Abstract
Cytochromes P450 (P450s) contribute to the metabolic activation and inactivation of various endogenous substrates. Despite years of research, the physiological role of CYP2S1 remains unknown. CYP2S1 has demonstrated NADPH P450-reductase-independent metabolism of cyclooxygenase (COX)-derived prostaglandins [e.g., prostaglandin G2 (PGG2)] at nanomolar concentrations. Arachidonic acid is converted to prostaglandin precursors [PGG2 and prostaglandin H2 (PGH2)] through COX. These precursors are used to synthesize numerous prostanoids, including PGE2. Prostaglandin E2 (PGE2) promotes cell proliferation and cell migration and inhibits apoptosis. CYP2S1 metabolism of PGG2 presumably sequesters PGG2 and PGH2, making them unavailable for synthesis of prostanoids such as PGE2. Whether CYP2S1 contributes to prostaglandin metabolism and influences cell physiological remains to be determined. The purpose of this study was to evaluate the physiological role of CYP2S1, if any, in human bronchial epithelial cells [SV40-derived bronchial epithelial cell line (BEAS-2B)]. To do this, we used small interfering RNA to deplete CYP2S1 mRNA and protein by approximately 75% and evaluated the impact of CYP2S1 depletion on cell proliferation and migration. CYP2S1 depletion enhanced both cell proliferation and migration in BEAS-2B cells. Consistent with the proposed role of CYP2S1 in PGE2 synthesis, the reduction in CYP2S1 expression doubled intracellular PGE2 levels. Pharmacological administration of PGE2 enhanced cell proliferation in BEAS-2B cells but failed to promote migration. Our data reveal an important role for CYP2S1 in the regulation of cell proliferation and migration, occurring in part through modulation of prostaglandin synthesis.
Introduction
Cytochromes P450 (P450s) are heme-containing monooxygenase enzymes capable of metabolizing various endogenous or exogenous compounds. CYP2S1 is one the most recently characterized members of the P450 family (Rylander et al., 2001). Its expression is restricted to extrahepatic epithelial cells (Rivera et al., 2002) and is significantly up-regulated in response to inflammatory disease. CYP2S1 expression is significantly elevated in psoriatic plaques characterized by inflammation and cell proliferation (Smith et al., 2003).
Expression data also suggest that CYP2S1 may be linked to carcinogenesis: elevated CYP2S1 immunoreactivity is observed in human epithelial colorectal (Kumarakulasingham et al., 2005), metastatic ovarian (Downie et al., 2005), breast (Murray et al., 2010), and squamous cell carcinomas (Saarikoski et al., 2005) and correlates with poor prognosis in colorectal, ovarian, and breast cancer (Downie et al., 2005; Kumarakulasingham et al., 2005; Murray et al., 2010). Better understanding of how alterations in CYP2S1 expression influence endogenous metabolism and of the cellular consequences associated with this regulation is an essential first step in determining the impact of elevated CYP2S1 expression, if any, in disease.
Despite the identification of potential endogenous substrates (e.g., all-trans-retinoic acid (Bui and Hankinson, 2009) and eicosanoids (Bui et al., 2011), the proposed metabolic mechanism and the relevance of CYP2S1-mediated metabolism remain controversial (Nishida et al., 2010; Xiao et al., 2011). Bui and colleagues identified CYP2S1-mediated metabolism of potential endogenous substrates that include retinoic acid (Bui and Hankinson, 2009) as well as members of the cyclooxygenase (COX) and lipoxygenase (LOX)-derived eicosanoids (Bui et al., 2011), using a codon-optimized synthetic CYP2S1 (Bui and Hankinson, 2009; Bui et al., 2009, 2011). Metabolism of endogenous substrates was shown to be independent of NADPH P450 reductase, because it requires peroxide utilization for metabolism (Bui and Hankinson, 2009; Bui et al., 2011). Of the endogenous substrates tested, COX-derived prostaglandins [e.g., prostaglandin G2 (PGG2) and prostaglandin H2 (PGH2)] were predicted as likely endogenous substrates for CYP2S1 isomerase activity (Bui et al., 2011). Arachidonic acid (AA) is converted to PGG2 and PGH2 via the COX enzymes. PGH2 is further metabolized to bioactive prostanoids, including prostaglandin E2 (PGE2). Using peroxide cofactors, CYP2S1 was able to metabolize PGG2 (Km = 270 nM) and PGH2 (Km = 11 μM) into numerous metabolites. The authors predicted that CYP2S1 expression may effectively divert synthesis of bioactive prostanoids and demonstrated depletion of PGE2 and prostaglandin D2 (PGD2) in mammalian cells overexpressing CYP2S1 when supplemented with the PGH2 precursor. Although it suggests that CYP2S1 influences PGE2 synthesis, whether this modulation is physiologically relevant remains to be determined (Nishida et al., 2010; Xiao et al., 2011) and is the subject of this investigation.
PGE2 is the most well studied COX-derived prostanoids. The physiological effects of PGE2 are mediated through PGE2 activation of its cognate G protein-coupled E prostanoid receptors (EP1–EP4) (reviewed in Wang and Dubois, 2006). In epithelial cells, PGE2 stimulates cell proliferation (Pai et al., 2002) and cell migration (Buchanan et al., 2003) while inhibiting apoptosis (Munkarah et al., 2002).
The purpose of this study was to identify the physiological significance, if any, of CYP2S1 in human bronchial epithelial cells [SV40-derived bronchial epithelial cell line (BEAS-2B)] by selectively depleting its expression and evaluating the cellular consequences. Our study reveals that the CYP2S1 expression and presumably changes in endogenous metabolism alter cell migration and proliferation in human lung cells. Cell migration and proliferation observed in CYP2S1-depleted cells appear to be mediated through disparate actions of distinct intracellular pathways. CYP2S1-depleted cells have twice the level of intracellular PGE2. Our results suggest that elevated PGE2 levels may contribute to enhanced cell proliferation, but not migration, in BEAS-2B cells. These data are consistent with the proposed physiological role for CYP2S1-mediated metabolism of PGG2 (Bui et al., 2011), supporting the idea that PGG2 may be a physiologically relevant substrate in bronchial epithelial cells. In addition, our data hint that CYP2S1-mediated metabolism influences other, as of yet unidentified, endogenous pathways that influence cell migration.
Materials and Methods
CYP2S1 Depletion Using Short Hairpin RNA.
