Abstract
Regulation of gene transcription is controlled in part by nuclear receptors that function coordinately with coregulator proteins. The human constitutive androstane receptor (CAR; NR1I3) is expressed primarily in liver and regulates the expression of genes involved in xenobiotic metabolism as well as hormone, energy, and lipid homeostasis. In this report, DAX-1, a nuclear receptor family member with corepressor properties, was identified as a potent CAR regulator. Results of transaction and mutational studies demonstrated that both DAX-1's downstream LXXLL and its PCFQVLP motifs were critical contributors to DAX-1's corepression activities, although two other LXXM/LL motifs located nearer the N terminus had no impact on the CAR functional interaction. Deletion of DAX-1's C-terminal transcription silencing domain restored CAR1 transactivation activity in reporter assays to approximately 90% of control, demonstrating its critical function in mediating the CAR repression activities. Furthermore, results obtained from mammalian two-hybrid experiments assessing various domain configurations of the respective receptors showed that full-length DAX-1 inhibited the CAR-SRC1 interaction by approximately 50%, whereas the same interaction was restored to 90% of control when the DAX-1 transcription silencing domain was deleted. Direct interaction between CAR and DAX-1 was demonstrated with both alpha-screen and coimmunoprecipitation experiments, and this interaction was enhanced in the presence of the CAR activator 6-(4-chlorophenyl)imidazo[2,1-b]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO). Results obtained in primary human hepatocytes further demonstrated DAX-1 inhibition of CAR-mediated CITCO induction of the CYP2B6 target gene. The results of this investigation identify DAX-1 as a novel and potent CAR corepressor and suggest that DAX-1 functions as a coordinate hepatic regulator of CAR's biological function.
Introduction
Nuclear receptors (NRs) are transcription factors that play an essential role in the regulation of gene expression, and their activity involves a complex interplay of various proteins with diverse functions. NRs are composed of several functional regions that dictate their activity. Most NRs contain an A/B domain that includes an activator function (AF-1). The C domain contains the DNA-binding portion, which is linked to the ligand binding domain E via a hinge region (domain D). Domain E also contains the interface for NR dimerization and a transactivation AF-2 function that mediates coregulator binding (Aranda and Pascual, 2001). Agonist binding induces an AF-2 conformational change, which provides a “coregulator cleft” for coactivator binding and subsequent activation of transcription. In the absence of agonist or the presence of antagonist, the position of AF-2 exposes a corepressor binding site, causing transcriptional repression (Perissi et al., 1999).
The interaction of coregulators with NRs is accomplished through α-helical structures that contain consensus motifs. In general, NR coactivators contain LXXLL sequences (Heery et al., 1997), also known as NR boxes, whereas NR corepressors have the consensus motif LXXI/HIXXXI/L or CoRNR box (Hu and Lazar, 1999). Numerous coactivators exist, and previous studies show that they exhibit distinct NR preferences (Ding et al., 1998; Heery et al., 2001). Furthermore, the specificity of the interaction between NRs and these motifs is governed by the sequences that flank the N and C termini of the NR box (Chang et al., 1999). The agonist bound also may influence the recruitment of NR coactivators depending on the degree of AF-2 conformational change that it induces (Togashi et al., 2005). These findings suggested the intriguing possibility that there are multiple layers that dictate the recruitment of coregulatory proteins and thus influence the action NRs in a given milieu.
The constitutive androstane receptor (CAR; NR1I3) is an atypical NR in that it does not have an A/B domain and the reference form (CAR1) does not require exogenous ligand for activity (Baes et al., 1994). This constitutive activity is thought to result from a shortened AF-2 domain and hydrogen bonding interactions that stabilize the AF-2 domain in the active conformation (Xu et al., 2004). Alternative mRNA splicing results in CAR variants, including CAR2 and CAR3 (Auerbach et al., 2003), and unlike the constitutively active CAR1, CAR2 and CAR3 require ligand for activation and exhibit differences in ligand selectivity (Auerbach et al., 2005; Dekeyser et al., 2009; Dekeyser et al., 2011). CAR and CAR variant transcripts are detectable in many tissues but are expressed primarily in the liver, where they act as xenosensors, controlling the expression of phase I and phase II metabolic enzymes and phase III transporter (Timsit and Negishi, 2007). More importantly, CAR now is recognized as a regulator of endogenous physiological processes, such as energy, glucose, and lipid homeostasis and bile acid elimination (Guo et al., 2003; Kodama et al., 2004; Konno et al., 2008; Masson et al., 2008).
With regard to its interactions with nuclear coregulatory proteins, CAR interacts with coactivators in the SRC family, GRIP1, PGC1α, and TIF2 (e.g., Kim et al., 1998; Muangmoonchai et al., 2001; Min et al., 2002; Shiraki et al., 2003. However, only SRC family members and PGC1α appear to enhance CAR transcriptional activity. For example, SRC-1 enhances CAR-mediated induction of CYP2B1 in rat hepatocytes (Muangmoonchai et al., 2001). Furthermore, although all SRC family members enhance mouse CAR activity, SRC-3 seems to be the most important mediator (Chen et al., 2011). The human reference CAR was crystallized in the presence of SRC-1 NR box peptides (Xu et al., 2004). With respect to corepressor protein interactions, NCoR and SMRT have been shown to interact with both human and mouse CAR (Dussault et al., 2002; Lempiainen et al., 2005), and NCoR inhibits their transactivation in reporter assays (Lempiainen et al., 2005). SHP (NR0B2) interacts with mouse CAR in pull-down and yeast two-hybrid assays (Seol et al., 1996; Park et al., 2004) and represses mouse CAR activity approximately 50% in transactivation experiments (Bae et al., 2004).
