Abstract
Heparin is the most widely used pharmaceutical to control blood coagulation in modern medicine. A health crisis that took place in 2008 led to a demand for production of heparin from non-animal sources. Since Chinese hamster ovary (CHO) cells are capable of producing heparan sulfate (HS), a related polysaccharide naturally, and heparin and HS share the same biosynthetic pathway, we hypothesized that heparin could be produced in CHO cells by metabolic engineering. We developed stable human N-deacetylase/N-sulfotransferase (NDST2) and mouse heparan sulfate 3-O-sulfotransferase 1 (Hs3st1) expressing cell lines based on the expression of endogenous enzymes in the HS/heparin pathways of CHO-S cells. Both activity assay and disaccharide analysis showed that engineered HS attained heparin-like characteristics but not identical to pharmaceutical heparin, suggesting that further balancing the expression of transgenes with the expression levels of endogenous enzymes involved in HS/heparin biosynthesis might be necessary.
Keywords: Chinese hamster ovary cells, LC-MS, anticoagulant, heparin, metabolic engineering, transcriptional regulation, translational regulation
Heparin is a highly sulfated, glycosaminoglycan (GAG) that consists of repeated disaccharides, L-iduronic acid (IdoA) or D-glucuronic acid (GlcA) and N-acetyl-D-glucosamine (GlcNAc) with or without sulfo groups (Fig. 1). Heparin is widely used clinically as an anticoagulant, particularly for surgery and kidney dialysis.1 Greater than 100 tons of heparin are used annually, with a market value of ~$7 billion.2 Virtually all clinically available heparin is animal-derived, primarily from porcine intestinal mucosa. It is estimated that only three doses of heparin can be obtained from one animal.3 In early 2008, there was a marked increase in serious adverse events associated with heparin therapy, with nearly 100 associated deaths in the United States alone. After thorough investigation, it was discovered that the heparin injected into patients had been contaminated by oversulfated GAGs,4 highlighting the potential risks of contamination from the current methods of heparin production, extraction from animal tissues. While chemoenzymatic approaches to developing a bioengineered heparin have been reported,5-8 the yields are still quite low, creating an impetus for other approaches to create a bioengineered heparin.
Figure 1.
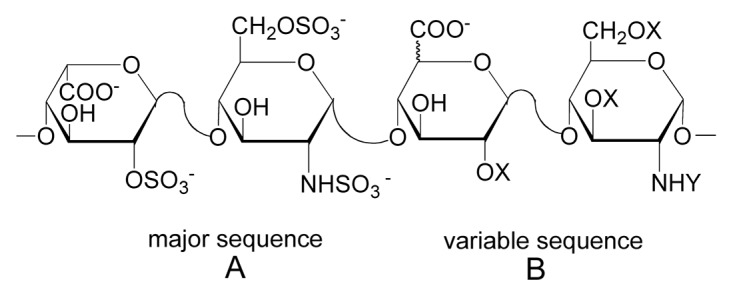
The structures of the major (A) and variable (B) repeating disaccharides comprising heparin, where X = SO3- or H and Y = SO3- or COCH3. Fully sulfated heparin chains are composed of uniform, repeating sequences of trisulfated disaccharides whereas heparin chains having intermediate level of sulfation (2.5 sulfo groups per disaccharide) are composed of long segments of fully sulfated sequences with intervening undersulfated domains.
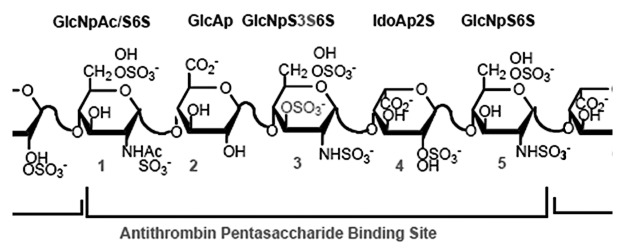
Chinese hamster ovary (CHO) cells, the most widely used cells for therapeutic protein production, are good candidates for production of a bioengineered heparin. They express many of the enzymes involved in glycosylation; they are relatively safe from biological contamination such as viruses, and they can be adapted to suspension culture and easily scaled up. More importantly, CHO cells produce heparan sulfate (HS), a less sulfated GAG that has same basic disaccharide units as heparin. HS also has considerably lower anticoagulant activity than heparin due to lacking a unique pentasaccharide motif, an antithrombin (ATIII) binding site (Fig. 2).9-11 Similarities and differences between HS and heparin are summarized in Table 1.