BEAS-2B cells were plated in six-well plates and transfected with pLKO.1 short hairpin RNA (shRNA) plasmids (Sigma MISSION shRNA; Sigma-Aldrich, St. Louis, MO) bearing 21 nucleotide sequences directed against either CYP2S1 or the nontargeting scrambled control (SCHSCRAM, referred to as SCRAM). shRNA sequences targeting CYP2S1 were directed toward exon 3 (SCH00984, referred to as 984), and the 3′-untranslated region (UTR) (SCH000759, referred to as 759). The nontargeting SCRAM was used as a control. Stable individual colonies expressing the SCRAM, 759, and 984 shRNA were identified and isolated using puromycin (Sigma P8833; Sigma-Aldrich) selection. Stable colonies were analyzed for CYP2S1 expression using quantitative real-time polymerase chain reaction (qRT-PCR) and Western analysis, and the colonies with the greatest knockdown (SCRAM#1, 759#7, and 984#1) were used in our study.
qRT-PCR Analysis.
Cells were grown in replicates of three wells in six-well plates until confluent and were rinsed with phosphate-buffered saline (PBS). RNA was isolated according to the manufacturer's protocol (QIAGEN RNeasy; QIAGEN, Valencia, CA) and eluted in 40 μl of RNase-free water. Total RNA was then quantified using nano-drop spectrometric analysis as well as the Bioanalyzer nano-RNA protocol (Agilent Technologies, Santa Clara, CA). cDNA was synthesized from 1 μg of total RNA using iScript cDNA synthesis (Bio-Rad Laboratories, Hercules, CA) and diluted 1:5 in nuclease-free H2O and stored at −20°C. qRT-PCR was conducted using IQ SYBR green Supermix (Bio-Rad Laboratories). The real-time polymerase chain reactions were performed in a final volume of 10 μl using 1 μl of cDNA and 500 nM primers. qRT-PCR was conducted on a Bio-Rad CFX9600 programming (Bio-Rad Laboratories) in primer efficiency using standard curves obtained from plasmid amplicons. Primers were designed using Beacon Designer (Biosoft, Milltown, NJ) and Roche software suites (Roche Applied Science, Indianapolis, IN). The polymerase chain reaction primers for CYP2S1 gene amplification were 5′-AGGCGTTCCTGCCCTTCTCC-3′ (sense) and 5′-CAGTGGGACGGACTTGCAGC-3′ (antisense); ACTB gene amplifications were 5′-GACAACGGCTCCGGCATGTGCA-3′ (sense) and 5′-TGAGGATGCCTCTCTTGCTCTG-3′ (antisense). Five additional housekeeping genes were used to normalize CYP2S1 expression; their sequences (Supplemental Table 1) and results (Supplemental Fig. 1) are referred to in the table in supplemental materials.
Western Blot Analysis.
BEAS-2B cells (CLR-9609; American Type Culture Collection, Manassas, VA) were grown in 75-cm2 flasks until confluent, rinsed with PBS and isolated using the NE-PER isolation kit (Thermo Fisher Scientific, Waltham, MA). Cytoplasmic proteins were quantified using the (bicinchoninic acid) BCA protein assay kit (Thermo Fisher Scientific), according to the manufacturer's instructions, and were frozen at −80°C until analysis. Western analysis was performed using 50 μg of protein, which was reduced with DDT and loading buffer and boiled before loading on a 12% Bis-Tris gel (Invitrogen, Carlsbad, CA). The protein was run and transferred to a nitrocellulose membrane at 100 V for 1 h in transfer buffer. The membrane was then rinsed, blocked, and incubated overnight with CYP2S1 antibody (kindly provided by Dr. Roland Wolf, University of Dundee, Dundee, Scotland). We also used the commercially available antibody, CYP2S1 (C-19) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The protein was then visualized using the Femto chemiluminescent detection kit (Millipore, Billerica, MA). Dr. Oliver Hankinson (UCLA Jonsson Comprehensive Cancer Center, Los Angeles, CA) kindly provided the CYP2S1 protein loading control. The chemiluminescent signal was visualized using the Chemidoc XRS system (Bio-Rad Laboratories). Protein loading was controlled using the polyclonal anti-rabbit glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Sigma G9545; Sigma-Aldrich) antibody.
Wound-Healing Assay.
Bronchial epithelial cells were plated in replicates of three in six-well plates at a 300,000 cells/well for 24 h or until confluence was reached. The horizontal scratch was made in the center of the well using a p10 pipette tip. Immediately after the scratch, the cells were washed with PBS, and new media was added. Vertical lines were drawn across the scratch as reference markers for imaging. Images were taken at different time points t = 0 and 24 h using the Zeiss Axioscope II (Carl Zeiss, Inc., Thornwood, NY). Images from each time point were aligned using Photoshop CS3 Professional software suite (Adobe Systems, San Jose, CA), and migrating cells were counted to determine invasive characteristics of cell lines. Cells were counted in replicates of three independent biological experiments.
Cell Proliferation Assays.
Cell numbers were calculated using the hemocytometer. BEAS-2B cells were plated in a 96-well plate at 2000 to 4000 cells/well in replicates of at least six per treatment or genotype. Cells were allowed to incubate overnight at 5% CO2 at 37°C before the addition of alamarBlue (Invitrogen) was added to a final concentration of 10%. Media alone and in the presence of fully reduced alamarBlue (media containing autoclaved alamarBlue) was used to establish the background and 100% reduced alamarBlue. Percentage reduction of alamarBlue was calculated, and these values were used to compare cells. Care was taken to follow only the wells (n = 3 or more) that had similar fluorescent readings between samples at t = 0. These experiments were conducted at least three times in this manner. alamarBlue data were analyzed using a one-way analysis of variance in Prism statistical software (GraphPad Software, Inc., San Diego, CA), analyzing linear growth rates, and differences in growth were assessed at 24 and 48 h by post hoc t tests. All data were normalized to blank media wells (alamarBlue background fluorescence levels). For 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assays, Cells were plated as described above (See Cell Proliferation Assays introduction). MTT assays were performed according to the manufacturer's instructions (Sigma-Aldrich), and the absorbance was monitored at 560 nm using the BioTek FL600 plate reader (BioTek Instruments, Winooski, VT).
PGE2 Enzyme-Linked Immunosorbent Assy.
PGE2 level was measured according to the Prostaglandin E2 Biotrak Enzymeimmunoassay system (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). In brief, BEAS-2B cells were plated at 50,000 cells/well in a 96-well plate to reach confluence. The next day, total cellular PGE2 was assessed according to manufacturer's instructions (GE Healthcare). After lysis, total protein content was determined using the BCA protein assay kit (Thermo Fisher Scientific). PGE2 concentration was normalized to total protein content and was represented as picograms of PGE2 per microgram of total protein. Statistical analysis was performed using Student's t test.