DAX-1 (NR0B1) is an atypical NR whose gene (dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1) was identified as responsible for adrenal hypoplasia congenita (Zanaria et al., 1994). Similar to SHP, DAX-1 is unusual in that it lacks the typical NR DNA-binding domain (DBD) and its N-terminal region contains three LXXL/ML motifs typically found in NR coactivators. However, DAX-1 appears to function as a transcriptional corepressor of many NRs, including SF-1 (Ito et al., 1997), ER (Zhang et al., 2000), AR (Holter et al., 2002), Nurr77, (Song et al., 2004), PPARγ (Kim et al., 2008), LRH-1 (Sablin et al., 2008), GR (Zhou et al., 2008), HNF4α, (Nedumaran et al., 2009), and LXRα (Nedumaran et al., 2010). In addition, DAX-1 plays an important role in steroidogenesis, adrenal and reproductive development, and maintenance of stem cell pluripotency (Lalli and Sassone-Corsi, 2003; Jeong and Mangelsdorf, 2009). Recent studies indicated a further role for DAX-1 in hepatic lipogenesis and gluconeogenesis (Nedumaran et al., 2009, 2010).
Because CAR mediates xenobiotic responses and has a role in endogenous homeostatic processes, different coregulator interactions may direct CAR's various functional activities. Although several CAR coregulators have been identified, mostly with mouse CAR, the aim of this study was to identify new potential coregulators of human CAR and CAR splice variants. Screening a library of estrogen receptor interacting peptides (Chang et al., 1999) led to the identification of DAX-1 as a novel and potent repressor of CAR activity.
Materials and Methods
Chemicals and Reagents.
General chemicals, dimethyl sulfoxide (DMSO; CAS no. 67-68-5), 6-(4-chlorophenyl)imidazo[2,1-b]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (CITCO; CAS no. 338404-52-7), 5α-androstan-3α-ol (ANDRO; CAS no. 7657-50-3), anti-mouse IgG-horseradish peroxidase (HRP) conjugate, anti-FLAG M2 affinity resin, and mouse monoclonal anti-FLAG M2-HRP were purchased from Sigma-Aldrich (St. Louis, MO). Phenobarbital (PB; CAS no. 50-06-0) was obtained from the Drug Services Division of the University of Washington (Seattle, WA). The human DAX-1 and SHP cDNA clones in the pCMV6-AC expression vector were purchased from Origene (Rockville, MD). Primers were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Anti-DAX-1 antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Plasmid Constructs and Generation of Mutants.
The pM vectors expressing GAL4-DBD-LXXLL peptide motifs were a gift from Dr. Donald P. McDonnell (Duke University) (Chang et al., 1999). The vectors CMV2-CAR1, CMV2-CAR2, 3.1-RXRα, and 2B6-XREM-PBREM luciferase reporter were described previously (Auerbach et al., 2007). The CMV2-CAR3, 3XFLAG-CAR1, and mammalian two-hybrid vectors also were reported previously (Auerbach et al., 2005). Full-length DAX-1 was amplified from the pCMV6-DAX-1 expression vector using the primers shown in Table 1 and sublconed into the pM (Gal4-DBD) and pVP16 (AD) mammalian two-hybrid vectors (Clontech, Mountain View, CA). The DAX-1 mutants are represented in Fig. 1 and were generated using the pCMV6-AC DAX-1 cDNA clone and QuikChange Lightning mutagenesis kit according to the manufacturer's instructions (Agilent Technologies, Santa Clara, CA). Mutagenesis primers are shown in Table 1 and were designed using the QuikChange primer design tool. The truncated DAX1-TSD expression plasmid was generated by amplifying bases 1 to 1325 of the pCMV6-AC-DAX-1 clone using primers shown in Table 1. The truncated DAX-1 product was the inserted into an empty pCMV6-AC vector. This truncated version of DAX-1 was subcloned into the mammalian two-hybrid vectors pM and pVP16. For the creation of the pCDH-dCMV-DAX1 construct, the dual promoter pCDH-CMV-MCS-EF1-copGFP (System Biosciences, Mountain View, CA) first was modified by replacing EF1-copGFP with CMV-copGFP (excised from pSIH-H1-copGFP; Systems Biosciences), resulting in a dual CMV promoter vector, pCDH-CMV-MCS-CMV-copGFP (pCDH-dCMV). This was done because previous experiments suggested that the EF1 promoter did not function efficiently in primary hepatocytes (T. Chen and C. J. Omiecinski, unpublished observations). DAX1 then was excised from pCMV6-AC-DAX1 and subcloned into pCDH-dCMV to create a construct in which two separate CMV promoters drive DAX1 and copGFP expression.
TABLE 1.
