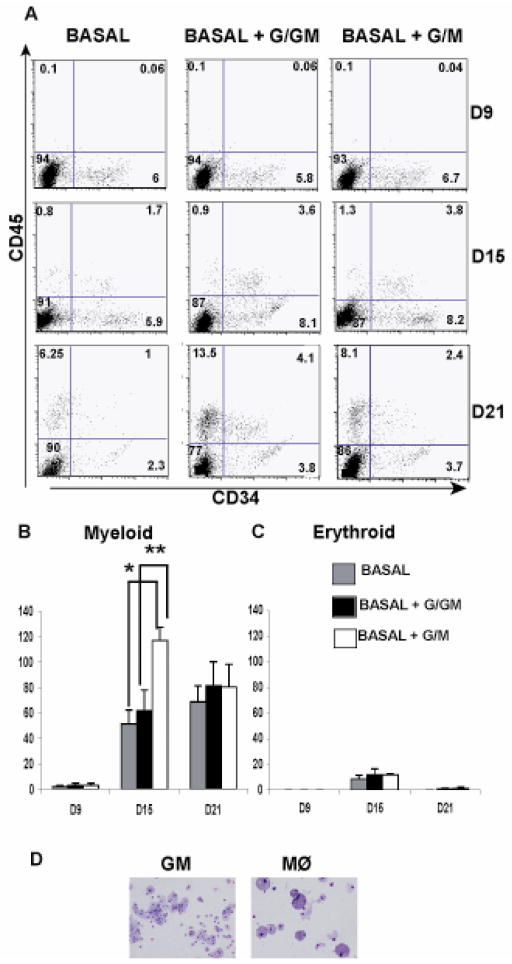
Based on our initial observation that H1 EB cultures result in higher yields of CD14 expressing cells, we next sought to optimize our EB culture conditions to obtain higher numbers of hematopoietic progenitors with maximum myeloid potential. We compared three different combinations of cytokines as shown in Table 1. BASAL medium served as the primary cytokine cocktail and contained BMP-4, IL-3, IL-6, SCF and FLT-3L [8]. Addition of G-CSF (Granulocyte colony- stimulating factor) and GM-CSF (Granulocyte macrophage colony-stimulating factor) to the BASAL medium yielded the second cocktail (BASAL +G/GM) while the addition of G-CSF and M-CSF (macrophage colony-stimulating factor yielded the third (BASAL +G/M). BMP-4, a strong ventral mesoderm inducer, is essential for hematopoietic progenitor differentiation [8]. SCF and FLT-3L, the ligand for the receptor tyrosine kinase Flk-2, are known to play a key role in the survival and development of hematopoietic progenitors isolated from bone marrow and cord blood [25, 26]. IL-6 is involved in myeloid cell differentiation [27, 28]. IL-3 determines the progression from pluripotent stem cell state to a definitive myeloid-restricted progenitor[29]. GM-CSF and M-CSF are intermediate acting growth factors and give rise to monocyte/macrophage progeny [29]. To compare the efficiencies of these three combinations, H1 cells were cultured as EB’s with the three different cocktails and the cells were analyzed from 9–21 days for hematopoietic marker expression by flow cytometry. We also compared their ability to form colonies in methylcellulose. As observed in Figure 2A the expression of CD34 was at its peak at day 15 and dropped at day 21 in all three culture conditions. Further the expression of CD45 increased over time in all three conditions.
Figure 2.
BASAL+G/M medium yields the highest numbers of myeloid colonies in culture: A) H1 cells were cultured as EB’s in 3 different media conditions as described in Table 1. The cultures were dissociated at various time points (Day9, Day15 and Day 21) and analyzed for cell surface expression of CD34 and CD45 by flow cytometry. The figure is representative of 3 independent experiments. (B, C) The H1 EB’s were dissociated and plated in methylcellulose medium at various time points to determine hematopoietic colony potential. Both myeloid and erythroid colonies were scored at 2 weeks post plating. The data is representative of 3 independent experiments. Asterisks represent values that are significantly different between the different time points of culture (*, P=0.002, **, P<0.001). P values were determined using the Students’s t-test. (D) The colonies were stained with Wright-Giemsa Stain to determine cellular morphology. The figure depicts a Mφ and a Granulocyte-Macrophage (GM) colony. The magnification is 200× for the images captured with an Olympus IX51 inverted microscope.
The yields of CD34+ cells were higher at both day 15 and day 21 in the BASAL + G/GM and BASAL + G/M cultures compared to the BASAL medium cultures. Expression of CD45 also showed the same trend. Between the BASAL +G/GM and BASAL +G/M cultures, the number of cells expressing CD34 was slightly higher in the BASAL+G/GM at both day 15 and 21. We then compared the myeloid and erythroid colony forming potential among all the three culture conditions at the above mentioned time points. Although the yields of CD34 expressing cells as determined by flow cytometry appear similar at day 15 of culture between the BASAL + G/GM and BASAL + G/M culture, the colony forming potential differs significantly between these 2 conditions with the latter producing approximately 2.5 fold more myeloid colonies (Figure 2B). At day 21 of culture the total colony forming potential did not significantly differ between the 3 culture conditions. In addition, in all the three culture conditions, the myeloid colonies outnumber the erythroid colonies (Figure 2B, C). The cellular morphology of the colonies obtained in culture was determined by Wright-Giemsa stains (Figure 2D). Based on our observations that the myeloid colony potential is significantly different and peaks at day 15 in BASAL + G/M as compared to the other 2 conditions, we conclude that 15 days of culture in the BASAL +G/M medium is optimal for hematopoietic progenitor derivation.