Results
CYP2S1 shRNA-Reduced CYP2S1 mRNA and Protein Expression in Bronchial Epithelial BEAS-2B Cells.
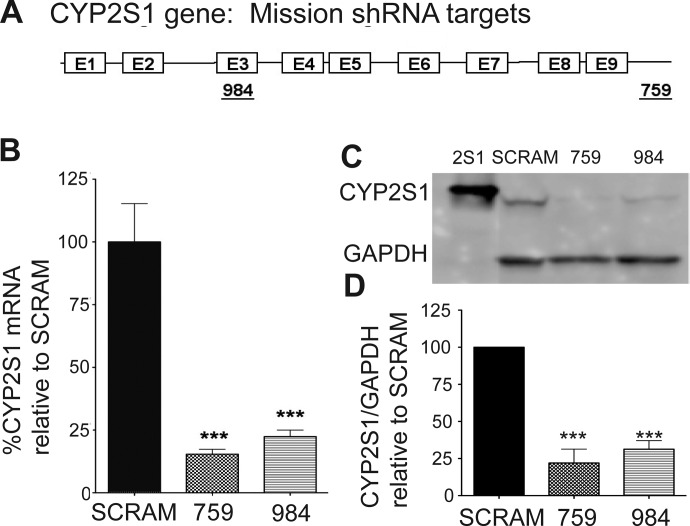
To determine whether changes in CYP2S1 gene expression influences bronchial epithelial cell physiology, we evaluated the shRNA sequences targeting the CYP2S1 mRNA. Sigma MISSION shRNA plasmids (Sigma-Aldrich) bearing 21 nucleotide sequences directed toward exon 3 (SCH00984, referred to as 984) as well as the 3′-UTR (SCH000759, referred to as 759) of CYP2S1 were used to deplete CYP2S1 expression (Fig. 1A). The nontargeting SCRAM was used as a control. We transfected each of the plasmids into BEAS-2B cells and isolated stable colonies derived from both 759 and 984. Stable colonies demonstrating the most significant difference in CYP2S1 expression relative to SCRAM controls were used for subsequent experiments. CYP2S1 mRNA expression was analyzed using qRT-PCR and normalized to the β-actin (ACTB) housekeeping gene. CYP2S1 mRNA was reduced by approximately 75% in both CYP2S1-depleted 759 (crosshatched bars) and CYP2S1-depleted 984 (horizontal lined bar) compared with the SCRAM (black bar) (Fig. 1B). This data were also normalized to five additional housekeeping genes with similar reductions in CYP2S1 mRNA (see Supplemental Fig. 1). Western analysis was performed using both the human CYP2S1 antibody (provided by Dr. Roland Wolf) and the commercially available antibody (see Materials and Methods). Each CYP2S1 antibody produced similar results (data not shown). BEAS-2B cells displayed a single band at 50 kDa, which was consistent with the positive CYP2S1 protein control (2S1; provided by Dr. Oliver Hankinson). Western (Fig. 1C) and densitometric analysis (Fig. 1D) revealed marked depletion of CYP2S1 protein levels by approximately 75 to 70% in 759 and 984, respectively, compared with the SCRAM. These CYP2S1 mRNA and protein reductions were stable through multiple passages as well as from older (multiple years) frozen stocks.
Fig. 1.
Small interfering RNA (siRNA)-mediated depletion of CYP2S1 expression in BEAS-2B cells. A, cartoon depiction of the CYP2S1 gene. Boxes indicate exons (1–9), and lines indicate introns as well as the 3′-UTR. siRNA target sequences, and their positions within the gene are indicated by the underlined numbers 759 (targeting the 3′-UTR) and 984 (targeting exon 3). B, qRT-PCR quantification of CYP2S1 mRNA transcripts in stably transformed cell lines expressing either the 759 (crosshatched bars) or 984 (horizontal line bars) shRNA plasmids was compared with that of SCRAM (closed bars). CYP2S1 mRNA derived from three independent experiments was normalized to the β-actin (ACTB) housekeeping gene and is represented as percentage of knockdown relative to SCRAM. C, representative Western blot of stably transformed cell lines bearing the control (SCRAM) and CYP2S1 siRNA (759 and 984). Top, CYP2S1 immunoreactivity. Bottom, GAPDH immunoreactivity. D, densitometric analysis for CYP2S1 immunoreactivity was performed on five independent Western blots and expressed as a percentage of density corresponding to SCRAM. Differences in protein loading are accounted for by normalization to GAPDH immunoreactivity. Statistical significance was determined using a Student's t test in both qRT-PCR (B) and densitometric (D) analysis. Data are represented as mean ± S.D. ***, p < 0.001.
Effects of CYP2S1 Depletion on Bronchial Epithelial Cell Migration and Proliferation.
Because CYP2S1 expression is elevated in epithelial-derived cancers (Downie et al., 2005; Kumarakulasingham et al., 2005; Murray et al., 2010), we tested the possibility that altering CYP2S1 expression in human bronchial epithelial cell would influence cellular pathways that either promote or inhibit cell proliferation and/or migration. To determine whether CYP2S1 expression alters cell proliferation and migration in human lung cells, we compared cell proliferation and migration in stably transformed SCRAM to cells with significantly reduced CYP2S1 mRNA and protein (759 and 984) (Figs. 2 and 3).
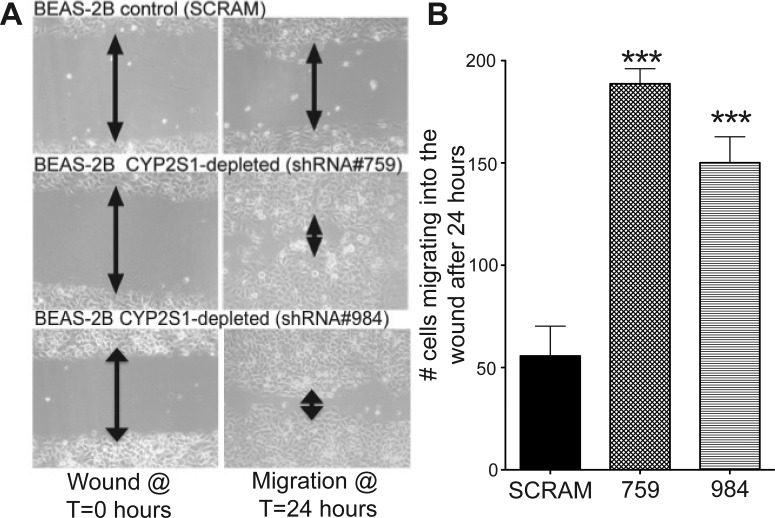
Fig. 2.
