Abstract
Prostaglandin reductase 1 (PTGR1) is a highly inducible enzyme with enone reductase activity. Previous studies demonstrated the role of rat PTGR1 in the activation of acylfulvene analogs, a class of antitumor natural product derivatives. Of these, hydroxymethylacylfulvene (HMAF) was in advanced clinical development for the treatment of advanced solid tumors, including prostate, ovarian, and pancreatic cancers. However, the efficiency of human PTGR1 in activating acylfulvenes and its potential to enhance therapeutic efficacy have remained uncharacterized. In this study, human PTGR1 was polymerase chain reaction-cloned and purified. Conversion of HMAF to its cellular metabolite by the purified enzyme proceeded at a 20-fold higher rate than with the rat variant of the enzyme. The Km was 4.9 μM, which was 40-fold lower than for the rat variant and similar to the therapeutic dose. Human cell lines, including colon cancer lines, were transfected with a vector containing rat PTGR1 or human PTGR1, and cell viability was examined after dosing with HMAF. New data obtained in this study suggest that transfection with human PTGR1, or its induction in colon and liver cancer cell lines with 1,2-dithiol-3-thione, enhances susceptibility to the cytotoxic influences of HMAF by 2- to 10-fold. Furthermore, similar or enhanced enzyme induction and HMAF toxicity results from preconditioning cancer cells with the bioactive food components curcumin and resveratrol. The functional impact of PTGR1 induction in human cells and chemical-based strategies for its activation can provide important knowledge for the design of clinical strategies involving reductively activated cytotoxic chemotherapeutics.
Introduction
Chemotherapeutic agents, together with surgery, radiation, and immunotherapy, are key components of effective modern anticancer therapy. Among them, alkylating agents are highly effective tumor cytotoxins. These drugs elicit therapeutic responses by covalently binding to cellular nucleophiles, such as DNA and proteins, thereby leading to stalled DNA synthesis, inhibited enzyme function, and tumor cell death. Several classes of alkylating agents, including nitrogen mustards (i.e., cyclophosphamide and ifosphamide) and quinones (i.e., mitomycin C) gain a selective advantage in tumor cells on the basis of enzymatic bioactivation that unmasks chemical electrophiles (Powis, 1983). A detailed understanding of the chemical and biochemical mechanisms of bioactivation, including the identification of specific drug-metabolizing enzymes in human cells, can improve drug design and clinical therapeutic strategies.
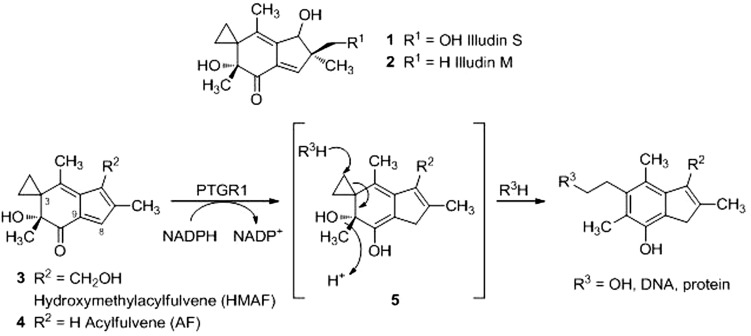
Acylfulvenes (AFs) are semisynthetic derivatives of the mycotoxin illudin S (1; Scheme 1) (Kelner et al., 1996; McMorris et al., 1996). Illudins (1 and 2; Scheme 1) are potent cytotoxins at nanomolar concentrations in several human cancer cell lines (Kelner et al., 1997), but have poor therapeutic indices and are associated with substantial systemic cytotoxicity in animal models (Kelner et al., 1987). AF (4; Scheme 1) and its analogs, including clinically tested hydroxymethylacylfulvene (HMAF, marketed as irofulven; 3; Scheme 1), are up to two orders of magnitude less cytotoxic than illudin S, but retain efficacy in tumor cell lines and xenografts (MacDonald et al., 1997; Gong et al., 2006). Outcomes from clinical trials with HMAF were mixed, ranging from little effects in non–small-cell lung cancer and gastric cancer to modest effects in ovarian and pancreatic cancer. The array of side effects included thrombocytopenia, nausea and vomiting, visual symptoms, and renal dysfunction (Tanasova and Sturla, 2012). These observations suggest a pressing need to better understand factors controlling toxicity with the aim of devising improved analogs or therapeutic strategies.
Scheme 1.
Bioactivation of acylfulvenes by PTGR1. The structures of illudin M (1), illudin S (2), HMAF (3), and AF (4) are shown. Reduction at the C8 position results in the unstable cyclohexadiene intermediate 5, which may react with water or other cellular nucleophiles (R3).
The metabolic enzyme prostaglandin reductase 1 (PTGR1)1 has been invoked as a key activating enzyme that significantly influences the activities of AF analogs (Dick et al., 2004; Gong et al., 2006, 2007). PTGR1 was originally identified as leukotriene B4 dehydrogenase (Yokomizo et al., 1993), and it transforms prostaglandins by enone reduction (Tai et al., 2002; Yu et al., 2006). PTGR1 was later discovered to be strongly induced by 1,2-dithiol-3-thione (D3T) in rat liver by nuclear factor E2-related factor 2 (Nrf2)-mediated gene activation (Primiano et al., 1996; Dick et al., 2001) and further characterized as alkenal/one oxidoreductase for its capacity to reduce endogenous and exogenous α,β-unsaturated aldehydes and ketones (Dick et al., 2001; Dick and Kensler, 2004; Gong et al., 2006, 2007; Yu et al., 2006; Neels et al., 2007).
Rat PTGR1 (rPTGR1) was shown to be a determining factor in the activation of AFs in vitro (Dick et al., 2004); however, the competence of human PTGR1 (hPTGR1) in HMAF bioactivation has not been defined. The primary sequences of rPTGR1 and hPTGR1 are 83% identical and 329 amino acids in length. Primary sequence differences include the NADPH binding domain (Supplemental Fig. 1; Hori et al., 2004). With NADPH as cofactor and AF analogs as substrates, enone reduction at the 8,9-double bond primes the AF cyclopropane for nucleophilic ring opening and results in aromatization of the six-member ring (5; Scheme 1) (Dick et al., 2004). Ectopic expression of rPTGR1 in human embryonic kidney 293 (HEK293) cells reduced the IC50 of HMAF 100-fold. Furthermore, reductase activity in NCI-60 cell line extracts correlates with their sensitivity to HMAF (Dick et al., 2004), but there is no direct information available regarding the bioactivation of HMAF by hPTGR1. Data obtained in this study establishes the role of hPTGR1 in HMAF bioactivation and explores strategies for sensitizing human tumor cells to the drug.