CD14 Expressing Cells Differentiated from H1 Embryoid Bodies Express Normal Phenotypic Markers
To determine whether cells differentiated from EB’s exhibit phenotypic markers characteristic of differentiated Mφ’s, we compared these cells to mature Mφ’s derived from PBMC cultured for the same duration of time. CD34+ progenitors derived from EB’s cultured in BASAL + G/M medium as previously described, were isolated and expanded in culture in the presence of IL-3, SCF and M-CSF for a week followed by further differentiation for 5–6 days in the presence of M-CSF alone. Mφ’s were differentiated from PBMC by adherence to tissue culture dishes. We first analyzed the cells for expression of the hematopoietic markers CD34 and CD45. We then determined expression of functional markers of mature Mφ’s including CD14, CD11c, HLADR, CD68, CD4 and CD209 (Figure 3). CD14 is the high affinity receptor for lipopolysaccharide (LPS) on Mφ’s. CD45 is the pan leukocyte marker on human cells. CD11c, an integrin family member [30], is involved in the phagocytosis of various pathogens as well as in the binding of cell adhesion molecules [31, 32], bacterial LPS [33] and complement protein [34]. HLADR is a class II MHC molecule expressed on antigen presenting cells. CD4 on T cells is the co-receptor in MHC class II induced antigen activation, however its function on human Mφ’s is not well understood. CD209, also known as DC-SIGN, is a C-type lectin that binds Human Immunodeficiency Virus [35] and other mannose-type carbohydrate expressing pathogens and is expressed selectively on subpopulations of Mφ’s in the tonsil, dermis of skin and in lymph nodes [36]. CD68 [37], an intracellular class D scavenger receptor (SR) is involved in low-density lipoprotein uptake [19] and in pathogen interactions. Some of the SR’s are also known to act as pattern recognition receptors [20] and hence the expression of this molecule is key in determining aspects of Mφ function involved in pathogen recognition and phagocytosis.
Figure 3.
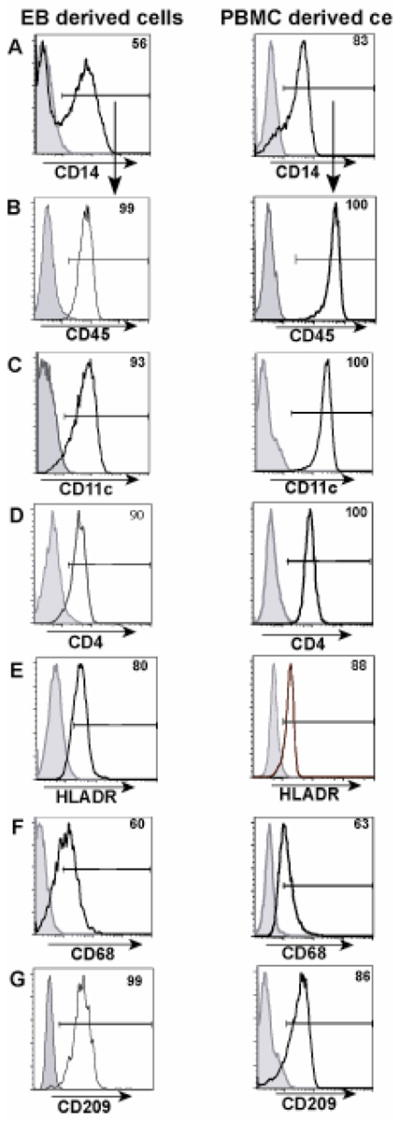
H1 derived macrophage cells exhibit similar phenotypic expression compared to PBMC derived macrophages. H1 cells were cultured as EB’s in BASAL+G/M medium and CD34 cells were sorted at 15 days of culture. The sorted cells were differentiated in medium containing IL3, SCF and M-CSF to obtain Mφ’s. Panel A represents CD14 expression on EB - derived and PBMC - derived cells. Subsequent panels B, C, D, E, F and G are all gated on the CD14 positive population of panel A.
CD34 expression was detectable in very low amounts (<1.5%) in the EB-derived cells (data not shown) suggesting that the progenitors were losing pluripotency and becoming committed to specific lineages. The majority of cells in both EB –derived as well as PBMC – derived cultures expressed CD45. The expression levels were very high on the EB-derived cells compared to the initial expression on CD34+ progenitors (Figure 1B) suggesting that the CD34+ progenitors are able to differentiate successfully in the presence of the utilized cytokines. The expression levels of Mφ lineage markers CD14, CD11c, HLADR and CD4, CD209 and CD68 were all similar in EB derived cells and in PBMC derived Mφ’s. In addition, the EB - derived cells showed increased adherence to tissue culture plates, fibroblastoid appearance and process extension (data not shown), which are morphological characteristics of mature Mφ’s. Hence, the EB cultures give rise to hESC derived Mφ’s that are morphologically and phenotypically similar to adult peripheral blood derived Mφ’s.