Depleting CYP2S1 expression in human lung cells promotes cell migration in BEAS-2B cells. A, representative images from the wound-healing assay, comparing cell migration in BEAS-2B cells with normal CYP2S1 expression (SCRAM, top) to two independent BEAS-2B clones with depleted CYP2S1 expression (759 and 984, middle and bottom, respectively). Images were acquired at two independent time points: time = 0 h (directly after wounding, left) and 24 h after wounding (right). B, the number of cells migrating into the wound were quantified 24 h after wounding in BEAS-2B cells stably transformed with SCRAM (closed bars) as well as CYP2S1-depleted BEAS-2B cells (759 and 984, crosshatched and horizontal line bars, respectively). Cell quantification is derived using three images each from three independent experiments. Data represent the mean ± S.D. ***, a significant increase of p < 0.001.
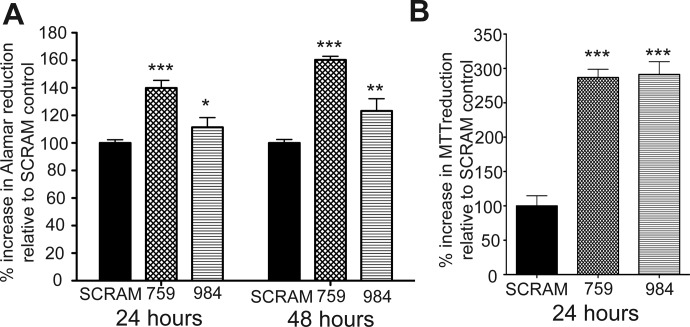
Fig. 3.
Depleting CYP2S1 expression in human lung cells promotes cell proliferation in BEAS-2B cells. A, alamarBlue proliferation assay was used to monitor cell proliferation. Proliferation was significantly increased in CYP2S1-depleted BEAS-2B cells (759 and 984, crosshatched and horizontal line bars, respectively) compared with SCRAM (closed bars). Data represent the mean ± S.D. of alamarBlue reduction at 24 and 48 h. One-way analysis of variance was used to determine differences between groups followed by post hoc t tests. B, the MTT was also assessed to evaluate differences in cell proliferation after 24 h of growth. Cell viability was also found to be significantly increased in CYP2S1-depleted BEAS-2B cells (759 and 984, crosshatched and horizontal line bars, respectively) compared with SCRAM (closed bars). Statistical analysis was performed using the Student's t test. *, a significance of p < 0.05; **, a significance of p < 0.01; and ***, a significance of p < 0.001.
To assess the effect of CYP2S1 depletion on bronchial epithelial migration, we performed the wound-healing assay. In brief, the two populations of cells were grown to confluence in six-well plates, and a wound was created using a 10-μl pipette tip. The progression of wound healing was visualized at different time points (0 and 24 h), and the number of cells invading the wound at 24 h was quantified (Fig. 2, A and B). This assay produced striking results. After 24 h, CYP2S1-depleted cells nearly covered the entire wound, whereas SCRAM hardly migrated. Quantification of these results revealed roughly 3-fold increase in cell migration in CYP2S1-depleted (∼150 cells) over SCRAM (∼50 cells) (Fig. 2B). Our results demonstrate that CYP2S1 depletion promotes cell migration, presumably by reducing CYP2S1-mediated metabolism of currently unknown endogenous substrates.
Differences in cell proliferation were assessed using two independent methods: alamarBlue and MTT cell-viability assays. The alamarBlue assay detects viable cells using the blue nonfluorescent dye (resazurin). Under conditions promoting cell proliferation, resazurin is reduced to its fluorescent form, resorufin, and detected by a fluorescent plate reader. MTT is a comparable cell-viability detection method that measures metabolic activity in viable cells through the reduction of the yellow tetrazole compound to a purple formazan that can be detected though absorbance changes at 562 nm.
CYP2S1-depleted cells (759 and 984) as well as SCRAM controls were plated at equal numbers (∼2000 cells) and allowed to proliferate for 24 h (alamarBlue and MTT) or up to 48 h (alamarBlue). Each cell type was assessed for increases in viability. Cell viability in CYP2S1-depleted cells was compared with that in SCRAM controls, which are designated as 100% growth (Fig. 3, A and B). CYP2S1-depleted cells exhibited statistically significant increases of 40 (759, crosshatched bars) and 15% (984, horizontal lined bars) in cell viability compared with SCRAM control (SCRAM, closed bars) at 24 h (Fig. 3A). After 48 h, 759 rose an additional 20 to 60% over SCRAM control, whereas 984 increased by roughly 5 to 20% increase over SCRAM control (Fig. 3A). It is interesting that the MTT assay demonstrated a more robust and consistent increase between the CYP2S1-depleted cell lines 759 (crosshatched bars) and 984 (horizontal lined bars). Both CYP2S1-depleted cells (759 and 984) exhibited roughly 300% increase over SCRAM control and were not statistically different from one another. We are uncertain as to what factors may contribute to the differences between these assays. However, these data clearly demonstrate that CYP2S1 depletion can enhance cell proliferation in human bronchial epithelial cells.
Elevated PGE2 in CYP2S1-Depleted Cells Promotes Cell Proliferation but Not Cell Migration.