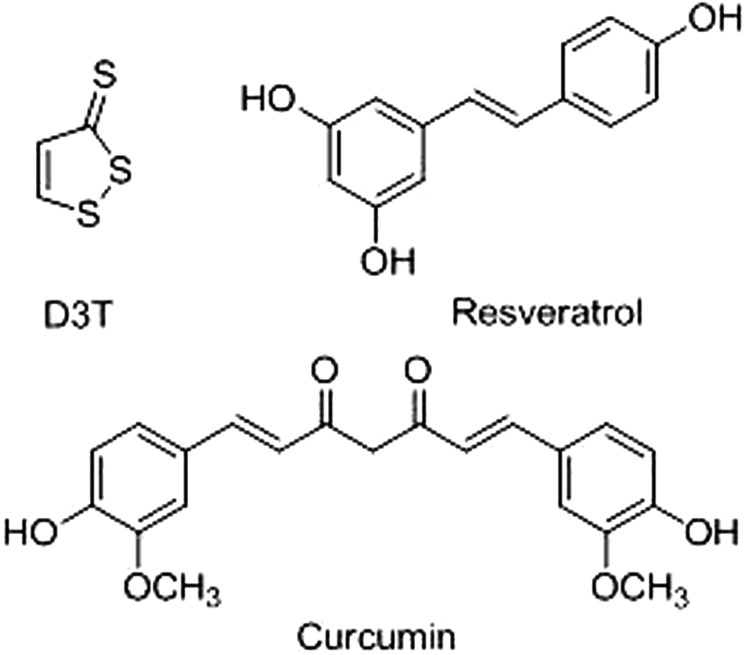
In response to a wide range of endogenous and exogenous signals, many enzymes are regulated by the transcription factor Nrf2, a central regulator in xenobiotic metabolism and cellular stress response. Nrf2 triggers include thiol-modifying chemicals, such as dithiolthiones and sulforaphane, and Michael-type acceptors, such as triterpenoids and retinoid acid derivatives, among others (Fig. 1) (Kwak et al., 2004). Reactions with cysteine residues on the protein Keap1, a negative regulator of Nrf2, lead to Nrf2 translocation from the cytoplasm into the nucleus. Coupled with cofactors in the nucleus, Nrf2 recognizes the antioxidant response element (ARE) in the promoter regions of its target genes and initiates transcriptional activation (Kwak et al., 2001). This molecular mechanistic paradigm offers possible options to modulate PTGR1 expression in human cells and, in addition, carries implications for potential anticancer therapies involving a PTGR1 inducer in combination with bioreductive drugs.
Fig. 1.
Structures of D3T (top left) and the dietary components curcumin [(1E,6E)-1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione[rsqb] (bottom) and resveratrol (3,4′,5-stilbenetriol) (top right), three examples of Nrf2 triggers used in this study.
Materials and Methods
Chemicals and Reagents.
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. All solvents and chemicals were of analytical reagent grade or higher. HMAF was provided by Eisai Co. (Tokyo, Japan).
Plasmid Construction.
hPTGR1 was amplified by polymerase chain reaction (PCR) from a human adult liver cDNA library (Clontech, Mountain View, CA), according to the National Center for Biotechnology Information GenBank sequence (accession NM_012212) using the following primer set: forward, 5′-GTCGCGGAATTCAGCTTCAGGATGGTTCGTACTAAGACATGG and reverse, 5′-GTCGCGCTCGAGTTACTATCATGCTTTCACTATTGTCTTCCCC. PCR product was cleaned and ligated into pBlueScript (Agilent Technologies, Santa Clara, CA) between EcoRI and XhoI sites. DNA insert was confirmed by sequencing and subcloned into the episomal vector pCEP4.
For N-terminal FLAG-tagged recombinant protein expression, both rPTGR1 and hPTGR1 were PCR cloned by using the following primer sets to omit the starting ATG codon: rPTGR1 forward, GCGAATTCAGTACAAGCTAAGACCTGGAC; rPTGR1 reverse, GCTCTAGATCACGCTTTCACTATAGTCTTCC; hPTGR1 forward, GCGAATTCAGTTCGTACTAAGACATGGAC; and hPTGR1 reverse, GCTCTAGATCATGCTTTGACTATTGTCTTCC. PCR products were cleaned and ligated into p3×FLAG-myc-CMV-24 (Sigma-Aldrich,) at EcoRI and XbaI sites to produce N-terminal FLAG-tagged proteins. DNA insert was confirmed by sequencing.
Cell Culture and Transfection.
HEK293, HCT15, SW620, and HepG2 cells were obtained from the American Type Culture Collection (Manassas, VA) and grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and incubated at 37°C in a humidified atmosphere containing 5% CO2. Plasmids were transfected into cells with Lipofectamine 2000 reagents (Invitrogen) according to the manufacturer's recommendations (http://tools.invitrogen.com/content/sfs/manuals/lipofectamine2000_man.pdf).
Recombinant Protein Expression and Purification.
rPTGR1 was expressed in Escherichia coli as described previously (Dick et al., 2004). p3×FLAG-hPTGR1 was transiently transfected into HEK293 cells as described above. Cells were harvested 24 h later and resuspended in lysis buffer (50 mM Tris, pH 7.4, 1 mM EDTA, 150 mM NaCl, and 1% Triton X-100) at a concentration of 1.5 × 107 cells/ml buffer. Cell lysate was centrifuged for 15 min at 12,000g to eliminate cell debris. Supernatant was incubated with anti-FLAG agarose gel (Sigma-Aldrich) at a ratio of 1 ml per 200 μl of gel and mixed for 2 h at 4°C. Agarose gel was washed with five bed volumes of Tris buffer (50 mM Tris, pH 7.4, 1 mM EDTA, and 150 mM NaCl) three times and eluted three times with one bed volume wash buffer containing 500 μg/ml 3×FLAG peptide. The eluent was dialyzed against 25 mM Tris buffer, pH 7.4, and 1 mM dithiolthreitol at 4°C overnight, and aliquots were adjusted to 10% glycerol and stored at −80°C.
