Abstract
The five two domain stimulatory KIR genes carried by 100 random African Americans were characterized by DNA sequencing of genomic DNA covering the majority of coding exons. The frequency of individual loci was similar to that found in European Americans with the exception of a reduced frequency for KIR2DS1 in African Americans. New alleles were identified at the KIR2DS1 (*008), KIR2DS2 (*006), KIR2DS3 (*00104, *00105, *00106, *004), KIR2DS4 (*00103, *00104, *009, *011, *012, *013), and KIR2DS5 (*006, *007, *00801, *00802, *009) loci. The distribution of alleles at each locus was similar to that found in a European American population except for KIR2DS5. KIR2DS5 exhibits a single allele in European Americans; the same allele is found at reduced frequency (41% of gene positive individuals) accompanied by KIR2DS5*006 (18%), KIR2DS5*007 (26%), and six other alleles (25%) in African Americans.
Introduction
A balance of inhibitory and stimulatory signals determines the activation state of natural killer (NK) cells. These cells express a number of cell surface stimulatory receptors which detect ligands associated with abnormal cells: NKG2D detecting stress proteins; natural cytotoxicity receptor NKp46, viral components; and CD16 Fc receptor, antigen antibody complexes [1]. Also expressed by human NK cells are the stimulatory killer cell immunoglobulin-like receptors (sKIR). The six sKIR (KIR2DS1-KIR2DS5 and KIR3DS1) belong to a larger family of homologous receptors that also includes inhibitory receptors KIR2DL1-KIR2DL5 and KIR3DL1-KIR3DL3 [2]. The similarity of sKIR extracellular domains to those of inhibitory KIR (iKIR) suggests that sKIR recognize the same or similar ligands, i.e., major histocompatibility complex (MHC) class I molecules. The identification of ligands for KIR2DS1[3] [4] and KIR2DS4 [5] supports this although the binding to MHC molecules is weak suggesting that specific MHC-antigenic peptide complexes might be required or that other pathogen-associated MHC-like ligands might exist [6].
Like several other stimulatory receptors, sKIR have a short cytoplasmic domain and signal through an associated immunotyrosine activation motif (ITAM)-bearing adaptor protein, DAP12 for the sKIR. When antibodies directed to sKIR are used to trigger these receptors on NK cells, cytotoxicity is increased and cytokines (interferon gamma, tumor necrosis factor) are produced [7-9].
The genes encoding all KIR are clustered on chromosome 19 [10] and many haplotypes encoding variable numbers of KIR genes segregate in the population [11]. Haplotypes designated as “A” carry only KIR2DS4 as their sKIR in combination with iKIR KIR2DL1, KIR2DL3, KIR3DL1, and KIR3DL2. Haplotypes with other sKIR loci are designated as “B”; KIR2DS4 is found on some of the B haplotypes. Some haplotypes may encode no surface expressed KIR2DS loci [12]; others encode several. The allelic products of one sKIR locus, KIR2DS3, do not appear to be surface expressed and the locus may be nonfunctional [13]. In addition to the diversity in receptor gene repertoire, the KIR loci themselves are polymorphic [14]. For the majority of sKIR, most populations studied have one or two predominant alleles at a locus with small numbers of individuals carrying other alleles [15-17]. The exception is KIR2DS4 which is more polymorphic. The KIR allelic variation may impact the level of cell surface expression and/or the affinity of interaction with ligand [16,18]. Another level of diversity is the expression of specific KIR on cell surfaces of individual NK clones. KIR are expressed by the CD56 dim subset of NK cells and individual cells in this subset express receptors encoded by subsets of their KIR genes. Some cells may express a single KIR while others might express two or more. The specific receptors expressed depends, in part, on the HLA ligands expressed [16] with the overall repertoire appearing to balance the reactivity to missing self ligands [19].
The goal of this study was to examine the allelic diversity in the two domain sKIR genes carried by a random population of African Americans and to contrast their diversity with that observed in other populations.
Materials and Methods
Population studied
This study used Epstein Barr Virus-transformed B-cell lines from the National Institute of General Medical Sciences (NIGMS) Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/nigms/). Genomic DNA was isolated from 100 unique and unrelated African Americans from the human variation panel using a QIAamp® DNA Blood Mini Kit (Qiagen, Valencia, CA) following manufacturer’s instructions.