Figure 2.
Heparin’s antithrombin binding site. Anticoagulant activity of heparin is mediated through the interaction of an antithrombin III (ATIII) binding site with ATIII, a serine-protease inhibitor.
Table 1. Comparison of heparan sulfate and heparin characteristics.
| Characteristic | Heparan sulfate | Heparin |
|---|---|---|
| Size |
10–70 kDa |
10–12 kDa |
| Sulfo groups/disaccharide |
0.8–1.8 |
1.8–2.4 |
| GlcN N-sulfate |
40–60% |
> 85% |
| Iduronic acid (IdoA) |
30–50% |
> 70% |
| Core protein |
Syndecan and Glypican |
Serglycin |
| Site of synthesis |
Virtually all cells |
Mast cells |
| Binding to antithrombin (ATIII) | 0–0.3% | ~30% |
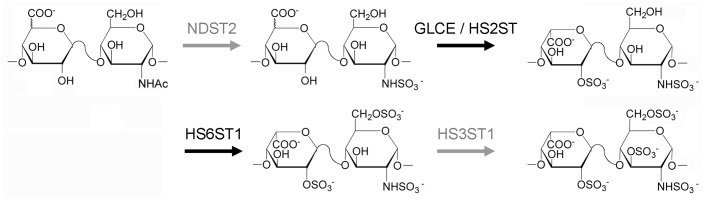
Extensive studies by Lindahl and coworkers have shown that heparin and HS are biosynthesized through a similar pathway. After formation of a common tetrasaccharide (xylose-galactose-galactose-glucuronic acid),12 the chains are polymerized by the sequential addition of basic disaccharides (GlcA-GlcNAc) followed by a series of modification including N-deacetylation and N-sulfonation of GlcNAc residues, epimerization of GlcA to IdoA, and finally, the introduction of O-sulfo groups at different positions of the glucosamine (GlcN) and uronic acid residues (Fig. 3).13 Complete or nearly complete modification of this nascent GAG chain results in a highly N-sulfo, O-sulfo, IdoA-rich GAG commonly referred to as heparin, which serves as the source of material for further fractionation to generate pharmaceutical heparin. HS is characterized by partial modification of the chains resulting in O-sulfo poor and GlcNAc, GlcA-rich chains.14,15 Since heparin and HS share a common biosynthetic pathway, we hypothesized that CHO cells could be metabolically engineered to produce heparin. In a recent work, we evaluated the expression levels of metabolic enzymes and isozymes involved in the biosynthetic pathway of HS/heparin and identified missing functionalities in CHO cells.16 We then constructed stable cell lines that expressed the relevant enzymes by genetic engineering. Finally, the structure and activity of the engineered HS was characterized.
Figure 3.
Biosynthetic pathway of HS/heparin. Enzymes shown in gray are not expressed in CHO-S host cells. NDST, N-deacetylase/N-sulfotransferase; GLCE, glucuronyl C5-epimerase; HS2ST, heparan sulfate 2-O-sulfotransferase; HS6ST, heparan sulfate 6-O-sulfotransferase; HS3ST, heparan sulfate 3-O-sulfotransferase.