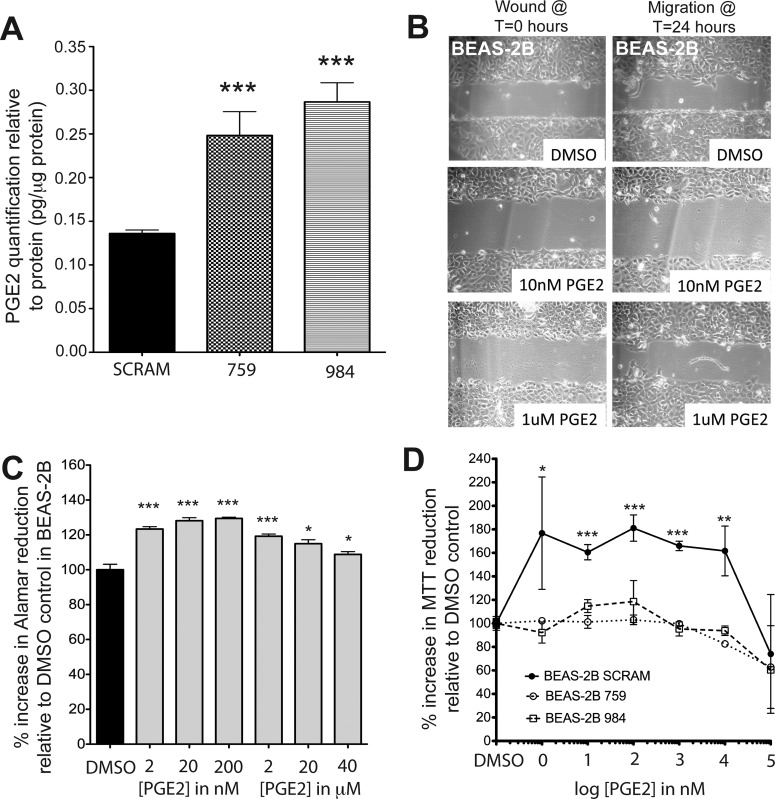
CYP2S1 was shown to metabolize bioactive lipids derived from the COX and LOX pathways in the absence of P450 reductase. Substrates included prostaglandins PGG2 and PGH2 from the COX pathway as well as numerous hydroperoxyeicosatetraenoic acid derivatives from the LOX pathway (Bui et al., 2011). The authors proposed that CYP2S1 metabolism of PGG2 and PGH2 could effectively divert the AA-derived production of PGE2 to CYP2S1-derived metabolites 12(S)-hydroxyheptadeca-5Z,8E,10E-trienoic acid (12-HHT) and thromboxane A2 (TXA2) (Bui et al., 2011). If true, CYP2S1 depletion should increase PGE2 levels (see schematic in Fig. 5). PGE2 enzyme-linked immunosorbent assay was performed to test whether there were differences in PGE2 levels between CYP2S1-depleted cells (759 and 984) and control (SCRAM). PGE2 levels in SCRAM were approximately 6 pg/well, or 0.15 pg/μg protein (Fig. 4A). This is similar to PGE2 concentrations reported previously (Cowan et al., 2006). In contrast, CYP2S1-depleted cells had nearly double the concentration of PGE2 (12 pg/well, or 0.3 pg/μg protein) in both 759 and 984 cell lines (Fig. 4A). Because PGE2 is known to promote cell migration and proliferation in a number of different cell types (Sheng et al., 2001), we tested its ability to enhance cell migration (Fig. 4B) and proliferation (Fig. 4C) in BEAS-2B. BEAS-2B cells were exposed to PGE2 at varying concentrations of PGE2. However, none of the concentrations of PGE2 tested was able to promote cellular migration. Thus, the migration observed in CYP2S1-depleted cells (Fig. 2) cannot be attributed to elevated PGE2 levels.
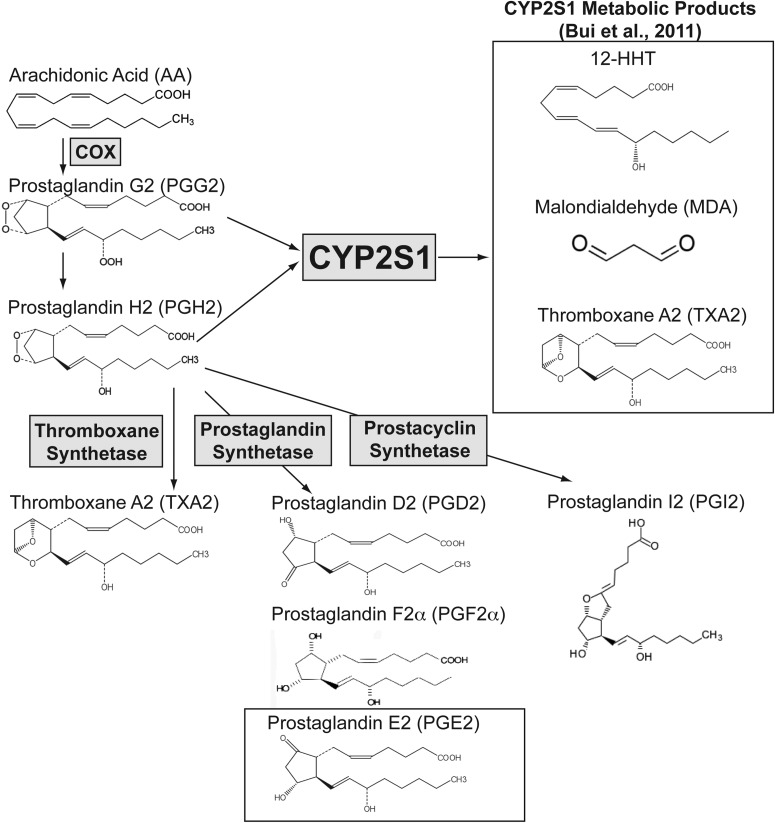
Fig. 5.
Proposed model for CYP2S1-mediated modulation of prostanoids, including PGE2. Free AA is converted to PGG2 and PGH2 via COX enzymes. PGH2 is converted to prostanoids through thromboxane synthetase (TXA2), prostaglandin synthetases (PGD2, prostaglandin F2α, and PGE2), and prostacyclin synthetase (prostaglandin I2). In vitro metabolic studies indicate that CYP2S1 can metabolize PGG2 (Km = 270 nM) and PGH2 (Km = 11 μM) to numerous products including the following: 12-HHT, MDA, and TXA2 (Bui et al., 2011). This could potentially divert synthesis away from PGE2. Consistent with this proposed role for CYP2S1 in prostaglandin metabolism, CYP2S1 depletion may elevate PGG2 and PGH2 precursors, increasing their availability for synthesis of downstream prostanoids, including PGE2. Enzymes are represented by names enclosed in gray boxes. Open boxes contain either CYP2S1 metabolites (12-HHT, MDA, and TXA2) or PGE2.
Fig. 4.