Cell Viability Assay.
pCEP4-, pCEP4-rPTGR1-, and pCEP4-hPTGR1-transfected HEK293 cells were maintained in DMEM supplemented with 10% FBS and 100 μg/ml hygromycin B (Invitrogen). Cells were plated in 96-well culture plates 18 h before treatment at a density of 4000 cells/well. Toxicity studies were conducted by replacing medium with the complete medium containing HMAF [stock solutions prepared in 100% dimethyl sulfoxide (DMSO)] at the desired concentrations. Cell viability was measured 24 h later by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (Invitrogen). HCT15, SW620, and HepG2 cells were maintained in DMEM with 10% FBS. pCep4-green fluorescent protein or pCep4-PTGR1-transfected HCT15 cells were seeded in 96-well plates at 8000 cells/well 6 h before treatment. Toxicity studies were conducted as mentioned above. Cell viability was measured 18 h later via Cell Titer-Blue assay (Promega, Madison, WI). SW620 and HepG2 cells were incubated with 20 μM D3T, 10 μM curcumin, or 15 μM resveratrol in DMEM + 10% FBS for 48 h before seeding in the 96-well plates. After washing out inducers with 1× phosphate-buffered saline (PBS), cytotoxicity assay was carried out as described for HCT15 cells. Survival fraction of each cell population was calculated by normalizing against the vehicle-treated population within each experimental arm. IC50 values were calculated via mathematical simulations of nonlinear regression based on data obtained from three or more independent experiments. Regression and statistical analyses were performed by using SigmaPlot (version 10.0; Systat Software, Inc., San Jose, CA).
Enzyme Kinetic Studies.
Because of the low yield of hPTGR1 from mammalian cells compared with rPTGR1 from bacterial cells, a very conservative protocol needed to be established for hPTGR1. In the process of developing the protocol for hPTGR1, rPTGR1 was used to assess the method, and results highly comparable with the standard rPTGR1 protocol were obtained. Initial velocities were measured by monitoring the decay of the absorbance at 420 nm (2210 cm−1 · M−1 for HMAF) (Gong et al., 2006) at 37°C with a Cary 100 Bio UV-visible spectrophotometer (Agilent Technologies) or a Beckman DU 800 spectrophotometer (Beckman Coulter, Fullerton, CA). Stock solutions of analytes were prepared in DMSO (50 mM) and diluted to a final concentration between 0.05 and 0.4 mM. The percentage of DMSO in each experiment was below 0.2% of the total volume. A saturating concentration of NADPH was used (150 μM) in 100 μl of reaction buffer (0.5× PBS supplemented with 0.01% Triton X-100 and 0.05% bovine serum albumin) with 1 μg of recombinant enzyme for hPTGR1 and rPTGR1 in 1 ml of reaction buffer with 5.1 μg of recombinant enzyme. Cell lysates (100 μg) were used to calculate the reaction rate for transfected cells. The UV-visible absorbance spectrum was recorded for 4 min. Km and Vmax values were calculated by using SigmaPlot (version 12.2).
Promoter Analysis.
Consensus sequences for transcription factor binding sites in the promoter region of PTGR1 were analyzed with Genomatix Gene2promoter software (www.genomatix.com). Sequences of PTGR1 with experimentally verified 5′ transcripts from database ElDorado 12-2010 (Genomatix) were used. Up to 1000 base pairs upstream of the transcription starting site was analyzed, and consensus transcriptional factor binding sites were identified.
Immunoblot Analysis.
Cultured cells were seeded in six-well culture plates at 1 × 106 cells/well 18 h before treatment with 20 μM D3T, 10 or 20 μM curcumin, 15 or 40 μM resveratrol, or DMSO (vehicle control) in the growth medium. After 48-h preconditioning, cells were washed with ice-cold 1× PBS and collected by centrifugation after trypsinization. After seeding for viability assay, the remaining cells were resuspended in radioimmunoprecipitation assay buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS). The suspension was vortex-mixed, and lysate was collected by centrifugation at 12,000g. Protein extracts were analyzed by SDS-polyacrylamide gel electrophoresis and electrotransferred to a nitrocellulose membrane with iBlot (Invitrogen). Immunodetection was performed by using mouse anti-FLAG antibody (Sigma-Aldrich) and anti-PTGR1 antibody (leukotriene B4 reductase polyclonal antibody; Abnova, Walnut, CA,) followed by horseradish peroxidase-conjugated rabbit-anti-mouse serum (Thermo Fisher Scientific, Waltham, MA). Electrochemiluminescence reagents (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) were used for chemiluminescent detection. Densitometry analyses were performed by using ImageJ software (National Institutes of Health, Bethesda, MD).
Quantitative Reverse Transcription-PCR.
Total RNA was purified by using TRIzol reagent (Invitrogen). Total RNA (1 μg) was reverse-transcribed in a 50-μl reaction volume with TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). A 5-μl solution of the reverse-transcribed cDNA was PCR-amplified according to Applied Biosystems' recommendations (http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_041280.pdf). TaqMan probes (Assay-on-Demand) were purchased Applied Biosystems. Three replicate reactions were run for each RNA sample.
Statistical Analysis.
Statistical comparison was performed by using pairwise t test for single pairs and analysis of variance for multiple groups and SigmaPlot (version 10.0).
Results
Cloning and Expression of hPTGR1.
hPTGR1 was PCR-amplified from a human adult liver cDNA library according to the National Center for Biotechnology Information GenBank sequence. The recombinant expression was not successful in modified E. coli strain BL21/codon+ or Drosophila melanogaster cell line S2. Therefore, protein expression was carried out in HEK293 cells, which were selected for their high transfection efficiency. Plasmid containing 3×FLAG-tagged hPTGR1 was transiently transfected, and the cells were allowed to grow for an additional 48 h. Cell lysate was purified by affinity tag techniques and analyzed by gel electrophoresis with hPTGR1 and FLAG antibody (Supplemental Fig. 2). Furthermore, the eluted hPTGR1 was verified to be pure by Coomassie staining. Typical yields were 150 μg of recombinant hPTGR1 per 1 × 108 cells.
hPTGR1 Activates HMAF and Improves Drug Efficacy.
With the successful expression and purification of hPTGR1, it was possible to directly address the competence of the enzyme in catalyzing the bioactivation of HMAF. Kinetic parameters of hPTGR1-catalyzed HMAF metabolism were measured, and the comparison between performances of rPTGR1 versus hPTGR1 is shown in Table 1. hPTGR1 was more efficient than rPTGR1. The 20-fold higher Vmax/Km value is largely attributable to a more than 40-fold lower Km for HMAF and a similar catalytic rate.
TABLE 1.