Metabolic Engineering Strategies
We used rat mast cells, natural producers of heparin, to identify the required expression of HS/heparin biosynthetic enzymes and compared it to that of CHO-S cells (Invitrogen). Based on the RT-PCR and western blotting results, we determined that CHO-S cells did not express Ndst2 and showed minimal expression of Hs3st1, which are known to be critical for anticoagulant heparin biosynthesis (Fig. 3). Ndst2 plays an important role in the introduction of N-sulfo groups into GlcNAc, which in turn, is important for subsequent sulfonation of the growing HS chain.17 Absence of Ndst2 expression can explain the low level of N-sulfo groups in CHO-S HS. Hs3st1 is responsible for the formation of the unique ATIII binding pentasaccharide, which makes heparin an important anticoagulant therapeutic molecule. A recent study of CHO-K1 genome and transcriptome sequences verified that these enzymes were not expressed in CHO cells.18 Therefore, NDST2 and Hs3st1 were engineered into the CHO-S cells. Two stable cell lines (Dual-3 and Dual-29) were selected and HS from these cells and cultured media was isolated for further characterization. Culture media consisted of CD-CHO (Invitrogen) supplemented with 8 mM GlutaMAXTM (Invitrogen) and 15 ml of hypoxanthine/thymidine solution per 500 ml of medium (HT, Mediatech, Manassas, VA). In addition, 1 mg/ml of Geneticin® and 500 mg/ml of ZeocinTM were added to the medium for dual-expressing cell lines.
Characterization of Engineered HS
The engineered HS was isolated from cell pellets and analyzed by an anti-factor Xa anticoagulation assay to determine whether the engineered HS showed increased anticoagulant activity. Since cell-membrane-bound HS proteoglycans can be shed through the action of proteases,19,20 the engineered HS was also purified from the culture medium and analyzed by the anti-factor Xa assay. As shown in Figure 4, the HS extracted from Dual-3 and Dual-29 cell pellets shows significantly increased anticoagulation activity (7.5-fold and 6.8-fold, respectively) compared with the HS from CHO-S host cells. Unfortunately, the anticoagulant activity of the dual-expressing cells is still considerably lower than that of the pharmaceutical heparin. However, the pharmaceutical heparin has been fractionated to obtain high anticoagulant activity, whereas the bioengineered HS used in the activity assay was unfractionated, which may explain some of the difference in activity. The HS purified from culture media of Dual-3 and Dual-29 also shows increased anticoagulation activity (52.9-fold and 97.2-fold, respectively) compared with CHO-S cells.
Figure 4.
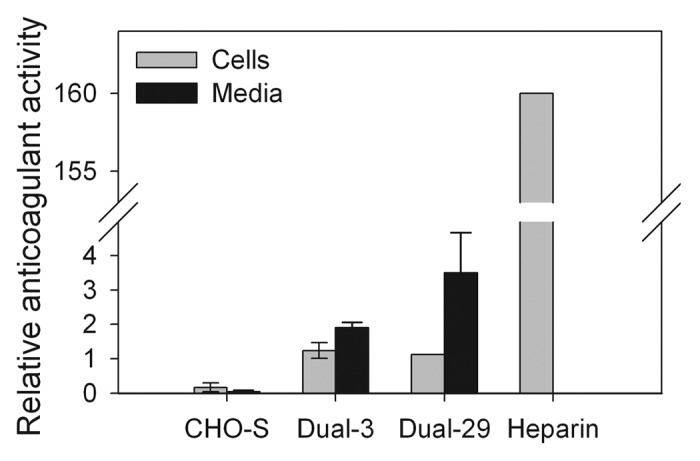
Factor Xa assay of dual NDST2 and Hs3st1-expressing cell lines. Pharmaceutical heparin was used as a positive control. Error bars represent 95% confidence intervals.
The total GAGs were isolated from culture media and analyzed by reverse-phase ion-pairing ultra-performance liquid chromatography mass spectrometry (RPIP-UPLC-MS).21 The amount of total HS/heparin and ratio of disaccharide components are summarized in Table 2 and Figure 5. The amount of HS/heparin secreted by the engineered cells was increased nearly 10-fold in comparison with the wild-type (a smaller increase was observed in the cell-associated HS/heparin). A substantial increase in sulfation was also observed, primarily N-sulfation, confirming the activity of NDST2. While this represents a significant improvement, it is still quite different from the trisulfated species that characterize the pharmaceutical heparin (Fig. 5). We could not verify the presence of a 3-O-sulfo group containing disaccharide because the action of the Hs3st1 enzyme affords a tetrasaccharide that is not cleavable by heparin lyases.22
Table 2. Amount of total heparan sulfate/heparin isolated from culture media or cell pellets.
| Sample | Amount of HS/heparin (µg/5 x 107 cells) |
|---|---|
| CHO-S media |
19.5 ± 1.48 (n = 2) |
| Dual-3 media |
79.4 ± 8.27 (n = 2) |
| Dual-29 media |
168.75 ± 6.29 (n = 2) |
| CHO-S cell pellet |
9.5 ± 0.65 (n = 3) |
| Dual-3 cell pellet |
8.8 ± 1.19 (n = 3) |
| Dual-29 cell pellet | 13.8 ± 3.02 (n = 3) |
Values are mean ± SD for the number of biological replicates shown. Analytical variation is < 5%.