KIR polymerase chain reaction amplification
Testing for the presence or absence of specific stimulatory 2D KIR used polymerase chain reaction (PCR) primers and reaction conditions previously described [15,20,21]. Amplified DNA products were visualized by agarose gel electrophoresis.
To identify alleles at each locus, sKIR genes were amplified from genomic DNA by PCR with two pairs of primers designed to yield overlapping amplicons covering most of the exons as previously described [15]. One exception was that the antisense exon 9 primer for KIR2DS4 was replaced to accommodate a novel polymorphism that impacted primer annealing. The new primer, 5′TGAAATGGAGAATTGTGGGCTAAG [22], anneals in the 3′ untranslated region. Amplicons were purified using AmPure magnetic beads (Agencourt Bioscience, Beverly MA) according to the manufacturer’s protocol.
Nested PCR was used to isolate KIR2DS4 alleles with and without a known exon 5 deletion in heterozygotes as previously described [15]. When required, PCR products were cloned using the TOPO TA cloning kit (Invitrogen). Qiagen HaploPrep reagents 488A (KIR2DS2) and 744d22 (KIR2DS4) (Valencia, CA) were used to isolate alleles [23] in some cases following manufacturer’s instructions.
KIR sequencing and analysis
Sequencing primers were positioned to obtain the sequence of both strands of each amplicon [15]. Sequencing was performed using Applied Biosystems’ BigDye Terminator Ready Reaction mix according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA). The reactions were purified using CleanSEQ (Agencourt Bioscience) according to the manufacturer’s protocol. Sequencing products were detected using an Applied Biosystems 3730XL DNA analyzer. Sample files were analyzed using Sequencher (Genecodes Corp., Ann Arbor, MI) and Assign SBT 3.2.7 (Conexio Genomics, Applecross, Western Australia) software. The sequences were compared to locus specific databases (ImmunoPolymorphismDatabase (IPD)-KIR Release 2.0.0) of known KIR sequences [24] created using Library Builder software (Conexio Genomics) to identify alleles. In this report, the numbering of nucleotides and codons is based on IPD-KIR unless noted. Sequences were submitted to GenBank and novel allele designations were assigned by the KIR subcommittee of the World Health Organization Nomenclature Committee for Factors of the HLA System [25]. Confirmatory sequences of previously described alleles were submitted to the IPD-KIR database and include: KIR2DS1*004 (GenBank FJ374884), KIR2DS1*006 (EU915286), KIR2DS2*00102 (FJ374880), KIR2DS5*003 (FJ374879), KIR2DS5*004 (FJ374881), KIR2DS5*005 (FJ374882).
Results
The percent of African Americans carrying specific two domain stimulatory genes is similar to other populations
The presence or absence of specific two domain stimulatory KIR genes in a population of 100 unrelated African Americans is shown in Table 1. The percentages are similar to that previously reported for 58 random African Americans from a Los Angeles blood donor population [26]. Since many African Americans trace their ancestry to slaves originating from the west coast of Africa [27], the percentages were compared also to percentages found in two west African populations (n=62 [28], n=90 [29]). The results are comparable with the exception of KIR2DS1 which was lower in a population from Senegal (27% vs 13%) and KIR2DS5 which was lower in both populations from west Africa (39% vs 24% (or 30%)). The lower percentage for KIR2DS5 is not explained by a failure in the earlier studies of primers to anneal to the novel alleles we are describing in this study. In a comparison with a European American population (n=77), percentages are similar except for KIR2DS1 where 27% of African Americans carried the gene in comparison with 42% of European Americans [15]. KIR2DS1. Like most populations studied [15-17], the predominant allele in African Americans is KIR2DS1*00201 found in 81% of gene positive individuals (Table 1). The previously described KIR2DS1*004, previously reported in a single Black individual [30], was identified in two individuals and the sequence was extended to include all of the leader sequence (GenBankFJ374884). KIR2DS1*006, previously found in an European American [15], was observed for a second time. One new allele, KIR2DS1*008, was detected in three individuals (Tables 1, 2). It differs by three nucleotides from KIR2DS1*00201, sharing codon 90 with KIR2DS1*004 (GTG/val; other alleles have TTG/leu) but has a unique substitution of two nucleotides in codon 123 altering asn (AAT) to ser (AGC). Codon 123 is located in the second extracellular domain and the serine substitution alters a potential N-linked glycosylation site. KIR2DS2. Like European Americans [15], the predominant allele is KIR2DS2*00101 although KIR2DS2*00102 was observed in five of 44 gene-positive individuals (Table 1). The latter confirms an allele previously identified in an individual of mixed race [31]. A new allele, KIR2DS2*006, differs from KIR2DS2*00102 by a single nucleotide substitution at codon 250 altering AAC (asn) to GAC (asp) in the transmembrane region; the same codon is found in KIR2DS2*005. At the protein level, the African American population is more homogeneous than the European American population, expressing two KIR2DS2 receptors compared to three [15], but more diverse than the Japanese population with one expressed receptor [16]. KIR2DS3. The predominant allele is KIR2DS3*00103 (Table 1). KIR2DS3*002 was observed twice, being previously found in Caucasians. Four new alleles were detected (Table 2). Silent variants of the predominant allele were designated KIR2DS3*00104, KIR2DS3*00105, and KIR2DS3*00106. KIR2DS3*004 differs at codon 131 from the other alleles at the locus (TGG/trp was altered to CGG/arg) and at codon 262 (a silent substitution). The amino acid substitution alters the second extracellular domain. Like the other alleles at this locus, KIR2DS3*004 has greatly reduced surface expression [13].