Kinetic parameters for HMAF metabolism by PTGR1
| Km | Vmax | Vmax/Km | |
|---|---|---|---|
| μM | μmol · mg−1 · min−1 | min−1 | |
| rPTGR1 | 215.1 | 1.2 | 5.3 |
| hPTGR1 | 4.9 | 0.6 | 111.7 |
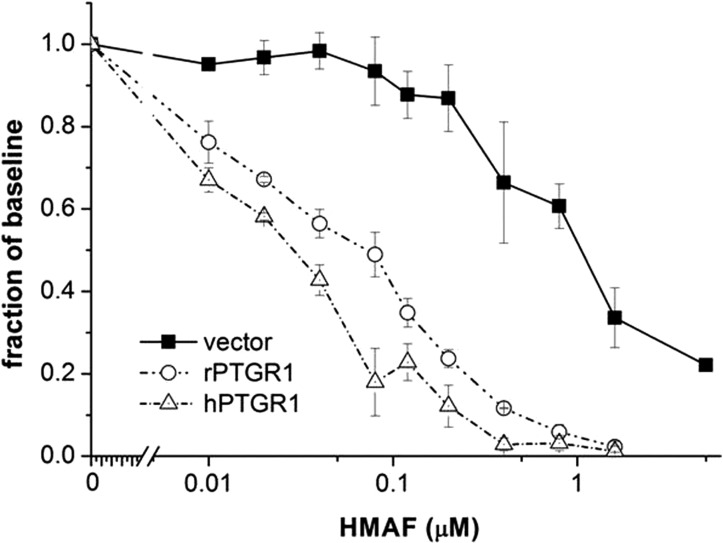
HEK293 cells transfected with blank episomal vector pCEP4, rPTGR1-expressing vector pCEP4-rPTGR1, or hPTGR1-expressing vector pCEP4-hPTGR1 were challenged with graded concentrations of HMAF, and cell viability was determined by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide assay. Data obtained from three independent studies (Fig. 2) demonstrated that both rPTGR1 and hPTGR1 transfection enhanced the cytotoxicity of HMAF more than 15-fold relative to the vector control (IC50 ∼1 μM). In accordance with the enzymatic data, HMAF consumption by total cell lysate from hPTGR1-transfected cells exhibited a more than 14-fold higher conversion rate, 247.6 versus 17.5 nmol/min/mg (Table 2). Consequently, hPTGR1 was more effective at sensitizing cells to HMAF, further reducing the IC50 to 25 from 54 nM in rPTGR1-transfected cells (Table 2; P < 0.05). These observations regarding a higher kinetic proficiency of hPTGR1 in a cell-free system, greater conversion by total cell lysate, and associated enhanced cellular sensitivity to HMAF are in accordance with a role for hPTGR1 in HMAF bioactivation.
Fig. 2.
hPTGR1 markedly sensitized HEK293 cells toward HMAF. HEK293 cells transfected with pCEP4, pCEP4-rPTGR1, or pCEP4-hPTGR1 were challenged with HMAF at the indicated concentrations. Cell viability was assessed 24 h later. Data were obtained from three independent experiments; error bars represent S.E.
TABLE 2.
HMAF metabolism and IC50 of HEK293 cells transfected with rPTGR1 vs. hPTGR1
| Enone Reductase Activity | IC50 | |
|---|---|---|
| nmol/min/mg | nM | |
| HEK293-rPTGR1 | 17.5 ± 7.0 | 54 ± 8 |
| HEK293-hPTGR1 | 247.6 ± 20.4 | 25 ± 6* |
, P < 0.05.
Overexpression of hPTGR1 in Colon Cancer Cell Line HCT15 Enhances HMAF Cytotoxicity.
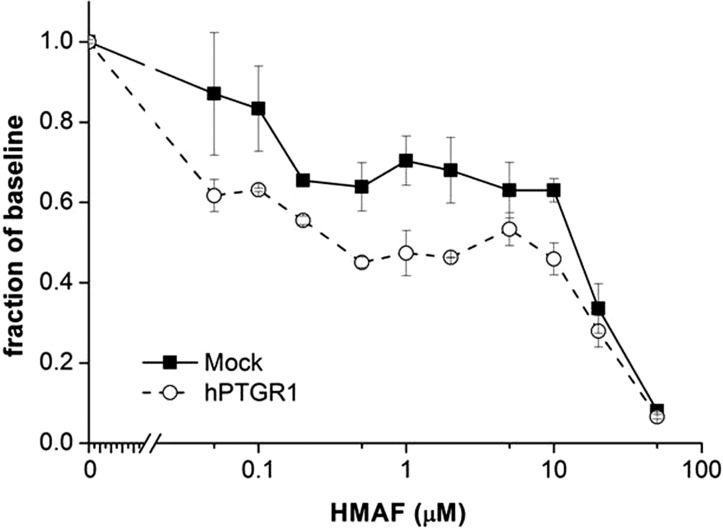
The results presented above involving HEK293 cells provided a proof of concept regarding the role of hPTGR1 in activating HMAF. We further examined the hypothesis that hPTGR1 up-regulation is associated with higher HMAF toxicity in more clinically relevant cancer cell models. Thus, an hPTGR1- or green fluorescent protein-containing vector was transiently expressed in the colon cancer cell line HCT15. After 48 h, transfected cells were challenged with graded concentrations of HMAF. Ectopic expression of hPTGR1 was 1.9-fold higher than the endogenous level, as determined by densitometry after immunoblots (Table 3; Supplemental Fig. 3a). The PTGR1 level increase was modest, but may be more physiologically relevant than the typical 25- to 100-fold increases of PTGR1 observed in transfected HEK293 cells (Dick et al., 2004). Moreover, the modest increase of hPTGR1 protein level reduced the IC50 value of HMAF, from 16.9 to 5.7 μM (P < 0.05; n = 3; Fig. 3; Table 3). This observation demonstrated for the first time that HMAF cytotoxicity can be enhanced by overexpression of hPTGR1 in a human cancer cell line.
TABLE 3.
Effects on HMAF IC50 by overexpression of hPTGR1 in HCT15 cells
| Transfection | hPTGR1 Fold Changea | HMAF IC50 | IC50 Fold Change |
|---|---|---|---|
| μM | |||
| Green fluorescent protein | - | 16.9 | - |
| hPTGR1 | 2 | 5.7* | 3* |
Fold change determined by densitometry of immunoblots.
, P < 0.05.
Fig. 3.
Overexpression of hPTGR1 in colon cancer cell line HCT15 significantly enhances HMAF cytotoxicity. A typical survival graph is shown. Error bars represent S.D. IC50 was calculated from three independent experiments by nonlinear regression analysis (P < 0.05).
Pretreatment of D3T Increases HMAF Cytotoxicity.
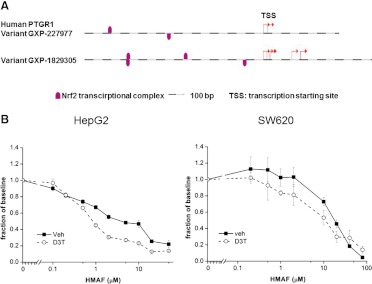
PTGR1 was discovered in a differential screening of D3T-treated rat liver, associating it with Nrf2-mediated gene activation (Primiano et al., 1996). Sequence analyses by the Genomatix gene2promoter algorithm revealed multiple Nrf2 binding sites 1000 base pairs upstream of the transcription starting site in both variants of PTGR1 (Fig. 4a). Thus, the colon cancer cell line SW620 was incubated for 48 h with 20 μM D3T before cell lysate was analyzed. Immunoblotting showed, as determined by densitometry, a 2-fold up-regulation of PTGR1 (Supplemental Fig. 4b). Furthermore, when these preconditioned cells were treated with HMAF, there was a reduction of IC50 value to 9.9 μM compared with 20.1 μM for vehicle-treated cells (P < 0.05; Fig. 4b; Table 4).