Figure 5.
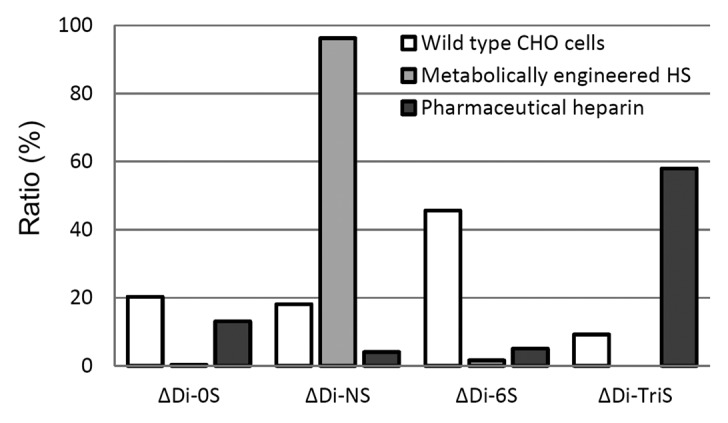
Disaccharide analysis of HS/heparin from culture media by RPIP-UPLC-MS.
Outlook: Balancing Expression of Enzymes Involved in HS/Heparin Synthesis
The relatively low anticoagulant activity of the engineered HS led us to investigate whether the engineered enzymes are properly targeted to the appropriate cellular compartment. We isolated Golgi and ER fractions and performed western blotting to detect the NDST2 and Hs3st1 enzymes. We found that NDST2 was localized in the Golgi complex, whereas Hs3st1 was detected in Golgi complex, ER and cytoplasm. This mistargeting of Hs3st1 may explain the low anticoagulant activity of engineered HS although Hs3st1 expression was high. There are two possible causes for the mistargeting. First, the dual cells could be expressing so much Hs3st1 enzymes that the excess amount cannot all be targeted to the Golgi complex. Second, Hs3st1 has a weak Golgi-targeting signal peptide. To address these possibilities, we have taken two approaches; (1) characterizing low Hs3st1 expressing clones, (2) introducing stronger Golgi-targeting signals.
The relative amounts of disaccharide species in dual-expressing cells show unexpected patterns compared with those of pharmaceutical heparin. The major species in dual-expressing cell lines are NS disaccharides while the major species in heparin are tri-sulfated disaccharides, suggesting that the expression level of NDST2 is too high, which overwhelms the actions of other enzymes. The mechanism that controls the HS/heparin biosynthetic pathway is also still unclear, but one widely supported hypothesis is that NDST is involved in the termination of sulfonation in HS/heparin.13 Therefore, overexpression of NDST might terminate the sulfonation of HS/heparin before other sulfotransferases were able to act on the HS chain, which means that it may be necessary to balance the expression levels of the transgenes with the endogenous genes. Our current study is focusing on balancing the metabolic engineering to produce an engineered HS identical to the pharmaceutical heparin in terms of structure and activity. This requires understanding the activity of the HS/heparin biosynthetic enzymes in the Golgi apparatus. Assays to establish the activity of GAG biosynthetic enzymes are under development in our laboratory.
Fine tuning of enzyme expression will also be necessary to precisely control the metabolic flux; this may be performed by either transcriptional or translational regulation of enzyme expression. In E. coli or S. cerevisiae, the expression levels of enzymes involved in metabolic pathways have been controlled by promoter engineering, gene titration or gene circuit engineering.23-25
In mammalian systems, the control and tuning of expression levels has been less widely practiced. This is likely due to a lack of genetic tools—common expression vectors for mammalian cells still utilize genetic elements [e.g., the human cytomegalovirus (CMV) promoter] that generate the highest levels of recombinant gene expression. Yet depending on the gene, product of interest, or desired cell phenotype, a high level of expression generated by an exogenous vector may prove to be suboptimal, physiologically irrelevant or even toxic. One approach to controlling expression levels is to use an inducible system. For example, with tetracycline-inducible systems,26 tetracycline or its analogs can be added to activate or deactivate synthetic transcription factors in a dose-dependent and tunable manner.