Table 1.
Percent of individuals positive for 2DS alleles
| Locus / Allele | Percent of total individuals |
Locus / Allele | Percent of total individuals |
|---|---|---|---|
| 2DS1* | 27 (27/100) | 2DS4* | 97 (97/100) |
| 00201 | 22 (22/100) | 00101a | 62 (62/100) |
| 004 | 2 | 00103b | 1 |
| 006 | 1 | 00104b | 5 |
| 008b | 3 | 003 | 24 |
| Other | 0 | 004 | 8 |
| 2DS2* | 44 (44/100) | 006b | 20 |
| 00101 | 36 (36/100) | 007b | 10 |
| 00102 | 5 | 009b | 5 |
| 006b | 3 | 011b | 2 |
| Other | 0 | 012b | 1 |
| 2DS3* | 23 (23/100) | 013b | 4 |
| 00103 | 19 (19/100) | Other | 0 |
| 00104b | 1 | 2DS5* | 39 (39/100) |
| 00105b | 1 | 00201 | 16 (16/100) |
| 00106b | 1 | 003 | 2 |
| 002 | 2 | 004 | 1 |
| 004b | 1 | 005 | 1 |
| Other | 0 | 006b | 7 |
| 007b | 10 | ||
| 00801b | 1 | ||
| 00802b | 1 | ||
| 009b | 3 | ||
| Other | 0 |
Alleles differing outside of coding regions were not resolved.
Novel allele described in this study.
Table 2.
Novel alleles
| Novel Allelea | Similar Allele | Codon (Amino Acid) Alteredb | GenBank Accession No. | Sample ID |
|---|---|---|---|---|
| 2DS1 | ||||
| 2DS1*008 | KIR2DS1*00201 | 90 TTG (L) > GTG (V), 123 AAT (N) > AGC (S) | EU277008 | GM17113 |
| 2DS2 | ||||
| 2DS2*006 | KIR2DS2*00102 | 250 AAC (N) > GAC (D) | FJ457922 | GM17148 |
| 2DS3 | ||||
| 2DS3*00104 | KIR2DS3*00103 | 157 AAC (N) > AAT (N) | EU277007 | GM17109 |
| 2DS3*00105 | KIR2DS3*00103 | 178 TTC (F) > TTT (F) | EU915289 | GM17200 |
| 2DS3*00106 | KIR2DS3*00103 | 262 GCG (A) > GCA (A) | EU915290 | GM17187 |
| 2DS3*004 | KIR2DS3*00103 | 131 TGG (W) > CGG (R), 262 GCG (A) > GCA (A) | EU277009 | GM17114 |
| 2DS4-full length | ||||
| 2DS4*00103 | KIR2DS4*00101 | 162 GCC (A) > GCT (A) | EU277010 | GM17114 |
| 2DS4*00104 | KIR2DS4*00101 | 280 GTG (V) > GTA (V) | EU933934 | GM17127 |
| 2DS4*011 | KIR2DS4*00101 | 250 GAC (D) > AAC (N) | EU915288 | GM17165 |
| 2DS4 deletion | ||||
| 2DS4*009 | KIR2DS4*003 | 112 GCC (A) > GCA (A), 158c CTC (L) > TTC (F), 171c TCG (S) > TTG (L) |
EU277011 | GM17123 |
| 2DS4*012 | KIR2DS4*006 | 74 GCA (A) > GAA (E) | EU915291 | GM17199 |
| 2DS4*013 | KIR2DS4*006 | 4 GTC (V) > GCC (A), 149c TCA (S) > TCC (S), 179c ACG (T) > ACA (T), nucleotides 787-789 CCG > GCA 3′ after termination codon |
FJ374883 | GM17141 |
| 2DS5 | ||||
| 2DS5*006 | KIR2DS5*005 | 154 CCC (P) > ACC (T) | EU277005 | GM17102 |
| 2DS5*007 | KIR2DS5*005 | 216 GAA (E) > AAA (K) | EU277006 | GM17103 |
| 2DS5*00801 | KIR2DS5*005 | 1 CAT (H) > CGT (R), 224 CAC (H) > CAT (H) | EU277004 | GM17128 |
| 2DS5*00802 | KIR2DS5*005 | 1 CAT (H) > CGT (R) | FJ374885 | GM17183 |
| 2DS5*009 | KIR2DS5*00201 | 176 AGA (R) > ACA (T) | EU915287 | GM17148 |
Designation assigned by the WHO Committee for Factors of the HLA System [25].