Human Embryonic Stem Cell Derived Macrophages Demonstrate Functional Toll-Like Receptor Responses
A hallmark of Mφ function is their ability to recognize microbial components and initiate immune responses. Ability of Mφ’s to interact with nonopsonized bacteria is based on recognition of conserved microbial pathogen associated molecular patterns by pattern recognition receptors such as TLR’s [38]. Recognition of LPS by TLR-4 [39], one of the TLR family members, induces the activation of both nuclear factor-kappa B (NF-kB) and interferon (IFN) pathways leading to upregulation of co-stimulatory molecules such as CD86 and CD80 [40] on the cell surface and secretion of inflammatory cytokines [41]. To examine the response of EB derived Mφ’s to TLR ligands we treated the cells with LPS and examined the levels of CD86 and TNFα expression. LPS increased both number of cells expressing CD86 and the mean fluorescence intensity (MFI) in EB derived cells (Figure 4A). PCR analysis also showed LPS-induced increase in TNFα; expression in both EB and PBMC derived Mφ’s (Figure 4B). This was also confirmed by ELISA measurements (Figure 4D). Activation of the type I IFN induction pathway by LPS leads to production of IFN that binds to the type I IFN receptor and activates the STAT1 pathway. We hence examined phosphorylation of STAT1 in the EB derived cells. As shown in Figure 4C, treatment of cells with LPS led to increased STAT1 phosphorylation in both EB and PBMC derived Mφ’s. These results suggest that EB derived cells have functional TLR pathways similar to adult peripheral blood - derived Mφ’s.
Figure 4.
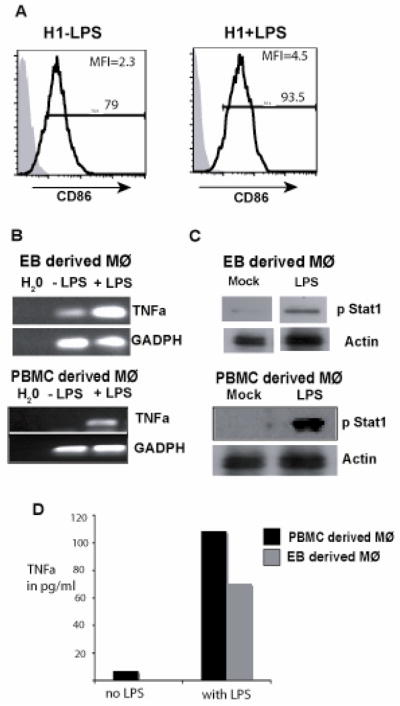
H1 derived macrophages show TLR specific responses similar to PBMC derived macrophages. (A) In order to determine TLR function, H1 EB - derived Mφ’s were stimulated with LPS for 18 hours. Following stimulation the cells were analyzed by flow cytometry for CD86 upregulation. Shaded histograms represent isotype controls. (B) Following LPS stimulation, the cells were assayed for TNFα expression by RT-PCR analysis. (C) Phosphorylation of STAT-1 was quantitated following stimulation in presence of LPS in both EB and PBMC - derived Mφ’s. Data are representative of 2 independent experiments. (D) EB and PBMC derived cells were subjected to TNFα ELISA following LPS stimulation for 6 hrs. The experiment was performed in duplicates.
Embryoid Body Derived Macrophages Efficiently Respond to Toll-Like Receptor Stimulation to Specifically Enhance Phagocytosis of Bacteria
To determine whether EB derived Mφ’s are functionally competent for phagocytic activity, we assayed their ability to ingest unopsonized Gram-negative E. coli expressing GFP. To confirm phagocytosis, cells were treated with polyinosinic acid (PI), a nonspecific polyanion inhibitor of bacterial phagocytosis by SR’s. The ingestion of bacteria was very minimal in the absence of LPS treatment. Following activation by LPS, we observed increased uptake of the bacteria in both EB derived cells and PBMC derived Mφ’s (Figure 5A, B).
Figure 5.