There are two potential drawbacks to using inducible systems: (1) turning an inducible system “off” generally leaves a level of leaky expression, which can still be higher than the optimized level one seeks; (2) optimization of multiple genes (where expression of one gene is independent of the other) requires multiple orthogonal inducible systems. A constitutive system that allows for expression level control can potentially avoid both of these drawbacks. Recently, Ferreira et al. developed a constitutive promoter library that is well suited for the tuning and optimization of genes exogenously expressed in mammalian cells.27 They performed site-directed mutagenesis at the CAAT and TATA box transcription factor binding sites and degenerate PCR mutagenesis to generate a total of 23 mutant CMV and EF1〈 promoters capable of an approximately 40-fold range in transcription levels. Furthermore, using a library of vectors employing these promoters, Ferreira et al. evaluated a broad range of oncogenic Ras expression levels.28 They were able to determine that the contribution of oncogenic Ras to imatinib drug resistance has an optimized, nonlinear behavior. A pre-B leukemia cell line proliferated optimally in the presence of imatinib when the exogenous oncogenic Ras level was approximately equivalent to the endogenous wild-type Ras expression level. Higher expression levels subsequently led to decreased proliferation rates. We anticipate that using a library of constitutive promoters to simultaneously optimize the expression levels of both Hs3st1 and NDST will increase both the yield and activity of recombinant HS.
Maximizing Total Expression of HS/Heparin
In addition to the challenges involved in obtaining HS/heparin with the correct structure for biological activity, maximizing the overall expression of heparin will also be necessary. In the dual-expressing cell lines, the amounts of HS/heparin in the media were increased significantly compared with HS/heparin extracted from the cells (4.5-fold to 9-fold), which implies that HS/heparin on the membrane are shed into the medium. Hence, we appear to have increased the metabolic flux through this pathway by metabolic engineering. Increased HS/heparin in the media also supports the hypothesis that bioengineered HS chains will still use the HS core proteins, syndecan and glypican, for extracellular targeting. Extracellular secretion using these HS core proteins will greatly simplify heparin purification as it will eliminate the necessity of cell lysis to recover the heparin. However, overexpression of the core proteins may be necessary to facilitate increased traffic of bioengineered HS/heparin. We are also currently exploring whether overexpression of Ext1 and Ext2 enzymes, which regulate HS/heparin chain polymerization, will lead to increased HS synthesis as has been observed in human embryonic kidney cells.29
Acknowledgments
This work was funded by a grant from the National Institutes of Health (R01GM090127).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/20902
References
- 1.Bigelow WG. Mysterious Heparin: The Key to Open Heart Surgery. Toronto-Montreal: McGraw-Hill Ryerson 1990. [Google Scholar]
- 2.Liu H, Zhang Z, Linhardt RJ. Lessons learned from the contamination of heparin. Nat Prod Rep. 2009;26:313–21. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linhardt RJ, Gunay NS. Production and chemical processing of low molecular weight heparins. Semin Thromb Hemost. 1999;25(Suppl 3):5–16. [PubMed] [Google Scholar]
- 4.Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, et al. Oversulfated chondroitin sulfate is a contaminant in heparin associated with adverse clinical events. Nat Biotechnol. 2008;26:669–75. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu R, Xu Y, Chen M, Weïwer M, Zhou X, Bridges AS, et al. Chemoenzymatic design of heparan sulfate oligosaccharides. J Biol Chem. 2010;285:34240–9. doi: 10.1074/jbc.M110.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, et al. Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins. Science. 