Numbering based on the IPD-KIR database [24]; most similar known allele listed first.
2DS4*009, *012, *013 carry the deletion found in 2DS4*003 and the numbering of codons and amino acids is based on alignment with 2DS4*003 following codon 130. Alleles with the deletion have a frameshift which dramatically alters the amino acid sequence beginning at codon 131 through a termination codon at 219.
KIR2DS4
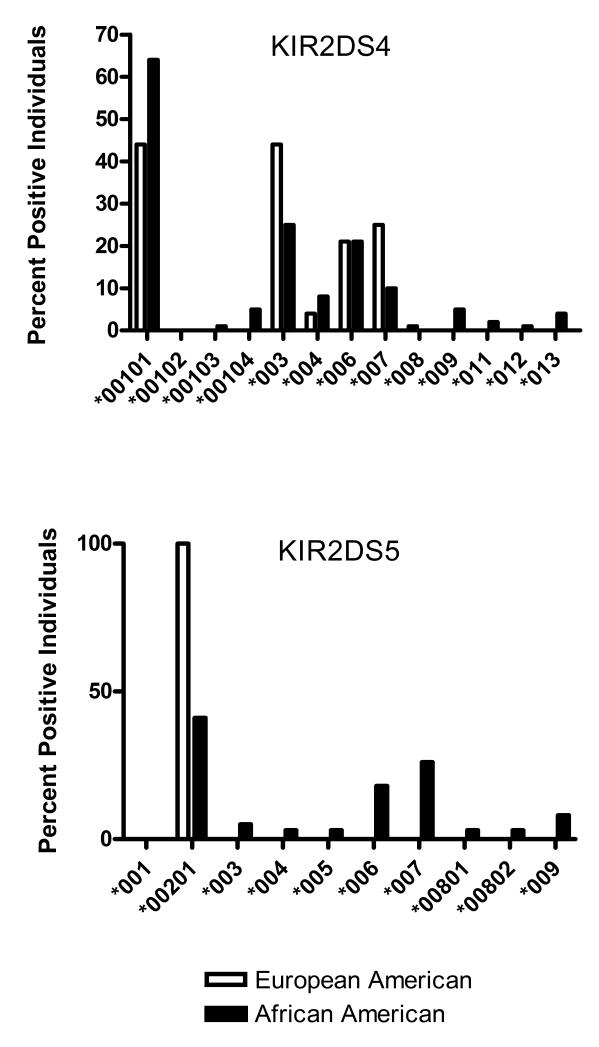
Alleles encoding all five known KIR2DS4-encoded polypeptides were found in African Americans (Table 1, Figure 1). KIR2DS4*00101 predominates as it does in a Japanese population and European Americans [15,16]. Other alleles are frequent; the second most common allele in European Americans, KIR2DL4*003 [17], is found in 25% of the gene positive African American panel. Six novel alleles were identified. Two novel alleles, KIR2DS4*00103 and KIR2DS*00104, differ at the nucleotide level from KIR2DS4*00101; their substitutions are unique. KIR2DS4*00104 was first identified by an inconsistency between the sequence-specific priming (SSP) results indicating the presence of at least two KIR2DS4 alleles differing for the known deletion and the initial DNA sequencing results yielding a single allele. The novel allele, KIR2DS4*00104, differs in the annealing site of our antisense primer for the B amplicon and was identified in five individuals. KIR2DS4*011, observed twice, differs from KIR2DS4*00101 at codon 250 in the transmembrane region altering the conserved GAC (asp) to AAC (asn).