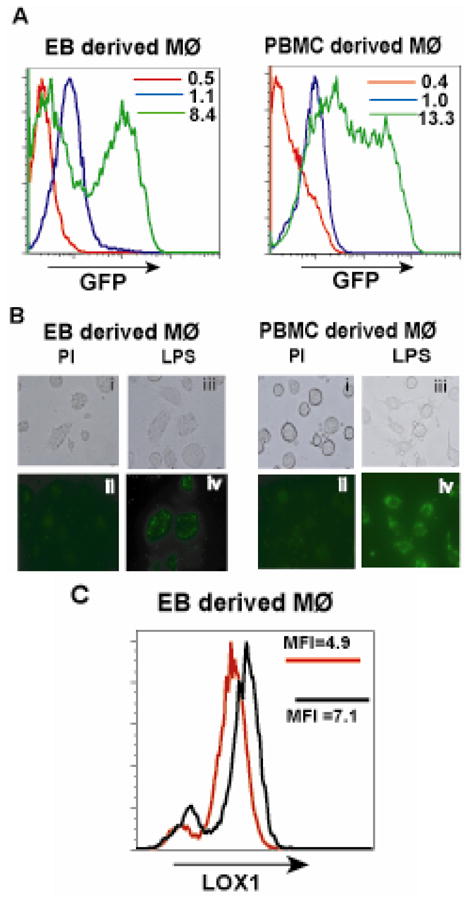
TLR ligands specifically increase phagocytosis of bacteria in EB derived macrophages similar to PBMC derived macrophages. (A) EB - derived Mφ’s and PBMC - derived Mφ’s were stimulated in the presence of LPS (green histograms) and in the absence of LPS (blue histograms) with media alone for 18hours. Following stimulation, the cells were challenged with E. coli expressing EGFP for 1.5 hrs and phagocytosis assayed by flow cytometry. Polyinosinic acid at a concentration of 0.5mg/ml was used as a negative control to inhibit phagocytic activity (red histograms). The MFI values are denoted in the histogram panels. (B), (C) The cells challenged with bacteria were analyzed by fluorescence microscopy. Panels (B) (i, ii) represent EB - derived Mφ’s treated with PI and (C) (i, ii) show PBMC - derived Mφ’s treated with PI. EB - derived Mφ’s treated with LPS are shown in panels (B) (iii, iv) while PBMC - derived Mφ’s are in panels (C) (iii, iv). Upper panels represent phase contrast images while lower panels represent fluorescent images. All images are at 200X magnification. (D) Upregulation of expression of LOX1 was analyzed by flow cytometry following treatment of cells with and without LPS. Data are representative of 2 independent experiments. Red histograms represent absence of LPS stimulation while black histograms represent cells treated with LPS. MFI values are shown within the histogram panels.
PI inhibited the uptake of E. coli significantly. Decreased phagocytosis in the presence of PI suggests that the uptake of the unopsonized bacteria is also dependent on the activity of SR’s many of which also serve as pattern recognition receptors.[20, 42] There are 8 independent structural classes of SR’s.[42] We analyzed the expression of LOX1, the class E SR, in EB derived cells in response to LPS treatment. LOX 1 is expressed in EB derived cells in the absence of LPS and expression increases on LPS treatment as observed by increased number of cells as well as higher mean fluorescence intensities (MFI) values (Figure 5C). The interaction of microbial components with Mφ’s can be either by direct binding to surface receptors like SR’s or mediated by opsonins that coat the particles and effect Fc gamma (Fcγ) receptor binding. To determine if the EB - derived cells were also functional in the context of Fcγ receptor specific phagocytosis we tested the cells with opsonized bioparticles derived from S. aureus, a gram-positive bacterium. Phagocytosis of the bioparticles was similar in both EB and PBMC derived Mφ’s (Figure 6C, D). Blocking Fcγ receptors by the addition of human serum inhibited the phagocytosis (Figure 6A, B) confirming that this functional response was indeed an Fcγ receptor specific process. We hence conclude that EB derived cells exhibit phagocytic activity similar to that displayed by Mφ’s differentiated from adult peripheral blood.
Figure 6.
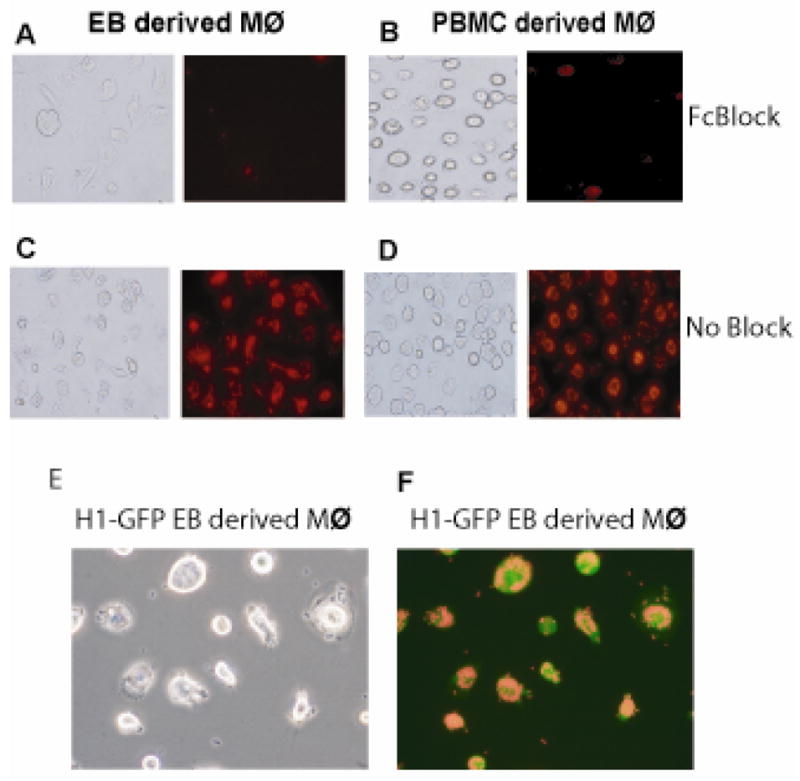
EB derived macrophages exhibit efficient Fcγ receptor specific phagocytic activity. EB - derived macrophages (A, C) and PBMC - derived macrophages (B, D) were treated with (A, B) or without (C, D) an Fcγ receptor blocking agent for 2 hours. Opsonized zymosan bioparticles were added to the cells, incubated for 1 hour and phagocytic activity determined by fluorescence microscopy using an Olympus fluorescent microscope. Images are at 200× magnification. The data is representative of 4 independent experiments. H1 cells transduced with EGFP differentiate into macrophages with normal phagocytic activity. H1 cells were transduced with a lentiviral vector expressing EGFP. The transduced cells were differentiated into macrophages and the functionality of the transduced cells was determined by assaying their ability to phagocytose opsonized zymosan bioparticles. The figure depicts the phase contrast (E) and fluorescent (F) images at 200× magnification.