Fig. 4.
a, identification of potential Nrf-2 binding sites (indicated by fuchsia marks) by promoter sequence analysis 1000 base pairs upstream from the transcription starting site of PTGR1. Both variant sequences of PTGR1 with experimentally verified 5′ transcripts from the database ElDorado 12-2010 were used (Genomatix Gene2promoter software). b, preconditioning with 20 μM D3T for 48 h induced hPTGR1 expression in colon cancer cell SW620 and liver cancer cell HepG2 and enhanced HMAF cytotoxicity. A typical survival graph is shown. Error bars represent S.D. IC50 was calculated from four independent experiments by nonlinear regression analysis (P < 0.05).
TABLE 4.
Pretreatment of HepG2 and SW620 cells sensitizes cells to HMAF
| Cells | Precondition | hPTGR1 Fold Changea | HMAF IC50 | IC50 Fold Change |
|---|---|---|---|---|
| μM | ||||
| HepG2 | Vehicle | - | 34.2 | - |
| HepG2 | D3T | 2 | 8.9* | 4 |
| HepG2 | Curcumin | 4 | 15.3* | 2 |
| HepG2 | Resveratrol | 3 | 7.4* | 5 |
| SW620 | Vehicle | - | 20.1 | - |
| SW620 | D3T | 2 | 9.9* | 2 |
| SW620 | Curcumin | 4 | 20.3 | - |
| SW620 | Resveratrol | 2 | 3.0* | 7 |
Fold change determined by densitometry of immunoblots.
, P < 0.05 compared with vehicle-treated population.
Hepatocytes are well established models for ARE-mediated enzyme inductions (Otieno et al., 2000). PTGR1 was previously shown to be highly inducible in rat liver by D3T (Primiano et al., 1996). Therefore the hepatocarcinoma cell line HepG2 was selected to test the impact of D3T preconditioning on HMAF toxicity. After incubation with D3T for 48 h, cells were washed and then subjected to HMAF at increasing concentrations, and viability was measured 18 h after HMAF treatment. Indeed, the IC50 value, which decreased from 31 to 3.0 μM, was more significantly reduced than it was for the colon cancer cells (Fig. 4b). Immunoblot analysis indicated 3-fold PTGR1 induction, also higher than for the colon cells, consistent with a positive correlation between PTGR1 levels and drug efficacy (Table 4; Supplemental Fig. 4).
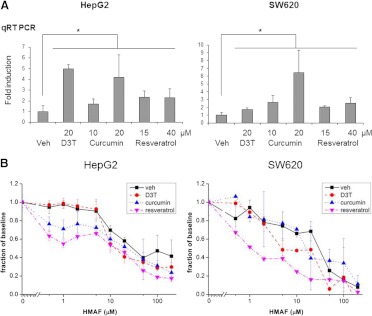
Curcumin and Resveratrol Up-Regulate hPTGR1 in HepG2 and SW620 Cells, Leading to Enhanced HMAF Toxicity.
Extensive data support the effectiveness of plant-based polyphenols, including dietary components such as curcumin (Fig. 1), a polyphenol in the spice turmeric, and resveratrol (Fig. 1), a stilbenoid in grape skins, in activating cytoprotective enzymes (Birringer, 2011). Such bioactive food components may be convenient, accessible, and efficient options for modulating desired enzymatic activities. RNA (24 h) and protein levels (48 h) of PTGR1 were examined in HepG2 and SW620 cells after treatment with 10 and 20 μM curcumin or 15 and 40 μM resveratrol. Compared with 20 μM D3T, these compounds similarly elevated PTGR1 levels (Fig. 5a). At 20 μM, curcumin was even more effective than the same dose of D3T in SW620 cells, elevating the RNA level of PTGR1 5-fold. Immunoblot indicated that the up-regulation at the protein level correlated well with mRNA quantitation (Supplemental Fig. 4).
Fig. 5.
a, up-regulation of PTGR1 by curcumin, resveratrol, or D3T. Total RNA (24 h) after treatment at indicated concentrations were analyzed via quantitative reverse transcription-PCR (qRT-PCR). Error bars represent S.E. from three independent experiments (*, P < 0.05). Veh, vehicle. b, effects of pretreatment with D3T (20 μM), curcumin (10 μM), or resveratrol (15 μM) on HMAF toxicity in SW620 and HepG2 cells. Error bars represent S.E. from three independent experiments (P < 0.05 with the exception of SW620 + 10 μM curcumin).
Effects of pretreatment on the toxicity of HMAF were examined after HepG2 and SW620 cells were incubated with 20 μM D3T, 10 μM curcumin, or 15 μM resveratrol for 48 h. With the exception of SW620 cells with 10 μM curcumin, all other pretreatments significantly enhanced cytotoxicity of HMAF and lowered the IC50 by 2- to 7-fold (Fig. 5b; Table 4).
Discussion
rPTGR1 has been expressed previously in E. coli as a recombinant protein. Difficulties in expressing the human enzyme in this system may be caused by the abundance of proline residues within the 3′ coding region of hPTGR1. Proline is an infrequently used amino acid in E. coli, accounting for 4% of total amino acids. Three of the four coding codons, CCC, CCU, and CCA, occur at an extremely low frequency of 3.5 to 7.5 per 1000 codons (Sharp et al., 1988). The presence of a proline-rich sequence, PLPPGPPP, within hPTGR1 (amino acids 250–257; Supplemental Fig. 1) and the corresponding coding sequence, CCA CTT CCC CCA GGC CCA CCC CCA that features six rare codons (underlined) in close proximity, is likely to stall E. coli translation machinery, leading to the degradation of the partially translated protein. Mutation of CCC and CCA codons to CCG, the proline codon used four times more frequently in E. coli (Sharp et al., 1988), may help to circumvent this obstacle; in the present study we successfully addressed the problem by expressing the protein in human cells.