CYP2S1 depletion enhances PGE2 production, which may account for enhanced proliferation but not migration effects observed in CYP2S1-depleted cells. A, PGE2 enzyme-linked immunosorbent assay detected elevated intracellular PGE2 in BEAS-2B cells depleted of CYP2S1 (759 and 984, crosshatched and horizontal line bars, respectively) compared with SCRAM (closed bars). PGE2 levels were normalized to cellular protein content (BCA protein assay kit) within each well. Error bars indicate variability between each well. B, PGE2 supplementation alone failed to promote migration of nontransformed (normal) BEAS-2B cells into the wound. Confluent BEAS-2B cells were wounded and allowed to migrate for 24 h with the addition of 10 nM and 1 μM PGE2. C, in contrast, PGE2 supplementation at nanomolar concentrations was sufficient to enhance cell proliferation in normal BEAS-2B cells. Data from three experiments are represented as percentage increases in alamarBlue reduction 24 h after t = 0 control. Statistical analysis was performed using a standard t-test where *** represents p < 0.001. D, MTT assay measuring the metabolic activity in the cells reveals an increase in proliferation at 24 h in response to PGE2 in SCRAM controls (solid line, closed circles), but not CYP2S1-depleted (759 and 984, dashed lines with open circles and open squares, respectively) BEAS-2B cells. Each experiment was performed at least three times. DMSO, dimethyl sulfoxide. Statistical analysis was performed using one-way ANOVA followed by Tukey Kramer post-hoc analysis. *, **, and *** indicate significant increases from vehicle controls of p < 0.05, p < 0.01, and p < 0.001, respectively.
Next, we tested whether PGE2 promotes cell proliferation in BEAS-2B. We performed alamarBlue studies, which are similar to the MTT assay because they are both indirect measurements of cell viability. BEAS-2B cells grown in the presence of the lowest concentration tested (2 nM) exhibited a significant increase (∼20%) in alamarBlue reduction compared with vehicle control (dimethyl sulfoxide) at 24 h. Increased cell proliferation appeared to peak at approximately 200 nM (∼25% increase) and, though still significantly different, began approaching dimethyl sulfoxide control levels at 40 μM (Fig. 4C). The increase in cell viability attributed to PGE2 at 24 h (∼25%) was within the range of alamarBlue reduction increases observed in response to CYP2S1 depletion in 759 (∼40%) and 984 (∼15%) (Fig. 3A). The PGE2 concentration required to promote cell proliferation was in the low nanomolar range. Once this initial increase in cell viability was attained, increasing PGE2 levels failed to promote further statistical increases in cell proliferation and at micromolar concentrations appeared to reduce PGE2's proliferative effects on normal (nontransformed) BEAS-2B cells. These results are consistent with PGE2's biphasic effect on cell proliferation in other cell types (Baylink et al., 1996; Sergeeva et al., 1997), albeit at higher concentrations.
To determine whether the increased levels of PGE2 observed in CYP2S1-depleted cells were sufficient to promote the cell proliferation observed with PGE2, we evaluated the effects of PGE2 on CYP2S1-depleted cells. Because the MTT assay was shown in our studies to be a more robust indicator of differences in cell proliferation after 24 h (Fig. 3), we performed the MTT assay to measure cell viability of both SCRAM controls and CYP2S1-depleted (759 and 984) BEAS-2B cells in response to varying concentrations of PGE2 (Fig. 4D). Consistent with effects observed in nontransformed normal BEAS-2B (Fig. 4C), the lowest level of PGE2 tested (1 nM) was sufficient to enhance cell viability by ∼60% in SCRAM controls. This increase is MTT reduction was sustained until PGE2 concentrations reached ∼100 μM. In contrast, cell proliferation was not enhanced in CYP2S1-depleted cells in response to exogenous PGE2 at any concentration. Though not statistically significant, we observe reduced viability in CYP2S1-depleted cells (759 in particular) at 10 μM rather than 100 μM concentrations in SCRAM controls. These data would be consistent with increased concentration of intracellular PGE2 observed in CYP2S1-depleted cells. Taken together, our data suggest that elevated intracellular levels of PGE2 contribute to increased cell proliferation observed in CYP2S1-depleted cells. These data are also consistent with the proposed role of CYP2S1 in metabolizing the PGE2 precursor PGG2, which would divert the cellular production of PGE2, and support the hypothesis that PGG2 is a physiologically relevant substrate for CYP2S1 in human bronchial epithelial cells.
Discussion
In our current study, we reveal an important contribution of the human CYP2S1 enzyme in the regulation of cell growth and migration in BEAS-2B cells, which are used as surrogates for normal bronchial epithelial cells. In particular, by depleting CYP2S1 expression and ultimately CYP2S1-mediated metabolism of endogenous substrates, cell proliferation and migration are enhanced. Proliferation and migration appear to be functionally divergent, suggesting that CYP2S1 depletion promotes each through distinct endogenous substrates, metabolites, or perturbation of downstream bioactive molecules.
Heterologous expression and metabolic studies using a synthetic CYP2S1 enzyme (Bui and Hankinson, 2009) have demonstrated metabolic activity toward potential endogenous substrates of the CYP2S1 enzyme, including the following: lipid products derived from the AA cascade (Bui et al., 2011) as well as all-trans-retinoic acid (Bui and Hankinson, 2009). Free AA is converted to bioactive eicosanoid metabolites via metabolism through either the LOX enzymes or the COX enzymes. Heterologous expression of the synthetic CYP2S1 enzyme has demonstrated P450 reductase-independent metabolic activity toward both LOX- and COX-derived eicosanoids. It is predicted that the most relevant CYP2S1-mediated metabolism may be of the COX-derived prostaglandin intermediate, PGG2, because of its low Km (270 nM). A summary of the AA cascade as well as the proposed role for CYP2S1 in prostanoid metabolism is depicted in Fig. 5. The first step in the COX pathway is to convert AA to PGG2 through COX peroxidase activity. PGG2 is then converted via COX enzymes or nonenzymatically to PGH2. PGH2, in turn, is metabolized via thromboxane, prostaglandin, and prostacyclin synthetases to TXA2, prostaglandins (PGD2, prostaglandin F2a, and PGE2), prostacyclin (prostaglandin I2) (reviewed in Kroetz and Zeldin, 2002; Wang and Dubois, 2006; Panigrahy et al., 2010). CYP2S1 is able to convert PGG2 (Km = 270 nM) and PGH2 (Km = 11 μM) to multiple products including 12-HHT, malondialdehyde (MDA), and TXA2 (Bui et al., 2011). If CYP2S1 plays an essential role in modulating prostanoid synthesis in human lung cells, we would predict that CYP2S1 depletion would enable more of the PGG2 precursor to be converted to PGH2 and its subsequent prostanoid products, including PGE2. Indeed, our results in BEAS-2B cells show a roughly 2-fold increase in intracellular PGE2 synthesis (Fig. 4A). On the other hand, elevated CYP2S1 expression in mouse hepatocyte (Hepa-1) cells reduced PGE2 and PGD2 levels (Bui et al., 2011). Our CYP2S1 depletion results are consistent with a modulatory role for CYP2S1 in the synthesis of COX-derived prostanoids and regulation of PGE2, in particular.