Primers used for creation of DAX-1 constructs
| Construct | Primer | Sequence |
|---|---|---|
| VP16-DAX1 | Forward (BamHI) | 5′-ATGCGGATCCTTGCGGGCGAGAACCACCAGTG-3′ |
| Reverse (XbaI) | 5′-ATGCTCTAGATTATATCTTTGTACAGAGCATTTCC-3′ | |
| pCMV6AC-DAX1-M1 | Sense | 5′-GGGCAGCATCCTCTACAACGCGGCTATGAGCGCGAAGCAAACG-3′ |
| Antisense | 5′-CGTTTGCTTCGCGCTCATAGCCGCGTTGTAGAGGATGCTGCCC-3' | |
| pCMV6AC-DAX1-M2 | Sense | 5′-GGCAGCATCCTCTACAGCGCGGCGACGAGCGCAAAGCAAAC-3′ |
| Antisense | 5′-GTTTGCTTTGCGCTCGTCGCCGCGCTGTAGAGGATGCTGCC-3′ | |
| pCMV6AC-DAX1-M3 | Sense | 5′-GGGCAGCATCCTCTACAGCGCGGCCACTAGCTCAAAGCAAACG-3′ |
| Antisense | 5′-CGTTTGCTTTGAGCTAGTGGCCGCGCTGTAGAGGATGCTGCCC-3′ | |
| pCMV6AC-DAX1-PCF | Sense | 5′-GCCCTGCTTCCAGGCGGCGCCCCTGGACCAGC-3′ |
| Antisense | 5′-GCTGGTCCAGGGGCGCCGCCTGGAAGCAGGGC-3′ | |
| pCMV6AC-DAX1-TSD | Forward (BamHI) | 5′-ATGCGGATCCTTGCGGGCGAGAACCACCAGTG-3′ |
| Reverse (XhoI) | 5′-GTCTCGAGTTATTGGCATTGATGAATCTCAGCAGG-3′ |
Fig. 1.
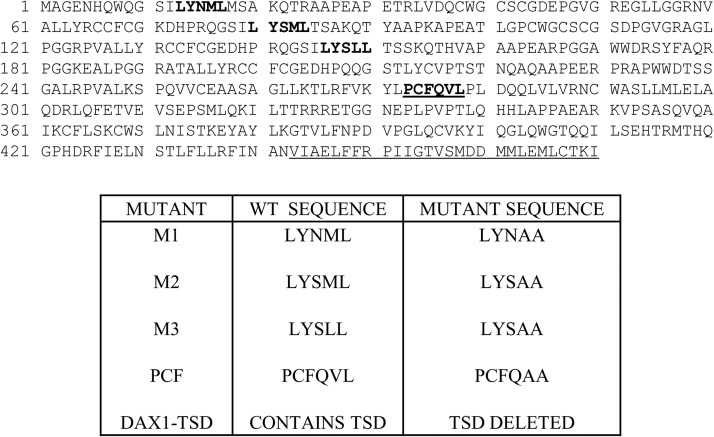
DAX-1 protein sequence showing the locations of the three LXXL/ML NR boxes (bold), the PCFQVL NR interacting sequence (bold and underlined), and the TSD (underlined).
Transactivation and Mammalian Two-Hybrid Assays.
Culture conditions for the maintenance of COS-1 cells (American Type Culture Collection, Manassas, VA) were published previously (Dekeyser et al., 2009). COS-1 cells were used because they are devoid of endogenous CAR expression/activity as demonstrated in previous reports (Auerbach et al., 2005, 2007). For transfection and chemical treatments, the same medium was used except dextran/charcoal-treated fetal bovine serum (HyClone Laboratories, Logan, UT) replaced normal fetal bovine serum. All of the transfections and chemical treatments for luciferase reporter and mammalian two-hybrid assays were performed in a 48-well format in triplicate or quadruplicate and repeated at least one time. Transfections for transactivation and mammalian two-hybrid assays were preformed as described previously (Dekeyser et al., 2009). All of the test compounds were diluted in DMSO, and the levels never exceeded 0.2% (v/v). CITCO was used as a positive control for CAR activation (Maglich et al., 2003). Because CAR1 is constitutively active, ANDRO (10 μM), a mouse CAR (Forman et al., 1998), and human CAR1 (Auerbach et al., 2007) inverse agonist are routinely included in assays to decrease CAR1 activity. This activity is restored in the presence of an agonist, which allows the study of agonist-induced CAR1 activity and for relative comparison of CAR1 inhibition. All of the chemical treatments were for 24 h, and luciferase assays were performed as described previously (Auerbach et al., 2007).
AlphaScreen Assays.
AlphaScreen assays were used to assess a direct interaction between DAX-1 and CAR1. AlphaScreen assays (PerkinElmer Life and Analytical Sciences, Waltham, MA) were performed as described for other NRs (Li et al., 2005). The human CAR1-LBD (residues 103–349) was expressed as a 6×His fusion protein from the Novagen expression vector pET24a (EMD Biosciences, San Diego, CA). The protein was purified from a nickel-nitrilotriacetic acid column followed by a Q Sepharose column. Human DAX-1 constructs a and b (residues 210–470 and 218–470, respectively) were expressed as glutathione transferase fusion proteins from the expression vector pGEX-4T-1. DAX-1 proteins were purified directly from a glutathione agarose column for the assays. The experiments were conducted with approximately 40 nM receptor ligand binding domain (LBD) each in the presence of 5 μg/ml donor and acceptor beads in a buffer containing 50 nM 4-morpholinepropanesulfonic acid, 50 mM NaF, 50 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, and 0.1 mg/ml bovine serum albumin, all adjusted to pH 7.4. AlphaScreen assays were performed in duplicate and repeated at least once.
Immunoprecipitation Assays.