Figure 1. Percentage of gene positive individuals with KIR2DS4 and KIR2DS5 alleles in two populations.
DNA sequencing was used to identify stimulatory KIR alleles in a panel of random African Americans (n=100) as compared to a panel of European Americans (n=77) [15].
Three other novel alleles carried the deletion observed in KIR2DS4*003 [12]. KIR2DS4*009 differs from KIR2DS4*003 by three nucleotide changes at previously conserved positions causing two amino acid substitutions at 158 (leu replaced by phe) and 171 (ser replaced by leu) in the portion of the unique polypeptide sequence introduced by the deletion. KIR2DS4*012 differs from KIR2DS4*006 by a single nucleotide substitution altering codon 74 from the conserved ala to glu in the first extracellular domain. The third novel deletion allele, KIR2DS4*013, differs from KIR2DS4*006 by five nucleotide changes. Four alter the nucleotide sequence following the deletion but do not impact the amino acid sequence. One at codon 4 (GTC (val) to GCC (ala)) alters a conserved amino acid in the first domain sequence. KIR2DS5. In contrast with studies in other populations [15-17] [32], KIR2DS5 is more diverse in African Americans. While KIR2DS5*00201 predominates, it is carried only by 41% of individuals carrying KIR2DS5 alleles (Table 1, Figure 1). In contrast, KIR2DS5*00201 is the only allele found in individuals of European [15,17] and Japanese ancestry [16]. Alleles KIR2DS5*003, KIR2DS5*004 and KIR2DS5*005, previously observed in African Blacks [31,32] and reported in single individuals, were found in African Americans. Five novel alleles were identified. An inconsistency between gene presence [33] or absence [20] using different locus specific primers signaled the presence of a novel allele, KIR2DS5*009, observed in 3 individuals. A substitution at codon 176 altered a conserved primer annealing site and introduced a new amino acid (thr instead of arg). Novel alleles KIR2DS5*006 and KIR2DS5*007 were found in multiple individuals including 18% and 26%, respectively, of the gene positive population. Both nonsynonymous substitutions alter previously conserved codons in the extracellular portion of the receptor. Amino acid substitutions in KIR2DS5*00801 and KIR2DS5*00802 alter the amino terminus of the mature receptor from his to arg. None of the novel KIR2DS5 alleles carry the substitutions known to result in the loss of surface expression observed for KIR2DS5*001 [34].
Discussion
The two domain sKIR carried by African Americans resemble that of European Americans [15]and other populations [16,17,28,29] at the level of gene presence or absence. One exception is KIR2DS1 which appears at a lower frequency in African Americans (this study and [26]) (27% in African Americans compared to 42% in European Americans) and this decrease is mirrored in African populations [28,29,35]. A second exception is KIR2DS5 being found in a smaller percent of individuals in west African populations compared to African Americans. Other loci are present in similar numbers of individuals (less than +/-10%) in African Americans and European Americans.
With the exception of KIR2DS5, the distribution of sKIR alleles at each locus is similar to European Americans. KIR2DS1, KIR2DS2, and KIR2DS3 all show the same predominant allele at each locus. KIR2DS4 is more polymorphic but, again, both population groups exhibit the same predominant alleles. The finding of a single predominant allele also applies to the single three domain sKIR. A single allele, KIR3DS1*013, accounts for 97% of the alleles identified in a study of 28 world-wide populations [36]. The exception is KIR2DS5 where all gene positive European Americans carry KIR2DS5*00201 but three alleles predominate in African Americans: KIR2DS5*00201 (41%), KIR2DS5*007 (26%), and KIR2DS5*006 (18%). These alleles encode receptors that differ from one another at three polymorphic positions in the extracellular portion of the receptor, two in the second domain and one just adjacent to the cell membrane. The substitutions are nonconservative (P154T, R157G, and E216K) and may impact the properties of the receptor.