Of the prostanoids, PGE2 is the most studied and has been implicated in cell proliferation and cell migration. PGE2 elicits myriad cellular effects through binding to and activation of its cognate EPs. Four G-protein-coupled EP receptor subtypes have been identified: EP1, EP2, EP3, and EP4. EP1 stimulates increased intracellular calcium though activation of phospholipase C. EP2 and EP4 are linked to Gαs, and it activates adenylate cyclase (AC) and increases cAMP synthesis. On the other hand, EP3 is linked to Gαi and decreases AC activity and cAMP synthesis. BEAS-2B cells were shown to express each EP receptor mRNA (Tavakoli et al., 2001; N′Guessan et al., 2007) and protein (N′Guessan et al., 2007), suggesting a full complement of activity mediated through PGE2 activation.
PGE2 stimulates proliferation in epithelial cell lines, including non-small-cell lung cancer cell lines. Our data reveal a previously undocumented role for PGE2 in stimulating cell proliferation within BEAS-2B cells (Fig. 4C). PGE2-stimulated proliferation occurs through EP1 (Krysan et al., 2005)-stimulated activation of extracellular signal-regulated kinase (ERK) signaling, EP4-stimulated signaling and transactivation of the epidermal growth factor receptor signaling (Pai et al., 2002), and downstream activation of the integrin-linked kinase (Zheng et al., 2009). Krysan et al. (2005) did not observe rapid stimulation of ERK phosphorylation in BEAS-2B cells, suggesting that the PGE2-mediated proliferation of BEAS-2B is not a consequence of EP1 activation of ERK signaling. Our proliferation data appear to be consistent with EP4 transactivation of epidermal growth factor receptor, which was identified to promote cell cycle progression in gastric epithelial cells, which was arrested in Go/G1 in response to a selective EP4 antagonist (Pai et al., 2002). Our results demonstrate that PGE2 is elevated in CYP2S1-depleted cells (Fig. 4A) and that the elevated PGE2 levels within these cells is sufficient to promote maximal cell viability, which, unlike SCRAM cells, is not further enhanced with exogenous PGE2 (Fig. 4D). However, the exact mechanism by which elevated PGE2 in CYP2S1-depleted cells promotes cell proliferation, and whether this is the only bioactive molecule responsible for increasing cell proliferation, is a subject of further investigation in our laboratory.
PGE2 promotes cell migration via EP4 activation of sarcoma (Src) signaling in human alveolar carcinoma cells (A549) (Kim et al., 2010). However, we were unable to phenocopy the enhanced migration phenotype observed in CYP2S1-depleted cells by exogenous application of PGE2, even at micromolar concentrations. These data suggest that although PGE2 levels are increased in CYP2S1-depleted cells (759 and 984), the cellular migration phenotype observed in these cells cannot be attributed to PGE2-mediated activation of EP4 and downstream Src signaling. PGE2 promotes cell migration in normal human bronchial epithelial cells (Savla et al., 2001); however, our results suggest that PGE2 alone is not sufficient to promote wound healing in BEAS-2B. This observation is consistent with published results from Cowan et al. (2006), whereby exogenous application of PGE2 fails to promote cell migration in BEAS-2B. It is possible that CYP2S1 depletion could influence the production of other eicosanoids or possibly novel endogenous substrate(s) and/or metabolites linked to cell migration. Our laboratory is actively pursuing the mechanism responsible for increased cell migration in response to CYP2S1 depletion.
In conclusion, this study is the first in vitro demonstration that CYP2S1 depletion promotes cell proliferation and migration in human lung cells. Our data provide further evidence supporting a modulatory role for CYP2S1 in regulating prostaglandin (specifically, PGE2) synthesis. It also demonstrates a functional role for PGE2 in enhancing cell proliferation in bronchial epithelial cells. More experiments are required to elucidate the EP receptors and signaling pathways responsible for promoting cell proliferation in CYP2S1-depleted cells. Future studies should reveal other potentially novel CYP2S1 endogenous substrates and/or metabolites responsible for the observed effects of CYP2S1 depletion on enhanced cell migration. Overall, our data suggest an important physiological role for CYP2S1 in regulating cell migration and proliferation and may ultimately have implications in carcinogenesis, hyperproliferative disease, and inflammatory disease.
Supplementary Material
This work was supported in part by the Society of Toxicology Colgate-Palmolive Postdoctoral Fellowship Award in In Vitro Toxicology (to A.M.R.) and by New Mexico State University startup funds. Graduate (to T.M.F.) and undergraduate (to N.B.) financial support was provided through the Minority Biomedical Research Support-Research Initiative for Scientific Enhancement (RISE)-New Mexico State University RISE to Excellence Grant, National Institutes of Health National Institute of General Medical Sciences [Grant R25-GM061222].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- P450
- cytochrome P450
- COX
- cyclooxygenase
- LOX
- lipoxygenase
- PGG2
- prostaglandin G2
- PGH2
- prostaglandin H2
- PGE2
- prostaglandin E2
- PGD2
- prostaglandin D2
- EP
- E prostanoid receptor
- shRNA
- short hairpin RNA
- UTR
- untranslated region
- SCRAM
- scrambled control small interfering RNA
- qRT-PCR
- quantitative real-time polymerase chain reaction
- PBS
- phosphate-buffered saline
- BCA
- bicinchoninic acid
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- AA
- arachidonic acid
- 12-HHT
- 12(S)-hydroxyheptadeca-5Z,8E,10E-trienoic acid
- TXA2
- thromboxane A2
- MDA
- malondialdehyde
- AC
- adenylate cyclase
- ERK
- extracellular signal-regulated kinase
- Src
- sarcoma
- siRNA
- small interfering RNA.
Authorship Contributions
Participated in research design: Madanayake, Fidler, and Rowland.
Conducted experiments: Madanayake, Fidler, Fresquez, and Bajaj.
Performed data analysis: Madanayake, Fidler, and Rowland.
Wrote or contributed to the writing of the manuscript: Madanayake, Fidler, and Rowland.