COS-1 cells were seeded at a density of 1.5 × 106 in 60-mm dishes. Cells were transfected with 2 μg of pCMV6-AC-DAX1 plus 2 μg of 3X-FLAG hCAR1 or 2 μg of 3X-FLAG empty with FuGENE 6 (1 μg of DNA per 3 μl of FuGENE 6) according to the manufacturer's directions. On the following day, the medium was replaced, and approximately 36 h after transfection, cells were treated with DMSO, CITCO (3 μM), PB (0.5 mM), or ANDRO (10 μM). After 6 h of treatment, the cells were washed with phosphate-buffered saline and harvested. Cell pellets were stored at −80°C. Cell pellets were resuspended in 500 μl of ice-cold lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% NP-40, and 1× protease inhibitor cocktail] and sonicated (10 pulses) using a Branson Sonifier 250 (VWR, West Chester, PA). The lysates were centrifuged at 14,000g for 20 min at 4°C, and the supernatants were retained. Protein concentrations were determined by bicinchoninic acid methods. Protein (300 μg) was incubated with 20 μl of prewashed and preblocked anti-FLAG M2 resin in a total volume of 500 μl of lysis buffer on a rotator overnight at 4°C. The tubes were centrifuged at 7500g to collect the resin, and the supernatant was removed. The resin then was washed five times with ice-cold wash buffer (lysis buffer with 0.4% Triton X-100 and 0.2% NP-40). Protein was eluted with the addition of 20 μl of 2× SDS-polyacrylamide gel electrophoresis sample loading buffer and heating at 95°C for 5 min. The samples then were analyzed by Western blot using analysis Bio-Rad Laboratories (Hercules, CA) reagents and equipment. The blots were incubated with anti-DAX1 [1:1000 in 2% nonfat dried milk (NFDM) and 1× Tween Tris-buffered saline (TTBS), 4°C overnight] followed by anti-rabbit HRP (1:5000 in 2% NFDM and 1× TTBS, 2 h at 24°C) and detection was with Pierce ECL chemiluminescent detection kit (Thermo Fisher Scientific, Waltham, MA). Blots were stripped using Pierce Restore reagent (Thermo Fisher Scientific) and CAR input was detected with anti-FLAG M2 HRP conjugate (1:2000 in 2% NFDM and 1× TTBS, 4°C overnight).
Culture and Transfection of Human Primary Hepatocytes.
Normal human hepatocytes in a 12-well format were obtained through the Liver Tissue Cell Distribution System (Pittsburgh, PA), funded by National Institutes of Health Contract N01-DK-7-0004/HHSN267200700004C. Cell culture conditions were published previously (Goyak et al., 2008). Upon receipt of the cells, medium was replaced every 24 h. For treatments of normal (untransfected) hepatocytes, chemical treatments (DMSO; phenobarbital, 500 μM; CITCO 2 μM; or DEHP 1 μM) were applied 3 to 5 days after the receipt of hepatocytes. After 24 h, hepatocytes were washed in 1× phosphate-buffered saline (pH 7.4), followed by the addition of 600 μl of TRIzol reagent (Invitrogen, Carlsbad, CA). The TRIzol solution was mixed by pipetting and then transferred to 1.5-ml tubes followed by immediate storage at −80°C. For experiments involving transfected primary human hepatocytes, cells were transfected 1 to 3 days after culture with 2.5 μg of pCDH-dCMV-DAX1 or PCDH-dCMV-empty using JetPEI hepatocyte transfection reagent (Polyplus Transfection, Inc., New York) at a 1:3.2 ratio (micrograms of DNA to microliters of JetPEI), according to the manufacturer's instructions. Approximately 18 h after transfection, the medium was removed and replaced with medium containing DMSO or 3 μM CITCO. Treatment continued 24 to 48 h, with fresh treatments prepared every 24 h. After treatment, cells were harvested as described above. RNA was prepared, and concentrations and purity were determined using a NanoDrop 2000 (Thermo Fisher Scientific). RNA integrity was assessed using a Bio-Rad Experion. RNA (2 μg) was used for the synthesis of cDNA using the High-Capacity Reverse Transcription cDNA kit (Applied Biosystems, Foster City, CA), and the remainder was stored at −80°C. The cDNA reaction was performed in a Bio-Rad C1000 thermocycler using the following conditions: 25°C 10 min, 37°C 2 h, 85°C 5 min, 4°C forever. Once the reactions had cooled to 4°C, cDNA was stored at −20°C until use.
Real-Time Polymerase Chain Reaction.
cDNA was thawed on ice and then diluted 5-fold in nuclease-free water to give a concentration of 20 ng/μl. Master mixes were prepared for each target. The volume of components per duplicate reaction was: 15 μl of PerfeCTa SYBR Green Supermix, ROX (Quanta Biosciences, Gaithersburg, MD), 0.6 μl of forward primer (final concentration 100 nM), 0.6 μl of reverse primer (final concentration 100 nM), 10.8 μl of nuclease-free water, and 3 μl of cDNA (final concentration 20 ng/μl). SYBR green primer sets are shown in Table 2. Duplicate aliquots from each master mix were transferred to a 96-well assay plate. The reactions were run on a Bio-Rad CFX96 real-time system equipped with a C1000 thermocylcer and CFX Manager Software, version 2. The reaction conditions were as follows: 45°C for 5 min, 95°C for 3 min, 95°C for 15 s, and 60°C for 1 min (40 cycles total). Melt curves were run from 65 to 95°C with an increment of 0.5°C after each run. Standard curves using serial dilutions of human primary hepatocyte cDNA were run for all of the targets to determine reaction efficiencies for each primer set. For DAX1, cDNA obtained from hepatocytes transfected with pCDH-CMV-DAX1 was used, and for CYP2B6, cDNA from hepatocytes treated with CITCO was used. Efficiencies and standard curve parameters are shown in Supplemental Table 1. Quantification data were corrected for reaction efficiencies. Expression values were determined using the ΔΔCt method with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal reference and expressed as fold change relative to empty vector transfected or untransfected controls.