Genetically Altered Embryoid Body Cultures Differentiate into Cells That Demonstrate Normal Phagocytic Responses
Use of the EB derived hematopoietic progenitors in therapeutic strategies or use of EB-derived Mφ’s to study gene function may require the introduction and maintenance of foreign genes in the cells. To determine if the introduction of a foreign gene perturbed the Mφ differentiation program, we genetically marked the hESC line H1 with a lentiviral vector which permits constitutive expression of the EGFP reporter gene under control of the EF1α promoter.[22] The H1-GFP transduced cells were differentiated as EB’s using BASAL +G/M medium as previously described. Mφ’s were differentiated from the CD34+ progenitors. We assessed expression of certain key phenotypic markers characteristic of Mφ’s such as CD14, CD11c, and CD45. The differentiated cells showed similar expression of all the markers analyzed (data not shown). In order to determine if the cells had functional phagocytic responses, they were tested with opsonized bioparticles as earlier described. We observed that the H1-GFP EB - derived cells were able to phagocytose the bioparticles (Figure 6E, F) and behaved similarly to the non-transduced cells (Figure 6C, D). Hence, the EB derived cells were able to maintain expression of the introduced foreign gene and perform characteristic functions of Mφ’s.
Discussion
Our studies have identified and characterized a highly efficient method for the differentiation of functional Mφ’s from hESC via differentiation to EB’s. The Mφ’s are derived from CD34+ hematopoietic progenitors that arise from EB cultures in the presence of various cytokines. The cytokines used greatly enrich the cell population for progenitors that yield primarily myeloid colonies in clonogenic progenitor assays. Utilizing this culture method, we have successfully circumvented the need for co-culture with murine feeders such as S17 and OP9. We also observed differences in hematopoietic marker expression between the two culture systems. It has been reported that hematopoietic progenitors in both early murine and human embryos lack CD45 expression [43, 44]. CD45 expression appears in more mature progenitors [8]. Interestingly, we observe that the CD34+CD45+ progenitors arising from the EB system are higher in numbers compared to CD34+CD45+ cells arising from OP9 cultures, suggesting that the EB culture system yields more mature CD34+ cells which may account for their higher efficiency in differentiation. As the CD34+ cells differentiate further in the presence of the myeloid specific cytokine M-CSF they acquire phenotypic markers characteristic of mature Mφ’s similar to adult PBMC - derived Mφ’s. Published work has shown that H1 derived Mφ’s obtained via co-culture on S17 cells are phenotypically similar to fetal liver - derived Mφ’s based on the expression of the surface markers CD14, CD4, CCR5, HLADR and CXCR4 [17]. This previous study did not examine the levels of CD45 expression in the Mφ’s obtained nor in the CD34+ progenitors differentiating from the S17 co-cultures. Hence further work involving gene expression profiling will be needed to determine if hESC derived Mφ’s are closer to fetal liver derived Mφ’s or adult Mφ’s before a conclusive decision can be made.
The main function of Mφ’s in immune responses is the detection of pathogens leading to receptor-mediated endocytosis, digestion and eventual presentation of foreign antigens in the context of MHC. A wide variety of receptors on Mφ’s are responsible for recognizing Gram-positive and Gram-negative bacteria via conserved pathogen-associated molecular patterns [15]. These receptors include SR’s, TLR’s and C-type lectins [15, 20]. The diverse receptors on the Mφ cell surface collaborate with each other to facilitate phagocytosis and/or signaling thereby providing the trigger for the adaptive immune response mechanisms. Our data show that EB derived Mφ’s respond to the TLR ligand LPS both by increasing expression of the surface proteins CD86 and LOX1 and showing increased phagocytic activity when exposed to GFP transduced E. coli as well as opsonized bioparticles. Functional responses were in addition characterized by the increase in expression levels of TNFα and the phosphorylation of STAT-1, molecules downstream of TLR signaling. These results suggest that the TLR pathways are functional in these cells. In addition phenotypically the EB derived Mφ cultures showed expression of characteristic markers of mature Mφ’s. In all these functional aspects the EB derived Mφ’s were very similar in behavior to PBMC derived Mφ’s. Hence, together these studies show that EB’s can be used to obtain large numbers of functional Mφ’s with normal physiological responses.