An aim of this study was to examine the role of hPTGR1 in HMAF activation based on the understanding that rPTGR1 overexpression was previously shown to greatly sensitize HEK293 cells toward AFs. In fact, we discovered that hPTGR1 exhibited an even higher catalytic activity toward HMAF compared with rPTGR1, leading to a higher cytotoxicity in HEK293 cells (Fig. 2). In vitro studies indicated that this enhancement is largely caused by the more than 40-fold lower Km of hPTGR1 toward HMAF (4.9 μM for hPTGR1 versus 215.1 μM for rPTGR1; Table 1). IC50 values of therapeutic HMAF concentrations tested in a NCI-60 cell line screen were between 0.2 and 80 μM (data provided by developmental therapeutic program at the National Cancer Institute). Therefore, hPTGR1 should have a profound impact on the activation of HMAF in vivo, because the lower Km value would allow hPTGR1 to operate at high efficiency within the drug's therapeutic window. As indicated in Table 2, cell lysate from HEK293 cells overexpressing hPTGR1 versus rPTGR1 displayed a much higher conversion rate of HMAF. Furthermore, a small increase in hPTGR1 level (2-fold in Table 3, Supplemental Fig. 3) resulted in a 3-fold HMAF IC50 reduction in HCT15 cells (Fig. 3; Table 3). Because one of the hurdles in multiple HMAF clinical trials was systemic toxicity, bioactivation by hPTGR1 could markedly lower the therapeutic dose of HMAF and therefore preserve efficacy while ameliorating unwanted side effects.
Like HMAF, many chemotherapeutic agents are administered as a prodrug, transformed in vivo by enzymatic bioactivation into proximal reactive species. Levels of drug-metabolizing enzymes can have pronounced effects on efficacy of therapeutics. Nitrosamine drugs can be significantly enhanced by increasing the level of CYP450 reductase. In both aerobic and hypoxic conditions, tiraparamine [3-amino-1,2,4-benzotriazine-1,4-dioxide (SR4233)] and 1-(2-nitro-1-imidazolyl)-3-aziridino-2-propanol (RSU 1069) were shown to be 2- to 10-fold more effective in multiple breast cancer cell lines when exogenous CYP450 reductase was introduced. Accordingly, drug IC50 values correlate with the expression of CYP450 reductase in tested primary tumors (Patterson et al., 1995a,b). Overexpression of aldo-keto reductase 1c3 in HCT116 cells enhanced PR104A cytotoxicity by 10-fold, and 1c3 expression correlated with drug sensitivity, as demonstrated by the screening of tissue arrays (Guise et al., 2010).
Various strategies have been explored to modulate the level of drug-activating enzymes to enhance efficacy, such as gene-directed enzyme prodrug therapy and subsequent diffusion of active metabolites into the solid tumor and its microenvironment (Roy and Waxman, 2006). However, limited efforts have focused on pharmacological means for enhancing activating enzymes, such as bioactive food components (Tanasova and Sturla, 2012). A series of studies by Begleiter and coworkers (Doherty et al., 1998; Wang et al., 1999; Begleiter et al., 2004; Digby et al., 2005) supports the feasibility of the approach. In addition, many relevant enzymes with ARE in their promoter region can be activated via the Nrf2 pathway (Kwak et al., 2004). Therefore, by using a chemical inducer such as D3T, efficacy may be enhanced by a strategy that would be less invasive than gene-directed enzyme prodrug therapy. For example, mitomycin C and EO9 efficacies were greatly enhanced in a number of cancer cell lines by elevating NAD(P)H:quinone oxidoreductase with D3T pretreatments of 50 to 100 μM (Doherty et al., 1998; Wang et al., 1999). Furthermore, such enhancement was also observed in vivo when mice were treated with the inducer dimethyl fumarate (Begleiter et al., 2004; Digby et al., 2005).
Similar to NAD(P)H:quinone oxidoreductase, PTGR1 was discovered to be highly inducible in rat liver by D3T (Primiano et al., 1996). The presence of Nrf2 binding sites within the PTGR1 promoter region (Fig. 4a) suggests activation by Nrf2-dependent inducers. In this study, pretreatment of SW620 and HepG2 cells with 20 μM D3T elevated PTGR1 levels (Supplemental Fig. 4b) and led to enhanced sensitivity to HMAF (Fig. 4b). Even more intriguingly, curcumin and resveratrol both demonstrated comparable or even greater enhancement of PTGR1 up-regulation (Fig. 5; Supplemental Fig. 4) as supported by RNA and protein levels. More importantly, preconditioning with 10 μM curcumin significantly enhanced HMAF cytotoxicity in HepG2 cells and 15 μM resveratrol in both HepG2 and SW620 cells (Fig. 5b; Table 4).
There is a lack of response of SW620 cells to HMAF after pretreatment with 10 μM curcumin, suggesting that other cellular processes, such as DNA repair (M. Tanasova, C. Otto, G. Spivak, P. C. Hanawalt, and S.J.S., unpublished work), could contribute to HMAF resistance. It has been reported that the activation of DNA repair pathways could lead to a diminished toxicity effect at higher drug doses in target cell population (Smith and Grisham, 1983). Such repair induction may account for some features of the survival curves measured in the present study, such as the slight plateau in the mid-dose range of HCT15 cell survival curves (Fig. 3). The DNA-alkylating nature of HMAF may lead to the activation of DNA repair once its concentration has reached a critical threshold. The dampened cytotoxic effect was observed in both control and PTGR1-transfected HCT15 cells, therefore, it does not seem to be PTGR1-related. In addition, curcumin- and resveratrol-treated cells responded in a somewhat biphasic fashion (Fig. 5b), such that there was an attenuation of toxicity response, again, in the midrange of dosage, similar to that observed for HCT15 cells (Fig. 3).
On the basis of features of the survival curves that suggested modest biphasic behavior for curcumin- or resveratrol-treated cells, we further analyzed the toxicity data in two separate phases. Independent linear regression analyses of the four lowest and four highest dose data points suggest that the preconditioned cells were significantly more sensitive to HMAF in the low-dose regime in all three experiments. For the high-dose regime, a similar differential response is apparent, however, only in two of three independent experiments. Such observation underscores the complexity of cellular response to toxins, and multiple independently regulated pathways can influence toxicity under different conditions. Furthermore, such dose-modulated responses by HMAF seem to be cell line-specific.
Previous reports suggest that for the precursor illudins preferential killing of tumor cells could result from increased cellular transport (Kelner et al., 1990); however, for HMAF preferential accumulation did not significantly account for enhanced toxicity in PC-3 and HT-29 cells (Woynarowska et al., 2000). Furthermore, microarray studies regarding cellular responses to D3T or resveratrol have failed to implicate alterations in membrane transport (Kwak et al., 2003; Whyte et al., 2007). Despite these data, transport and repair have not been explicitly evaluated in this study and cannot be excluded as possible factors contributing to cytotoxicity. Thus, although our data suggest that HMAF cytotoxicity can be enhanced by using dietary supplements to up-regulate hPTGR1, more studies are needed to elucidate the feasibility of such an approach.