TABLE 2.
SYBR Green PCR primer sets
| Target | Primer | Sequence | Amplicon Length | Reference |
|---|---|---|---|---|
| CYP2B6 | Forward | 5′-GGTGTGCCCCACATTGTCA-3′ | 94 | Li et al., 2008 |
| Reverse | 5′-GGAGAGCAGTGCTCAGGATGA-3' | |||
| DAX1 | Forward | 5′-AGGGCAGCATCCTCTACAAC-3′ | 193 | Nedumaran et al., 2009 |
| Reverse | 5′-TGGTCTTCACCACAAAAGCA-3′ | |||
| GAPDH | Forward | 5′-CCCATCACCATCTTCCAGGAG-3′ | 285 | Li et al., 2008 |
| Reverse | 5′-GTTGTCATGGATGACCTTGGC-3′ |
Statistical Analysis.
All of the statistical tests were performed using Prism Software, version 5.03 (GraphPad Software Inc., La Jolla, CA). For determining differences in the activation of CAR by various treatments and/or interacting proteins, two-way analyses of variance were performed, followed by a Bonferroni test for comparison to controls. A p value <0.05 was considered to be significant.
Results
McDonnell Panel Screening Assays.
Given the differences in CAR variant sequence and ligand selectivity, we reasoned that the CAR variants might have selective coregulator binding preferences. To investigate this possibility, we screened the McDonnell panel (Chang et al., 1999) of ER-interacting peptides that contain the LXXLL NR box motif for interaction with CAR variants in mammalian two-hybrid assays. Although only minor differences in peptide interactions with CAR variants were detected, all of the CAR variants revealed a relatively strong interaction with a clone designated as D48, containing the sequence SGWENSILYSLLSDRVSLD (Supplemental Table 2). This clone belongs to the class III peptides containing NR boxes that are found in RIP140 and also similar to those found in PGC-1, DAX-1, and SHP (Chang et al., 1999). A BLAST search (Altschul et al., 2005) of the D48 peptide sequence against the human nonredundant protein database revealed sequence homology with the three NR boxes found in human DAX-1 (Fig. 1).
Interaction between CAR1 and DAX-1 or SHP.
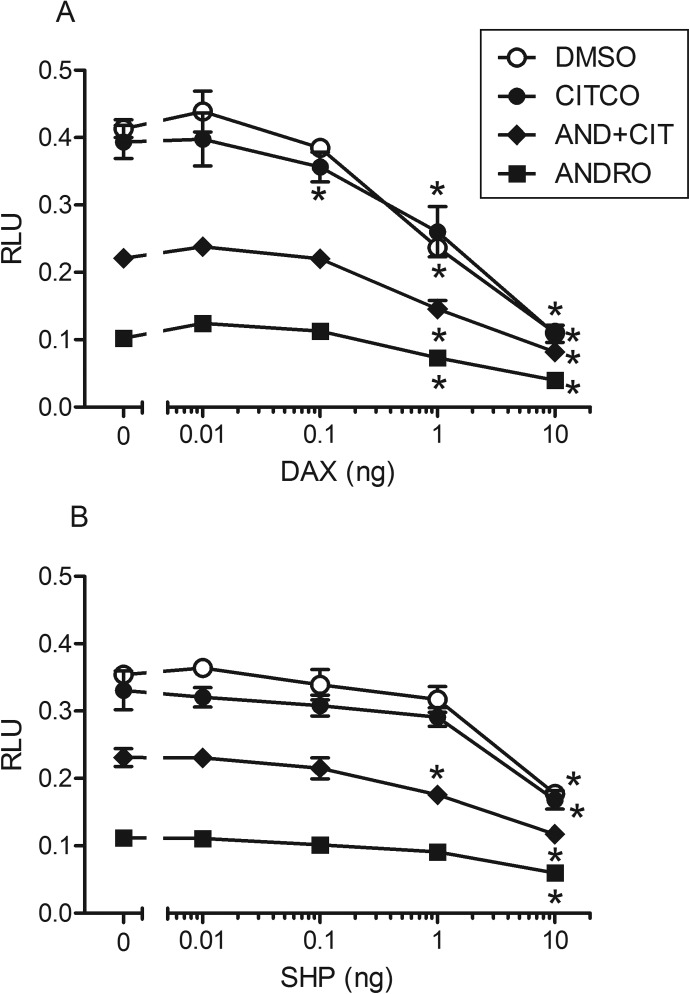
On the basis of the results of the McDonnell panel peptide screening and the fact that DAX-1 and SHP are related (Ehrlund and Treuter, 2012), we tested DAX-1 and SHP for interaction with CAR1 in reporter assays in COS-1 cells. Both DAX-1 and SHP markedly repressed the ability of CAR1 to activate the 2B6XREM luciferase reporter in a dose-dependent manner; however, DAX-1 exhibited much more potent inhibition (∼10×) than SHP (Fig. 2). These experiments also were conducted for the CAR2 and CAR3 splice variants of CAR1, with similar results obtained (Supplemental Fig. 1). In addition, DAX-1 was a potent repressor of pregnane X receptor activity in transactivation assays (Supplemental Fig. 2). For CAR1, the activity of DAX-1 was not dependent on the presence or absence of added ligand CITCO (agonist) (i.e., DAX-1 inhibited CAR1 activity in DMSO-treated cells). Interestingly, DAX-1 appeared to decrease CAR1 activity in ANDRO (inverse agonist)-treated cells. However, this reduction in activity in ANDRO-treated cells may be due to the repression of constitutive activity, because complete inhibition of CAR1 constitutive activity by DAX-1 is not achieved. These assays were repeated in hepatoma HepG2-C3A cells with analogous results (E. M. Laurenzana and C. J. Omiecinski, unpublished observations). Because of the apparent potent interaction between the CAR variants and DAX-1 in reporter assays, we further investigated the impact of DAX-1 on CAR activity.