Our data are unique in being able to show the ability to obtain functional Mφ’s from hematopoietic progenitors obtained from a stroma-free culture system. Further, the yields of our CD14+ cells are 4–5 fold higher from the EB culture system than from stromal cell co-cultures. In order to utilize hematopoietic progenitors in gene therapy strategies targeting the myeloid lineage, genetic manipulation of hESC may be necessary. Recent published work [18] has demonstrated the differentiation of Mφ’s from hESC derived EB cultures. In this system monocytes that can be differentiated into Mφ’s, are generated directly from the EB’s following cytokine culture. However, neither the EB’s nor the differentiated Mφ’s would be suitable for direct transplant into patients. As our Mφ’s are generated via hematopoietic progenitors, there is a potential for the progenitors to be used directly in clinical studies. Our results show that our approach is feasible as the differentiating EB cells retain expression of the transgene, as do the Mφ’s obtained from them. The transduced Mφ’s also show normal phenotype and functional responses. With further optimization utilizing inducible promoters this system could serve as a steady source of primary human Mφ’s for studying various aspects of human Mφ biology and relevant diseases involving Mφ’s such as tuberculosis and HIV.
Further, additional work is required to tease out the molecular signaling pathways directing efficient hematopoietic differentiation. In order to utilize these cells for transplantation purposes, the problems of immune rejection need to be overcome. The EB system of differentiation is devoid of animal cells and, with additional improvements, can be utilized to produce completely animal product-free cells. Major advances have been made recently in optimization of xeno-free conditions for hESC derivation and culturing [45]. The hESC can also be genetically modified, cultured for long periods of time and expanded to numbers suitable for clinical applications. Mφ’s are being utilized in cancer immunotherapy studies [46] and in the treatment of patients with lymphoma [47]. We can thus envision the use of EB derivatives in the future to surmount various genetic or metabolic disorders.
Acknowledgments
The authors would like to acknowledge Dr. Dimitrios Vatakis for help with preparation of the manuscript. This work was supported by the California Institute for Regenerative Medicine (CIRM) Training Grant T100005 awarded to AS, CIRM grant RCI-00149 awarded to JZ, NIH grants R01 AI036554 and P01 GM081621awarded to JZ, the Pendleton Foundation and the UCLA CFAR. BG is supported by a Special Fellowship from the Lymphoma and Leukemia Society. GC is supported in part by National Institutes of Health research grants R01 AI056154, R01 AI069120 and R37 AI47868.
Abbreviations
- hESC
Human Embryonic Stem Cells
- Mφ
Macrophage
- PBMC
Peripheral blood mononuclear cells
- EB
Embryoid bodies
- TLR8
Toll-like receptor 8
- MFI
Mean Fluorescence Intensity
- LPS
Lipopolysaccharide
- SR
Scavenger Receptor
Footnotes
Authorship
AS performed all research with human embryonic stem cell cultures, differentiation of macrophages and functional assessment, analyzed data and wrote the paper, B.G. carried out RNA and protein assays, MDM performed flow cytometry on macrophage cultures, ZG assisted with human embryonic stem cell cultures, SK assisted with flow cytometry on human embryonic stem cell cultures, AK provided technical help with human embryonic stem cell cultures, HJB assisted in writing and editing the manuscript, GC designed functional experiments with macrophages and JZ designed the research and assisted in writing and editing the manuscript.
References
- 1.Gordon S, Fraser I, Nath D, Hughes D, Clarke S. Macrophages in tissues and in vitro. Current Opinion in Immunology. 1992;4(1):25–32. doi: 10.1016/0952-7915(92)90119-y. [DOI] [PubMed] [Google Scholar]
- 2.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. Journal of Leukocye Biology. 2004;76(3):509–513. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orfanos Sergij Goerdtand Constantin E. Other Functions, Other Genes: Alternative activation of antigen-presenting cells. Immunity. 1999;10:137–142. doi: 10.1016/s1074-7613(00)80014-x. [DOI] [PubMed] [Google Scholar]
- 4.Iziksona Leonid, Kleinb Robyn S, Charoc Israel F, Weinera Howard L, Luster Andrew D. Resistance to Experimental Autoimmune Encephalomyelitis in Mice Lacking the CC Chemokine Receptor (CCR)2. Journal of Experimental Medicine. 2000;192(7):1075–1080. doi: 10.1084/jem.192.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witztum Christopher, Glass K, Joseph L. Atherosclerosis the Road Ahead. Cell. 2001;104(4):503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 6.Odorico Jon S, Kaufman DS, Thompson James A. Multilineage differentiation from Human Embryonic Stem Cell Lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 7.Kaufman Dan S, Hansondagger Eric T, Lewisdagger Rachel L, AuerbachDagger Robert, Thomsondagger James A. Hematopoietic colony-forming cells derived from human embryonic stem cells. Proceedings of the National Academy of Sciences. 2001;98(19):10716–10721. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadwick Kristin, Wang Lisheng, Li Li, Menendez Pablo, Murdoch Barbara, Rouleau Anne, Bhatia Mickie. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102(3):906–15. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- 9.Zhan X, Dravid G, Ye Z, Hammond H, Shamblott M, Gearhart J, Cheng L. Functional antigen-presenting leucocytes derived from human embryonic stem cells in vitro. Lancet. 2004;364(9429):163–171. doi: 10.1016/S0140-6736(04)16629-4. [DOI] [PubMed] [Google Scholar]