References
- Baylink TM, Mohan S, Fitzsimmons RJ, Baylink DJ. (1996) Evaluation of signal transduction mechanisms for the mitogenic effects of prostaglandin E2 in normal human bone cells in vitro. J Bone Miner Res 11:1413–1418 [DOI] [PubMed] [Google Scholar]
- Buchanan FG, Wang D, Bargiacchi F, DuBois RN. (2003) Prostaglandin E2 regulates cell migration via the intracellular activation of the epidermal growth factor receptor. J Biol Chem 278:35451–35457 [DOI] [PubMed] [Google Scholar]
- Bui P, Imaizumi S, Beedanagari SR, Reddy ST, Hankinson O. (2011) Human CYP2S1 metabolizes cyclooxygenase- and lipoxygenase-derived eicosanoids. Drug Metab Dispos 39:180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui PH, Hankinson O. (2009) Functional characterization of human cytochrome P450 2S1 using a synthetic gene-expressed protein in Escherichia coli. Mol Pharmacol 76:1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui PH, Hsu EL, Hankinson O. (2009) Fatty acid hydroperoxides support cytochrome P450 2S1-mediated bioactivation of benzo[a]pyrene-7,8-dihydrodiol. Mol Pharmacol 76:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan MJ, Coll T, Shelhamer JH. (2006) Polyamine-mediated reduction in human airway epithelial migration in response to wounding is PGE2 dependent through decreases in COX-2 and cPLA2 protein levels. J Appl Physiol 101:1127–1135 [DOI] [PubMed] [Google Scholar]
- Downie D, McFadyen MC, Rooney PH, Cruickshank ME, Parkin DE, Miller ID, Telfer C, Melvin WT, Murray GI. (2005) Profiling cytochrome P450 expression in ovarian cancer: identification of prognostic markers. Clin Cancer Res 11:7369–7375 [DOI] [PubMed] [Google Scholar]
- Kim JI, Lakshmikanthan V, Frilot N, Daaka Y. (2010) Prostaglandin E2 promotes lung cancer cell migration via EP4-betaArrestin1-c-Src signalsome. Mol Cancer Res 8:569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroetz DL, Zeldin DC. (2002) Cytochrome P450 pathways of arachidonic acid metabolism. Curr Opin Lipidol 13:273–283 [DOI] [PubMed] [Google Scholar]
- Krysan K, Reckamp KL, Dalwadi H, Sharma S, Rozengurt E, Dohadwala M, Dubinett SM. (2005) Prostaglandin E2 activates mitogen-activated protein kinase/Erk pathway signaling and cell proliferation in non-small cell lung cancer cells in an epidermal growth factor receptor-independent manner. Cancer Res 65:6275–6281 [DOI] [PubMed] [Google Scholar]
- Kumarakulasingham M, Rooney PH, Dundas SR, Telfer C, Melvin WT, Curran S, Murray GI. (2005) Cytochrome p450 profile of colorectal cancer: identification of markers of prognosis. Clin Cancer Res 11:3758–3765 [DOI] [PubMed] [Google Scholar]
- Munkarah AR, Morris R, Baumann P, Deppe G, Malone J, Diamond MP, Saed GM. (2002) Effects of prostaglandin E(2) on proliferation and apoptosis of epithelial ovarian cancer cells. J Soc Gynecol Investig 9:168–173 [PubMed] [Google Scholar]
- Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD. (2010) Profiling the expression of cytochrome P450 in breast cancer. Histopathology 57:202–211 [DOI] [PubMed] [Google Scholar]
- N′Guessan PD, Temmesfeld-Wollbrück B, Zahlten J, Eitel J, Zabel S, Schmeck B, Opitz B, Hippenstiel S, Suttorp N, Slevogt H. (2007) Moraxella catarrhalis induces ERK- and NF-kappaB-dependent COX-2 and prostaglandin E2 in lung epithelium. Eur Respir J 30:443–451 [DOI] [PubMed] [Google Scholar]
- Nishida CR, Lee M, de Montellano PR. (2010) Efficient hypoxic activation of the anticancer agent AQ4N by CYP2S1 and CYP2W1. Mol Pharmacol 78:497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai R, Soreghan B, Szabo IL, Pavelka M, Baatar D, Tarnawski AS. (2002) Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat Med 8:289–293 [DOI] [PubMed] [Google Scholar]
- Panigrahy D, Kaipainen A, Greene ER, Huang S. (2010) Cytochrome P450-derived eicosanoids: the neglected pathway in cancer. Cancer Metastasis Rev 29:723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera SP, Saarikoski ST, Hankinson O. (2002) Identification of a novel dioxin-inducible cytochrome P450. Mol Pharmacol 61:255–259 [DOI] [PubMed] [Google Scholar]
- Rylander T, Neve EP, Ingelman-Sundberg M, Oscarson M. (2001) Identification and tissue distribution of the novel human cytochrome P450 2S1 (CYP2S1). Biochem Biophys Res Commun 281:529–535 [DOI] [PubMed] [Google Scholar]
- Saarikoski ST, Rivera SP, Hankinson O, Husgafvel-Pursiainen K. (2005) CYP2S1: a short review. Toxicol Appl Pharmacol 207:62–69 [DOI] [PubMed] [Google Scholar]
- Savla U, Appel HJ, Sporn PH, Waters CM. (2001) Prostaglandin E(2) regulates wound closure in airway epithelium. Am J Physiol Lung Cell Mol Physiol 280:L421–L431 [DOI] [PubMed] [Google Scholar]
- Sergeeva MG, Gonchar MV, Mevkh AT, Varfolomeyev SD. (1997) Prostaglandin E2 biphasic control of lymphocyte proliferation: inhibition by picomolar concentrations. FEBS Lett 418:235–238 [DOI] [PubMed] [Google Scholar]
- Sheng H, Shao J, Washington MK, DuBois RN. (2001) Prostaglandin E2 increases growth and motility of colorectal carcinoma cells. J Biol Chem 276:18075–18081 [DOI] [PubMed] [Google Scholar]
- Smith G, Wolf CR, Deeni YY, Dawe RS, Evans AT, Comrie MM, Ferguson J, Ibbotson SH. (2003) Cutaneous expression of cytochrome P450 CYP2S1: individuality in regulation by therapeutic agents for psoriasis and other skin diseases. Lancet 361:1336–1343 [DOI] [PubMed] [Google Scholar]
- Tavakoli S, Cowan MJ, Benfield T, Logun C, Shelhamer JH. (2001) Prostaglandin E(2)-induced interleukin-6 release by a human airway epithelial cell line. Am J Physiol Lung Cell Mol Physiol 280:L127–L133 [DOI] [PubMed] [Google Scholar]
- Wang D, Dubois RN. (2006) Prostaglandins and cancer. Gut 55:115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Shinkyo R, Guengerich FP. (2011) Cytochrome P450 2S1 is reduced by NADPH-cytochrome P450 reductase. Drug Metab Dispos 39:944–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ritzenthaler JD, Sun X, Roman J, Han S. (2009) Prostaglandin E2 stimulates human lung carcinoma cell growth through induction of integrin-linked kinase: the involvement of EP4 and Sp1. Cancer Res 69:896–904 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.