Fig. 2.
Effect of DAX-1 or SHP on CAR1 activity. Results shown here represent single transfection experiments, with all of the treatments in quadruplicate. COS-1 cells were transfected with the CMV2-CAR1 and 3.1-RXRα expression vectors, the 2B6-XREM-PBREM reporter, the pRL-CMV vector for normalization of transfection efficiency, and varying amounts of PCMV6-DAX1 (A) or pCMV6-SHP (B). All of the treatments were for 24 h, and the data are represented as normalized luciferase values. Each data point represents the mean (±S.D.). Asterisks indicate significant difference from respective control (no DAX-1 or SHP) within a chemical treatment group (p < 0.01).
Mutation/Deletion Studies with DAX-1.
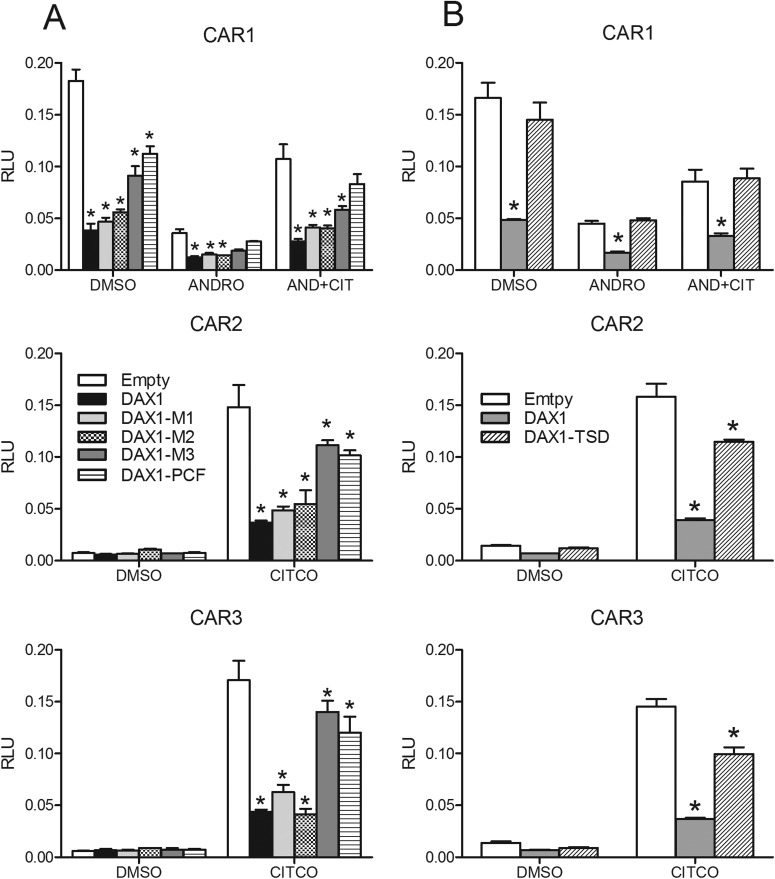
To provide further evidence of CAR and DAX-1 functional interaction and to examine the regions of DAX-1 that may contribute to the interaction, mutation analyses were performed. The three DAX-1 LXXLL/M motifs (Fig. 1) were mutated individually to produce the LXXAA mutants M1, M2, and M3. In addition, a fourth reported interacting PCFQVLP motif (Sablin et al., 2008) was mutated to yield PCFQAAP (PCF mutant). Most of the mutants significantly decreased CAR1 activity; however, wild-type DAX-1 and the DAX-1 M1 and M2 mutants were the most potent repressors, whereas the DAX-1 M3 and PCF mutants relieved DAX-1 repression of CAR1 activity to 50 to 75% of the empty vector control level, depending on the chemical treatment (Fig. 3A). Similar results were seen for the CAR2 and CAR3 splice variant receptors (Fig. 3A). A truncated version of DAX-1 (DAX1-TSD), where amino acids 442 to 469 corresponding to the TSD (Fig. 1) were deleted, also was constructed. This deletion is equivalent to a naturally occurring DAX-1 mutant that results in adrenal hypoplasia congenita (Ito et al., 1997). CAR1 activity was restored to 90 to 100% of control levels with the TSD-deleted DAX-1 construct across the various chemical treatments (Fig. 3B). Likewise, deletion of the DAX1-TSD restored CITCO-mediated activation of CAR2 and CAR3 to approximately 75% of control values. Experiments with these DAX-1 mutant constructs also were performed with pregnane X receptor. The results mirrored results seen with the CAR variants (Supplemental Fig. 3).
Fig. 3.