Two additional factors relevant to the metabolic induction as a sensitization strategy concern the selectivity of PTGR1 up-regulation in target tissues and the physiological relevance of inducer concentrations. In the present study, D3T was effective in HepG2 and SW620 cells, but not in HCT15, HCT116, or AGS cells (data not shown). Previous animal studies demonstrated that D3T induced ARE response genes in mouse forestomach, liver, colon, and brain (Ramos-Gomez et al., 2001; Burton et al., 2006; Osburn et al., 2007). Thus, it would be informative, in future studies, to profile the modulation of human PTGR1 in various tissues after curcumin or resveratrol treatment. Finally, the concentrations of curcumin and resveratrol used in this study were relatively low compared with other cellular studies commonly appearing in the literature, and moreover, in vivo data suggest that 10 to 20 μM curcumin and resveratrol may be achieved in the cells of human patients (Garcea et al., 2005; Boocock et al., 2007).
On the basis of preclinical data, drug bioactivation is a significant controlling factor for HMAF cytotoxicity and selectivity. However, mechanistic details regarding human physiological conditions remain to be elucidated. The data obtained in this study suggest that hPTGR1 effectively transforms HMAF into its active form at therapeutic concentrations and enhances its efficacy in cell-based models. Furthermore, data indicate that modulating hPTGR1 is feasible in vitro by food components. These observations establish a proof of principle regarding hPTGR1 in cytotoxicity and for clinical implementations of HMAF or related chemotherapeutic agents.
Supplementary Material
Acknowledgments
We thank Dr. Thomas W. Kensler and Dr. Olga Aprelikova for insightful guidance and discussions and Brian Hibler and Alana V. Rivera for critical reading of the manuscript.
This work was supported by the National Institutes of Health National Cancer Institute [Grant CA123007]; the Swiss National Science Foundation [Grant CRSII3_136247]; and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
 The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
1PTGR1, accession no. 012212 at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), was previously reported under the name leukotriene B4 reductase, dithiol-thione inducible gene 1, and alkenal/one oxidoreductase in others and our studies.
- AF
- acylfulvene
- AOR
- alkenal/one oxidoreductase
- ARE
- antioxidant response element
- D3T
- 1,2-dithiol-3-thione
- DMEM
- Dulbecco's modified Eagle's medium
- DMSO
- dimethyl sulfoxide
- FBS
- fetal bovine serum
- HEK293
- human embryonic kidney 293
- HMAF
- hydroxymethylacylfulvene
- Nrf2
- nuclear factor E2-related factor 2
- PBS
- phosphate-buffered saline
- PCR
- polymerase chain reaction
- PTGR1
- prostaglandin reductase 1
- rPTGR1
- rat PTGR1
- hPTGR1
- human PTGR1
- RSU 1069
- 1-(2-nitro-1-imidazolyl)-3-aziridino-2-propanol
- SR4233
- 3-amino-1,2,4-benzotriazine-1,4-dioxide.
Authorship Contributions
Participated in research design: Yu, Pietsch, Niederhuber, and Sturla.
Conducted experiments: Yu, Erzinger, Cervoni-Curet, and Whang.
Contributed new reagents or analytic tools: Yu and Pietsch.
Performed data analysis: Yu, Erzinger, Cervoni-Curet, and Whang.
Wrote or contributed to the writing of the manuscript: Yu, Erzinger, Pietsch, Niederhuber, and Sturla.
References
- Begleiter A, Leith MK, Thliveris JA, Digby T. (2004) Dietary induction of NQO1 increases the antitumour activity of mitomycin C in human colon tumours in vivo. Br J Cancer 91:1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birringer M. (2011) Hormetics: dietary triggers of an adaptive stress response. Pharm Res 28:2680–2694 [DOI] [PubMed] [Google Scholar]
- Boocock DJ, Faust GE, Patel KR, Schinas AM, Brown VA, Ducharme MP, Booth TD, Crowell JA, Perloff M, Gescher AJ, et al. (2007) Phase I dose escalation pharmacokinetic study in healthy volunteers of resveratrol, a potential cancer chemopreventive agent. Cancer Epidemiol Biomarkers Prev 16:1246–1252 [DOI] [PubMed] [Google Scholar]
- Burton NC, Kensler TW, Guilarte TR. (2006) In vivo modulation of the Parkinsonian phenotype by Nrf2. NeuroToxicology 27:1094–1100 [DOI] [PubMed] [Google Scholar]
- Dick RA, Kensler TW. (2004) The catalytic and kinetic mechanisms of NADPH-dependent alkenal/one oxidoreductase. J Biol Chem 279:17269–17277 [DOI] [PubMed] [Google Scholar]
- Dick RA, Kwak MK, Sutter TR, Kensler TW. (2001) Antioxidative function and substrate specificity of NAD(P)H-dependent alkenal/one oxidoreductase. A new role for leukotriene B4 12-hydroxydehydrogenase/15-oxoprostaglandin 13-reductase. J Biol Chem 276:40803–40810 [DOI] [PubMed] [Google Scholar]
- Dick RA, Yu X, Kensler TW. (2004) NADPH alkenal/one oxidoreductase activity determines sensitivity of cancer cells to the chemotherapeutic alkylating agent irofulven. Clin Cancer Res 10:1492–1499 [DOI] [PubMed] [Google Scholar]
- Digby T, Leith MK, Thliveris JA, Begleiter A. (2005) Effect of NQO1 induction on the antitumor activity of RH1 in human tumors in vitro and in vivo. Cancer Chemother Pharmacol 56:307–316 [DOI] [PubMed] [Google Scholar]
- Doherty GP, Leith MK, Wang X, Curphey TJ, Begleiter A. (1998) Induction of DT-diaphorase by 1,2-dithiole-3-thiones in human tumour and normal cells and effect on anti-tumour activity of bioreductive agents. Br J Cancer 77:1241–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcea G, Berry DP, Jones DJ, Singh R, Dennison AR, Farmer PB, Sharma RA, Steward WP, Gescher AJ. (2005) Consumption of the putative chemopreventive agent curcumin by cancer patients: assessment of curcumin levels in the colorectum and their pharmacodynamic consequences. Cancer Epidemiol Biomarkers Prev 14:120–125 [PubMed] [Google Scholar]
- Gong J, Neels JF, Yu X, Kensler TW, Peterson LA, Sturla SJ. (2006) Investigating the role of stereochemistry in the activity of anticancer acylfulvenes: synthesis, reductase-mediated bioactivation, and cellular toxicity. J Med Chem 49:2593–2599 [DOI] [PubMed] [Google Scholar]
- Gong J, Vaidyanathan VG, Yu X, Kensler TW, Peterson LA, Sturla SJ. (2007) Depurinating acylfulvene-DNA adducts: characterizing cellular chemical reactions of a selective antitumor agent. J Am Chem Soc 129:2101–2111 [DOI] [PubMed] [Google Scholar]