- 10.Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Molecular Medicine. 2000;6(2):88–95. [PMC free article] [PubMed] [Google Scholar]
- 11.Trounson Alan. The Production and Directed Differentiation of Human Embryonic Stem Cells. Endocrine Reviews. 2006;27:208–219. doi: 10.1210/er.2005-0016. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nature Biotechnology. 2005;23(6):699–708. doi: 10.1038/nbt1102. [DOI] [PubMed] [Google Scholar]
- 13.Wanga Lisheng, Menendeza Pablo, Cerdana Chantal, Bhatia Mickie. Hematopoietic development from human embryonic stem cell lines. Experimental Hematology. 2005;33(9):987–996. doi: 10.1016/j.exphem.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Keller Gordon. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes and Development. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 15.Vodyanik Maxim A, Thomson James A, Slukvin Igor I. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108(6):2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson MB, Miranda DE. Signal transduction pathways that contribute to myeloid differentiation. Leukemia. 2007;21:1363–1377. doi: 10.1038/sj.leu.2404690. [DOI] [PubMed] [Google Scholar]
- 17.Anderson JS, Bandi S, Kaufman DS, Akkina R. Derivation of normal macrophages from human embryonic stem (hES) cells for applications in HIV gene therapy. Retrovirology. 2006;3(24) doi: 10.1186/1742-4690-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlsson Karl R, Cowley Sally, Martinez Fernando O, Shaw Michael, Minger Stephen L, James William. Homogeneous monocytes and macrophages from human embryonic stem cells following coculture-free differentiation in M-CSF and IL-3. Experimental Hematology. 2008;36(9):1167–1175. doi: 10.1016/j.exphem.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramprasad MP, Terpstra V, Kondratenko N, Quehenberger O, Steinberg D. Cell surface expression of mouse macrosialin and human CD68 and their role as macrophage receptors for oxidized low density lipoprotein. Proceedings of the National Academy of Sciences. 1995;93:14833–14838. doi: 10.1073/pnas.93.25.14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Current Opinion in Immunology. 2002;14(1):123–128. doi: 10.1016/s0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- 21.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105(2):617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 22.Galic Z, Kitchen SG, Kacena A, Subramanian A, Burke B, Cortado R, Zack JA. T lineage differentiation from human embryonic stem cells. PNAS. 2006;103(31):11742–7. doi: 10.1073/pnas.0604244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle Sean E, O’Connell Ryan M, Miranda Gustavo A, Vaidya Sagar A, Chow Edward K, Liu Philip T, Suzuki Shinobu, Suzuki Nobutaka, Modlin Robert L, Yeh Wen-Chen, Lane Timothy F, Cheng Genhong. Toll-like Receptors Induce a Phagocytic Gene Program through p38. Journal of Experimental Medicine. 2003;199(1):81–90. doi: 10.1084/jem.20031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Li L, Shojaei F, Levac K, Cerdan C, Menendez P, Martin T, Rouleau A, Bhatia M. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immuity. 2004;20:31–41. doi: 10.1016/j.immuni.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immuity. 1995;3(1):147–161. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 26.Wodnar-Filipowicz Aleksandra. Flt3 Ligand: Role in Control of Hematopoietic and Immune Functions of the Bone Marrow. News in Physiological Sciences. 2003;18(6):247–251. doi: 10.1152/nips.01452.2003. [DOI] [PubMed] [Google Scholar]
- 27.Bernhard H, Lohmann M, Batten WY, Metzger J, Lohr HF, Peschel C, zum Buschenfelde KM, Rose-John S. The gp130-stimulating designer cytokine hyper-IL-6 promotes the expansion of human hematopoietic progenitor cells capable to differentiate into functional dendritic cells. Experimental Hematology. 2000;28:365–372. doi: 10.1016/s0301-472x(00)00126-0. [DOI] [PubMed] [Google Scholar]
- 28.Chomarat P, Banchereau J, Davoust J, Palucka AK. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nature Immunology. 2000;1(6):510–514. doi: 10.1038/82763. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa M. Differentiation and proliferation of hematopoietic stem cells. Blood. 1993;81(11):2844–2853. [PubMed] [Google Scholar]
- 30.Sadhu Chanchal, Ting Harold J, Lipsky Brian, Hensley Kelly, Garcia-Martinez Leon F, Simon Scott I, Staunton Donald E. CD11c/CD18: novel ligands and a role in delayed-type hypersensitivity. Jounal of Leukocyte biology. 2007;81:1395–1403. doi: 10.1189/jlb.1106680. [DOI] [PubMed] [Google Scholar]