Most individuals (>90%) who carry a locus exhibit a single allele although it is not possible to determine if they carry one or more copies without an analysis of haplotypes. For example, out of 27 individuals positive for KIR2DS1, 26 exhibit a single allele; single alleles are observed in all 44 KIR2DS2 gene positive individuals and 21 out of 23 KIR2DS3 gene positive individuals. This likely reflects the predominance of a single allele at these loci but even KIR2DS5 which is more polymorphic with three relatively frequent alleles exhibits 36 individuals with single alleles out of 39 individuals carrying the locus. In contrast, 46% (45 out of 97) of KIR2DS4 positive individuals are heterozygotes.
The 17 novel KIR2DS alleles described here impact 24 codons. The vast majority occur at previously conserved positions; 14 codon changes are nonsynonymous. The predominance of amino acid changes suggests a positive selection for protein variation. The majority of the nonsynonymous substitutions (12/14) impact the extracellular domains. Positive selection for amino acid substitutions in the extracellular domains has been reported previously for KIR3DL1/KIR3DS1 using tests that consider codon variation in the ratio of nonsynonymous to synonymous (dN/dS) substitutions [36].
The total number of sKIR loci present varies among individuals. Overall, only four out of 100 African Americans carry all five two domain sKIR. Multiple individuals carry four (15 individuals), three (25), and two (19) sKIR loci. Thirty seven individuals (37%) carry only one locus; the single locus is always KIR2DS4 suggesting that these individuals carry two haplotypes designated as “A”. Previous studies observed AA haplotypes in 35% of Africans [28], 30% of Caucasoids [37] and 59% of Japanese [16]. There are no individuals who fail to carry at least one sKIR gene but there are seven individuals who carry, as their only 2D sKIR, KIR2DS4 with an allele which lacks cell surface expression [12]. One further individual lacks a surface expressed KIR2DS4 allele and carries only the apparently nonexpressed KIR2DS3 locus [13]. Thus individuals run the gamut from carrying all five sKIR loci to those who carry a single locus encoding an apparently defective receptor.
While the sKIR profile in populations is still limited in the heterogeneity, the number of sKIR alleles that are discovered is increasing. Once the role of these receptors in the NK cell response is known, we can begin to evaluate the impact of this allelic diversity in health and in disease states.
Acknowledgments
We would like to thank Ian Belle and Noriko Steiner for their assistance. This research is supported by funding from the Office of Naval Research N00014-06-1-0726 (CH, JN). The views expressed in this article are those of the authors and do not reflect the official policy or position of the Department of the Navy, the Department of Defense, or any other agency of the U.S. government.
References
- 1.Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol Rev. 2006;214:73–91. doi: 10.1111/j.1600-065X.2006.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, Moretta L, Moretta A. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 4.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagne F, Ferracci G, Arosa FA, Moretta A, Sun PD, Ugolini S, Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katz G, Gazit R, Arnon TI, Gonen-Gross T, Tarcic G, Markel G, Gruda R, Achdout H, Drize O, Merims S, Mandelboim O. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173:1819–1825. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 6.French AR, Sjolin H, Kim S, Koka R, Yang L, Young DA, Cerboni C, Tomasello E, Ma A, Vivier E, Karre K, Yokoyama WM. DAP12 signaling directly augments proproliferative cytokine stimulation of NK cells during viral infections. J Immunol. 2006;177:4981–4990. doi: 10.4049/jimmunol.177.8.4981. [DOI] [PubMed] [Google Scholar]
- 7.Campbell KS, Cella M, Carretero M, Lopez-Botet M, Colonna M. Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur J Immunol. 1998;28:599–609. doi: 10.1002/(SICI)1521-4141(199802)28:02<599::AID-IMMU599>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiesa MD, Romeo E, Falco M, Balsamo M, Augugliaro R, Moretta L, Bottino C, Moretta A, Vitale M. Evidence that the KIR2DS5 gene codes for a surface receptor triggering natural killer cell function. Eur J Immunol. 2008;38:2284–2289. doi: 10.1002/eji.200838434. [DOI] [PubMed] [Google Scholar]