Effect of DAX-1 mutations on CAR variant activity. Results shown here represent single transfection experiments, with all of the treatments in quadruplicate. COS-1 cells were transfected with the 3.1-RXRα expression vector, the 2B6-XREM-PBREM reporter, the pRL-CMV vector for normalization of transfection efficiency, and varying amounts of CMV2-CAR1, -CAR2, or -CAR3 and PCMV6-empty, -DAX1 WT, -DAX1 M1, -DAX1 M2, -DAX1 M3, or -DAX1 PCF (A) or PCMV6-DAX1-TSD (B). All of the treatments were for 24 h, data are represented as normalized luciferase values, and each data point represents the mean (±S.D.). Asterisks indicate significant difference from respective control (no DAX1) within a chemical treatment group (p < 0.05).
Two-Hybrid Assays for CAR1 and DAX-1 Interaction.
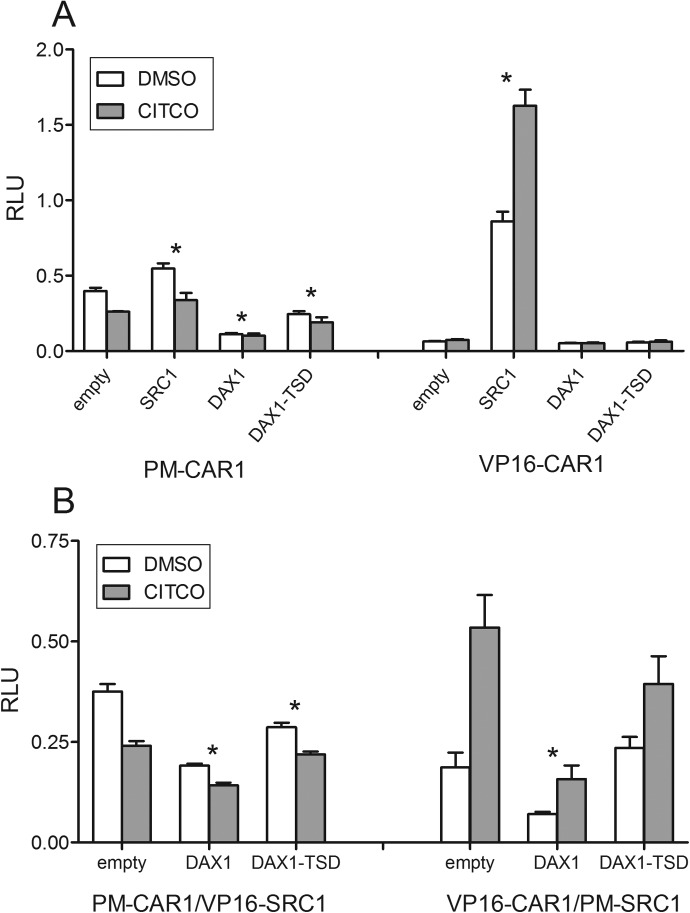
The interaction between CAR1 and DAX-1 was further assessed in mammalian two-hybrid assays. Because SRC1 is known to interact with human CAR (Auerbach et al., 2007), it was used as a positive control. The initial screening suggested no interaction between CAR1 and DAX-1, in either the presence or the absence of RXRα (E. M. Laurenzana and C. J. Omiecinski, unpublished observations). However, this negative result could manifest from the presence of the TSD in DAX-1 that itself likely suppresses transcription of the GAL4 luciferase reporter. Therefore, the DAX1-TSD mutant also was tested in the mammalian two-hybrid assay. These assays were performed in two different vector orientations (pm-CAR1 with VP16-test constructs and VP16-CAR with pm-test constructs). An interesting observation derived from the different vector orientations was that CITCO was slightly inhibitory of CAR1 interactions when CAR was in the pm vector, whereas it increased the interaction with SRC1 in the VP16 vector (Fig. 4A). The cause for this difference is unknown, but because the test constructs are expressed as fusion proteins with either the GAL4 DBD (pm vector) or the herpes virus transcription activation domain (VP16 vector), these different configurations may affect the respective conformational changes induced by an agonist and in turn affect the interaction between the test proteins. Another interesting observation was that pm-CAR1 exhibited a low basal activity (with VP16-empty), whereas VP16-CAR1 showed no basal activity with pm-empty. This result may extend from the CAR1-DBD fusion protein's ability to recruit endogenous transcriptional activators.
Fig. 4.
A, mammalian two-hybrid assays to assess CAR1 interaction with DAX-1. COS- cells were cotransfected with pmGAL4-CAR1-LBD and VP16-empty (negative control), VP16-SRC1 (positive control), VP16-DAX1, or VP16-DAX1 TSD, 3.1 RXRα-LBD, pFR-luciferase reporter, and pRL-CMV vector for transfection efficiency normalization (left side). On the right half of the graph, CAR1 was in the VP16 vector, and the test constructs were in the pmGAL4 vector. B, mammalian two-hybrid assay to assess the effect of DAX-1 on CAR1-SRC1 interaction. COS-1 cells were cotransfected with pmGAL4-CAR1 and VP16-SRC1 along with either VP16-empty, -DAX1, or -DAX1-TSD. The 3.1 RXRα-LBD, pFR-luciferase reporter, and pRL-CMV vectors were included in all of the transfections. On the right half of the graph, VP16-CAR1-LBD and pmGAL4-SRC1 were tested in the presence of pmGAL4-empty, -DAX1, or -DAX1-TSD. Both panels are representative single transfection experiments, with all of the treatments in quadruplicate, and each data point represents the mean (±S.D.). Asterisks indicate that each treatment was significantly different from its respective empty vector control (p < 0.05).