- 31.Diamond MS, Garcia-Aguilar J, Bickford JK, Corbi AL, Springer TA. The I domain is a major recognition site on the leukocyte integrin Mac-1 (CD11b/CD18) for four distinct adhesion ligands. Journal of Cell Biology. 1993;120:1031–1043. doi: 10.1083/jcb.120.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ihanus E, Uotila LM, Toivanen A, Varis M, Gahmberg CG. Red cell ICAM-4 is a ligand for the monocyte/macrophage integrin CD11c/CD18: characterization of the binding sites on ICAM-4. Blood. 2007;102:802–810. doi: 10.1182/blood-2006-04-014878. [DOI] [PubMed] [Google Scholar]
- 33.Ingalls RR, Golenbock DT. CD11c/CD18, a transmembrane signaling receptor for lipopolysaccharide. Jounal of Experimental Medicine. 1995;181:1473–1479. doi: 10.1084/jem.181.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malhotra V, Hogg N, Sim RB. Ligand binding by the p150,95 antigen of U937 monocytic cells: properties in common with complement receptor type 3 (CR3) European Journal of Immunology. 1986;16:1117–1123. doi: 10.1002/eji.1830160915. [DOI] [PubMed] [Google Scholar]
- 35.Soilleux EJ. DC-SIGN (dendritic cell-specific ICAM-grabbing non-integrin) and DC-SIGN-related (DC-SIGNR): friend or foe? Clinical Science (London) 2003;104(4):437–446. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 36.Granelli-Piperno Angela, Pritsker Alla, Pack Maggi, Shimeliovich Irina, Arrighi Jean-Francois, Park Chae Gyu, Trumpfheller Christine, Piguet Vincent, Moran Thomas M, Steinman Ralph M. Dendritic Cell-Specific Intercellular Adhesion Molecule 3-Grabbing Nonintegrin/CD209 Is Abundant on Macrophages in the Normal Human Lymph Node and Is Not Required for Dendritic Cell Stimulation of the Mixed Leukocyte Reaction. The Journal of Immunology. 2005;175:4265–4273. doi: 10.4049/jimmunol.175.7.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons CL, Holness DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81(6):1607–1613. [PubMed] [Google Scholar]
- 38.Takeda Kiyoshi. Toll-like receptors and their adaptors in innate immunity. Current Medicinal Chemistry. 2005;4:3–11. [Google Scholar]
- 39.Poltoraka Alexander, Smirnovaa Irina, Heb Xiaolong, Liuc Mu-Ya, Van Huffeld Christophe, Birdwella Dale, Alejosa Erica, Silvaa Maria, Du Xin, Thompsona Patricia, Chane Edward KL, Ledesmae Jessica, Roef Bruce, Cliftong Sandra, Vogelh Stefanie N, Beutlera Bruce. Genetic and Physical Mapping of theLpsLocus: Identification of the Toll-4 Receptor as a Candidate Gene in the Critical Region. Blood Cells, Molecules and Diseases. 1998;24(3):340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 40.Borriello Frank, Sethna Michael P, Boyd Scott D, Nicola Schweitzer A, Tivol Elizabeth A, Jacoby Douglas, Strom Terry B, Simpson Elizabeth M, Freeman Gordon J, Sharpe Arlene H. B7-1 and B7-2 Have Overlapping, Critical Roles in Immunoglobulin Class Switching and Germinal Center Formation. Immunity. 1997;6:303–313. doi: 10.1016/s1074-7613(00)80333-7. [DOI] [PubMed] [Google Scholar]
- 41.Vogel Marina, Dobrovolskaia A, Stefanie N. Toll receptors, CD14, and macrophage activation and deactivation by LPS. Microbes and Infection. 2002;4(9):903–914. doi: 10.1016/s1286-4579(02)01613-1. [DOI] [PubMed] [Google Scholar]
- 42.Pluddemann A, Mukhopadhyay S, Gordon S. The interactionof macrophage receptors with bacterial ligands. Expert reviews in molecular medicine. 2006;8(28):1–24. doi: 10.1017/S1462399406000159. [DOI] [PubMed] [Google Scholar]
- 43.Bertrand Julien Y, Giroux Sébastien, Golub Rachel, Klaine Michèle, Jalil Abdelali, Boucontet Laurent, Godin Isabelle, Cumano Ana. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proceedings of the National Academy of Sciences. 2004;102(1):134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavian B, Peault M. Hematopoietic Stem Cell Emergence in the Human Embryo and Fetus. Annals of the New York Academy of Sciences. 2003;(996):132–140. doi: 10.1111/j.1749-6632.2003.tb03241.x. [DOI] [PubMed] [Google Scholar]
- 45.Lei T, Jacob S, Ajil-Zaraa I, Dubuisson JB, Irion O, Jaconi M, Feki A. Xeno-free derivation and culture of human embryonic stem cells: current status, problems and challenges. Cell Research. 2007;17(8):682–688. doi: 10.1038/cr.2007.61. [DOI] [PubMed] [Google Scholar]
- 46.Pagès F, Lebel-Binay S, Vieillefond A, Deneux L, Cambillau M, Soubrane O, Debré B, Tardy D, Lemonne JL, Abastado JP, Fridman WH, Thiounn N. Local immunostimulation induced by intravesical administration of autologous interferon-gamma-activated macrophages in patients with superficial bladder cancer. Clinical and Experimental Immunology. 2002;127(2):303–309. doi: 10.1046/j.1365-2249.2002.01766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaperot L, Chokri M, Jacob M-C, Drillat P, Garban F, Egelhofer H, Molens J-P, Sotto J-J, Bensa J-C, Plumas J. Differentiation of antigen-presenting cells (dendritic cells and macrophages) for therapeutic application in patients with lymphoma. Leukemia. 2000;14(9):1667–1677. doi: 10.1038/sj.leu.2401888. [DOI] [PubMed] [Google Scholar]