- Guise CP, Abbattista MR, Singleton RS, Holford SD, Connolly J, Dachs GU, Fox SB, Pollock R, Harvey J, Guilford P, et al. (2010) The bioreductive prodrug PR-104A is activated under aerobic conditions by human aldo-keto reductase 1C3. Cancer Res 70:1573–1584 [DOI] [PubMed] [Google Scholar]
- Hori T, Yokomizo T, Ago H, Sugahara M, Ueno G, Yamamoto M, Kumasaka T, Shimizu T, Miyano M. (2004) Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop. J Biol Chem 279:22615–22623 [DOI] [PubMed] [Google Scholar]
- Kelner MJ, McMorris TC, Beck WT, Zamora JM, Taetle R. (1987) Preclinical evaluation of illudins as anticancer agents. Cancer Res 47:3186–3189 [PubMed] [Google Scholar]
- Kelner MJ, McMorris TC, Estes L, Wang W, Samson KM, Taetle R. (1996) Efficacy of HMAF (MGI-114) in the MV522 metastatic lung carcinoma xenograft model nonresponsive to traditional anticancer agents. Invest New Drugs 14:161–167 [DOI] [PubMed] [Google Scholar]
- Kelner MJ, McMorris TC, Montoya MA, Estes L, Rutherford M, Samson KM, Taetle R. (1997) Characterization of cellular accumulation and toxicity of illudin S in sensitive and nonsensitive tumor cells. Cancer Chemother Pharmacol 40:65–71 [DOI] [PubMed] [Google Scholar]
- Kelner MJ, McMorris TC, Taetle R. (1990) Preclinical evaluation of illudins as anticancer agents: basis for selective cytotoxicity. J Natl Cancer Inst 82:1562–1565 [DOI] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. (2001) Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol Med 7:135–145 [PMC free article] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. (2003) Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem 278:8135–8145 [DOI] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Kensler TW. (2004) Chemoprevention through the Keap1-Nrf2 signaling pathway by phase 2 enzyme inducers. Mutat Res 555:133–148 [DOI] [PubMed] [Google Scholar]
- MacDonald JR, Muscoplat CC, Dexter DL, Mangold GL, Chen SF, Kelner MJ, McMorris TC, Von Hoff DD. (1997) Preclinical antitumor activity of 6-hydroxymethylacylfulvene, a semisynthetic derivative of the mushroom toxin illudin S. Cancer Res 57:279–283 [PubMed] [Google Scholar]
- McMorris TC, Kelner MJ, Wang W, Yu J, Estes LA, Taetle R. (1996) (Hydroxymethyl)acylfulvene: an illudin derivative with superior antitumor properties. J Nat Prod 59:896–899 [DOI] [PubMed] [Google Scholar]
- Neels JF, Gong J, Yu X, Sturla SJ. (2007) Quantitative correlation of drug bioactivation and deoxyadenosine alkylation by acylfulvene. Chem Res Toxicol 20:1513–1519 [DOI] [PubMed] [Google Scholar]
- Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, Kensler TW. (2007) Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer 121:1883–1891 [DOI] [PubMed] [Google Scholar]
- Otieno MA, Kensler TW, Guyton KZ. (2000) Chemoprotective 3H-1,2-dithiole-3-thione induces antioxidant genes in vivo. Free Radic Biol Med 28:944–952 [DOI] [PubMed] [Google Scholar]
- Patterson AV, Barham HM, Chinje EC, Adams GE, Harris AL, Stratford IJ. (1995a) Importance of P450 reductase activity in determining sensitivity of breast tumour cells to the bioreductive drug, tirapazamine (SR 4233). Br J Cancer 72:1144–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AV, Zhang H, Moghaddam A, Bicknell R, Talbot DC, Stratford IJ, Harris AL. (1995b) Increased sensitivity to the prodrug 5′-deoxy-5-fluorouridine and modulation of 5-fluoro-2′-deoxyuridine sensitivity in MCF-7 cells transfected with thymidine phosphorylase. Br J Cancer 72:669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis G. (1983) Dose-dependent metabolism, therapeutic effect, and toxicity of anticancer drugs in man. Drug Metab Rev 14:1145–1163 [DOI] [PubMed] [Google Scholar]
- Primiano T, Gastel JA, Kensler TW, Sutter TR. (1996) Isolation of cDNAs representing dithiolethione-responsive genes. Carcinogenesis 17:2297–2303 [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A 98:3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P, Waxman DJ. (2006) Activation of oxazaphosphorines by cytochrome P450: application to gene-directed enzyme prodrug therapy for cancer. Toxicol In Vitro 20:176–186 [DOI] [PubMed] [Google Scholar]
- Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. (1988) Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res 16:8207–8211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Grisham JW. (1983) Cytotoxicity of monofunctional alkylating agents. Methyl methanesulfonate and methyl-N′-nitro-N-nitrosoguanidine have different mechanisms of toxicity for 10T1/2 cells. Mutat Res 111:405–417 [DOI] [PubMed] [Google Scholar]
- Tai H-H, Ensor CM, Tong M, Zhou H, Yan F. (2002) Prostaglandin catabolizing enzymes. Prostaglandins Other Lipid Mediat 68–69:483–493 [DOI] [PubMed] [Google Scholar]
- Tanasova M, Sturla SJ. (2012) Chemistry and biology of acylfulvenes: sesquiterpene-derived antitumor agents. Chem Rev 112:3578–3610 [DOI] [PubMed] [Google Scholar]
- Wang X, Doherty GP, Leith MK, Curphey TJ, Begleiter A. (1999) Enhanced cytotoxicity of mitomycin C in human tumour cells with inducers of DT-diaphorase. Br J Cancer 80:1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte L, Huang YY, Torres K, Mehta RG. (2007) Molecular mechanisms of resveratrol action in lung cancer cells using dual protein and microarray analyses. Cancer Res 67:12007–12017 [DOI] [PubMed] [Google Scholar]
- Woynarowska BA, Woynarowski JM, Herzig MC, Roberts K, Higdon AL, MacDonald JR. (2000) Differential cytotoxicity and induction of apoptosis in tumor and normal cells by hydroxymethylacylfulvene (HMAF). Biochem Pharmacol 59:1217–1226 [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Izumi T, Takahashi T, Kasama T, Kobayashi Y, Sato F, Taketani Y, Shimizu T. (1993) Enzymatic inactivation of leukotriene B4 by a novel enzyme found in the porcine kidney. Purification and properties of leukotriene B4 12-hydroxydehydrogenase. J Biol Chem 268:18128–18135 [PubMed] [Google Scholar]
- Yu X, Egner PA, Wakabayashi J, Wakabayashi N, Yamamoto M, Kensler TW. (2006) Nrf2-mediated induction of cytoprotective enzymes by 15-deoxy-δ12,14-prostaglandin J2 is attenuated by alkenal/one oxidoreductase. J Biol Chem 281:26245–26252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.