- 10.Martin AM, Kulski JK, Gaudieri S, Witt CS, Freitas EM, Trowsdale J, Christiansen FT. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene. 2004;335:121–131. doi: 10.1016/j.gene.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Hsu KC, Chida S, Dupont B, Geraghty DE. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunological Reviews. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 12.Middleton D, Gonzalez A, Gilmore PM. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum Immunol. 2007;68:128–134. doi: 10.1016/j.humimm.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 13.VandenBussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes and Immunity. 2008 doi: 10.1038/gene.2008.91. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson J, Waller MJ, Fail SC, Marsh SG. The IMGT/HLA and IPD databases. Hum Mutat. 2006;27:1192–1199. doi: 10.1002/humu.20406. [DOI] [PubMed] [Google Scholar]
- 15.Hou L, Steiner NK, Chen M, Belle I, Kubit AL, Ng J, Hurley CK. Limited Allelic Diversity of Stimulatory Two Domain Killer Immunoglobulin-Like Receptors. Human Immunology. 2008;69:174–178. doi: 10.1016/j.humimm.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59:145–158. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- 18.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 19.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 21.Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 22.Murdoch S, Seoud M, Kircheisen R, Mazhar B, Slim R. Detailed gene and allele content analysis of three homozygous KIR haplotypes. Tissue Antigens. 2006;68:72–77. doi: 10.1111/j.1399-0039.2006.00606.x. [DOI] [PubMed] [Google Scholar]
- 23.Dapprich J, Witter K, Gabel HW, Murphy NB, Albert ED. Identification of a new HLA-B allele (B*1576) by haplotype specific extraction. Hum Immunol. 2007;68:418–421. doi: 10.1016/j.humimm.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh SGE, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, Shilling H, Garcia CA, Hsu KC, Wain H. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens. 2003;62:79–86. doi: 10.1034/j.1399-0039.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 26.Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics. 2007;59:1–15. doi: 10.1007/s00251-006-0168-4. [DOI] [PubMed] [Google Scholar]
- 27.Curtin PD. The Atlantic Slave Trade: A Census. The University of Wisconsin Press; Madison: 1969. [Google Scholar]
- 28.Norman PJ, Carrington CVF, Byng M, Maxwell LD, Curran MD, Stephens HAF, Chandanayingyong D, Verity DH, Hameed K, Ramdath DD, Vaughan RW. Natural killer cell immunoglobulin-like receptor (KIR) locus profiles in African and South Asian populations. Genes and Immunity. 2002;3:86–95. doi: 10.1038/sj.gene.6363836. [DOI] [PubMed] [Google Scholar]
- 29.Denis L, Sivula J, Gourraud PA, Kerdudou N, Chout R, Ricard C, Moisan JP, Gagne K, Partanen J, Bignon JD. Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Reunion. Tissue Antigens. 2005;66:267–276. doi: 10.1111/j.1399-0039.2005.00473.x. [DOI] [PubMed] [Google Scholar]
- 30.Rajalingam R, Gardiner CM, Canavez F, Vilches C, Parham P. Identification of seventeen novel KIR variants: fourteen of them from two non-Caucasian donors. Tissue Antigens. 2001;57:22–31. doi: 10.1034/j.1399-0039.2001.057001022.x. [DOI] [PubMed] [Google Scholar]
- 31.Artavanis-Tsakonas K, Eleme K, McQueen KL, Cheng NW, Parham P, Davis DM, Riley EM. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J Immunol. 2003;171:5396–5405. doi: 10.4049/jimmunol.171.10.5396. [DOI] [PubMed] [Google Scholar]
- 32.Vilches C, Pando MJ, Rajalingam R, Gardiner CM, Parham P. Discovery of two novel variants of KIR2DS5 reveals this gene to be a common component of human KIR ‘B’ haplotypes. Tissue Antigens. 2000;56:453–456. doi: 10.1034/j.1399-0039.2000.560510.x. [DOI] [PubMed] [Google Scholar]
- 33.Gomez-Lozano N, Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: An update. Tissue Antigens. 2002;59:184–193. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- 34.Steiner NK, Dakshanamurthy S, VandenBussche CJ, Hurley CK. Extracellular domain alterations impact surface expression of stimulatory natural killer cell receptor KIR2DS5. Immunogenetics. 2008;60:655–667. doi: 10.1007/s00251-008-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Middleton D, Meenagh A, Moscoso J, Arnaiz-Villena A. Killer immunoglobulin receptor gene and allele frequencies in Caucasoid, Oriental and Black populations from different continents. Tissue Antigens. 2008;71:105–113. doi: 10.1111/j.1399-0039.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 36.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 37.Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 2001;52:195–205. doi: 10.1007/s002510000281. [DOI] [PubMed] [Google Scholar]