2011;334:498–501. doi: 10.1126/science.1207478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuko S, Bera S, Green DE, Weïwer M, Liu J, DeAngelis PL, et al. Chemoenzymatic synthesis of uridine diphosphate-GlcNAc and uridine diphosphate-GalNAc analogs for the preparation of unnatural glycosaminoglycans. J Org Chem. 2012;77:1449–56. doi: 10.1021/jo202322k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Yang B, Zhang Z, Ly M, Takieddin M, Mousa S, et al. Control of the heparosan N-deacetylation leads to an improved bioengineered heparin. Appl Microbiol Biotechnol. 2011;91:91–9. doi: 10.1007/s00253-011-3231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcum JA, Rosenberg RD. Anticoagulantly active heparan sulfate proteoglycan and the vascular endothelium. Semin Thromb Hemost. 1987;13:464–74. doi: 10.1055/s-2007-1003523. [DOI] [PubMed] [Google Scholar]
- 10.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::AID-ANIE390>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 11.Muszbek L, Bereczky Z, Kovács B, Komáromi I. Antithrombin deficiency and its laboratory diagnosis. Clin Chem Lab Med. 2010;48:67–78. doi: 10.1515/cclm.2010.368. [DOI] [PubMed] [Google Scholar]
- 12.Robinson HC, Horner AA, Höök M, Ogren S, Lindahl U. A proteoglycan form of heparin and its degradation to single-chain molecules. J Biol Chem. 1978;253:6687–93. [PubMed] [Google Scholar]
- 13.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–71. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 14.Casu B. Structure and biological activity of heparin. Adv Carbohydr Chem Biochem. 1985;43:51–134. doi: 10.1016/S0065-2318(08)60067-0. [DOI] [PubMed] [Google Scholar]
- 15.Nairn AV, Kinoshita-Toyoda A, Toyoda H, Xie J, Harris K, Dalton S, et al. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res. 2007;6:4374–87. doi: 10.1021/pr070446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baik JY, Gasimli L, Yang B, Datta P, Zhang F, Glass CA, et al. Metabolic engineering of Chinese hamster ovary cells: towards a bioengineered heparin. Metab Eng. 2012;14:81–90. doi: 10.1016/j.ymben.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUBMB Life. 2002;54:163–75. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–41. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–77. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 20.Bartlett AH, Hayashida K, Park PW. Molecular and cellular mechanisms of syndecans in tissue injury and inflammation. Mol Cells. 2007;24:153–66. [PubMed] [Google Scholar]
- 21.Yang B, Weyers A, Baik JY, Sterner E, Sharfstein S, Mousa SA, et al. Ultra-performance ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for heparin disaccharide analysis. Anal Biochem. 2011;415:59–66. doi: 10.1016/j.ab.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao W, Garron ML, Yang B, Xiao Z, Esko JD, Cygler M, et al. Asparagine 405 of heparin lyase II prevents the cleavage of glycosidic linkages proximate to a 3-O-sulfoglucosamine residue. FEBS Lett. 2011;585:2461–6. doi: 10.1016/j.febslet.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A. 2005;102:12678–83. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalodimitrakis K, Isalan M. Engineering prokaryotic gene circuits. FEMS Microbiol Rev. 2009;33:27–37. doi: 10.1111/j.1574-6976.2008.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peacock RWS, Sullivan KA, Wang CL. Tetracycline-Regulated Expression Implemented through Transcriptional Activation Combined with Proximal and Distal Repression. ACS Synthetic Biology. 2012;1:156–62. doi: 10.1021/sb200029a. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira JP, Peacock RWS, Lawhorn IEB, Wang CL. Modulating ectopic gene expression levels by using retroviral vectors equipped with synthetic promoters. Syst Synth Biol. 2011;5:131–8. doi: 10.1007/s11693-011-9089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira JP, Lawhorn IE, Peacock RW, Wang CL. Quantitative assessment of Ras over-expression via shotgun deployment of vectors utilizing synthetic promoters. Integr Biol (Camb) 2012;4:108–14. doi: 10.1039/c1ib00082a. [DOI] [PubMed] [Google Scholar]
- 29.Busse M, Feta A, Presto J, Wilén M, Grønning M, Kjellén L, et al. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem. 2007;282:32802–10. doi: 10.1074/jbc.M703560200. [DOI] [PubMed] [Google Scholar]