When pm-CAR1 was tested (Fig. 4A, left panel), the DAX1-TSD mutant exhibited higher activity with CAR1 than with full-length DAX-1, although not as robust as the interaction between SRC1 and CAR1. Furthermore, the pm-CAR1:DAX1-TSD activity was lower than that generated by the pmCAR1:VP16-empty vector, suggesting some suppression of CAR1 basal activity. When VP16-CAR1 was assayed with the pm-test constructs (Fig. 4A, right), an interaction was detected only when SRC1 was present. To help determine whether DAX-1 might compete with SRC1 for binding to CAR1, the effect of DAX-1 on the interaction between CAR1 and SRC1 was assessed. Again, the experiments were performed in both vector orientations. DAX-1 inhibited the interaction between CAR1 and SRC1 by approximately 50% compared with control. In contrast, when the DAX1-TSD mutant was used, the interaction between CAR1 and SRC1 was inhibited by only 10% compared with control (Fig. 4B).
AlphaScreen Assays.
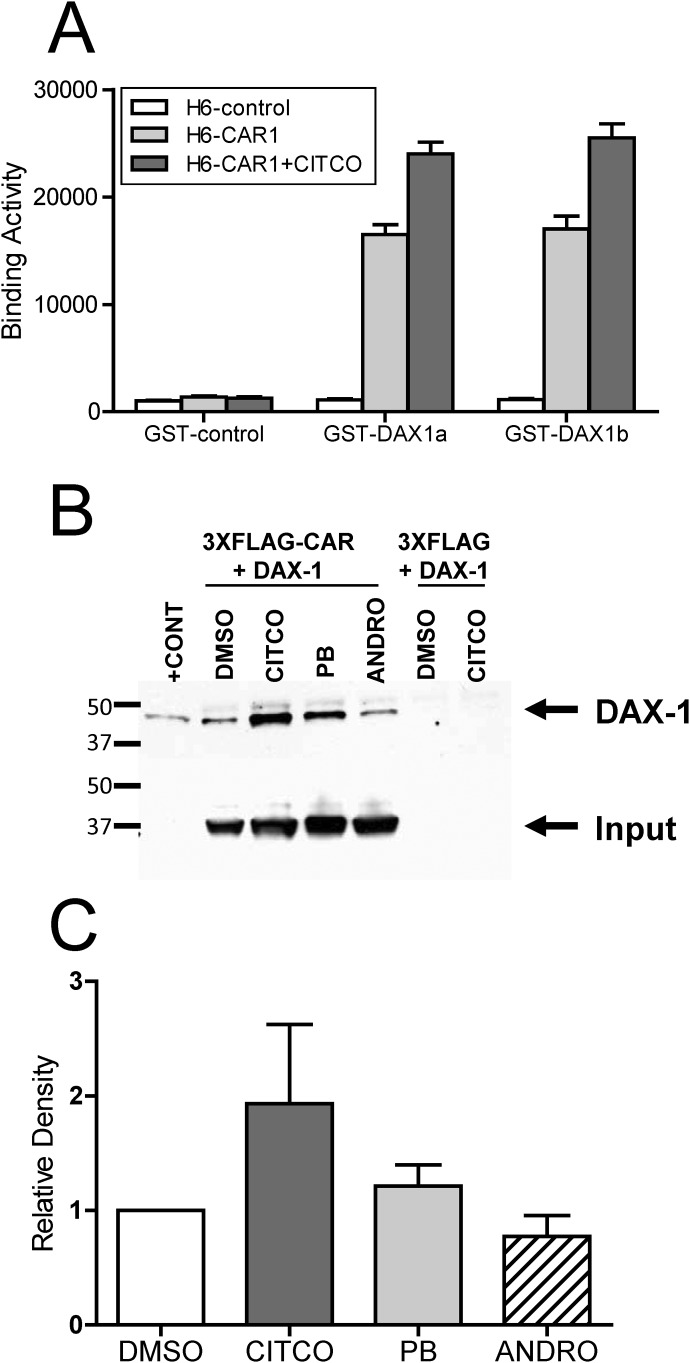
To obtain evidence of a direct interaction between CAR1 and DAX-1, AlphaScreen assays were performed. These assays demonstrated a direct interaction between CAR1 and two different DAX1 constructs (Fig. 5A). Furthermore, this interaction was enhanced significantly by the addition of the CAR ligand CITCO, suggesting that DAX-1 preferentially binds CAR1 in the ligand-bound state. It is noteworthy that both of the DAX-1 constructs contained only the PCFQVLP NR box, supporting the conclusion that this sequence is important for the interaction with CAR proteins (Fig. 6).
Fig. 5.
Direct interaction between CAR1 and DAX-1. AlphaScreen assay for CAR1 interaction with DAX-1 (A). Purified CAR1-LBB and DAX-1-GST (∼40 nM each) were incubated in the presence of 5 μg/ml donor and acceptor beads. The DAX-1a construct contains amino acids 210 to 470 and DAX-1b contains amino acids 218 to 470. Bars represent the mean (±S.D.) of two separate experiments, each performed in duplicate. Representative coimmunoprecipitation of CAR1 and DAX1 (B). COS-1 cells were cotransfected with pCMV6-DAX1 and p3XFLAG CAR or 3XFLAFG empty. After ∼40 h, the cells were treated with DMSO, CITCO, ANDRO, or PB for 5 to 6 h and then harvested. Cell lysates were immunoprecipitated with anti-FLAG beads, and precipitated protein was subjected to Western blot analysis with anti-DAX1 antibody, followed by anti-FLAG antibody. C, densitometry analysis of the Western blot data.
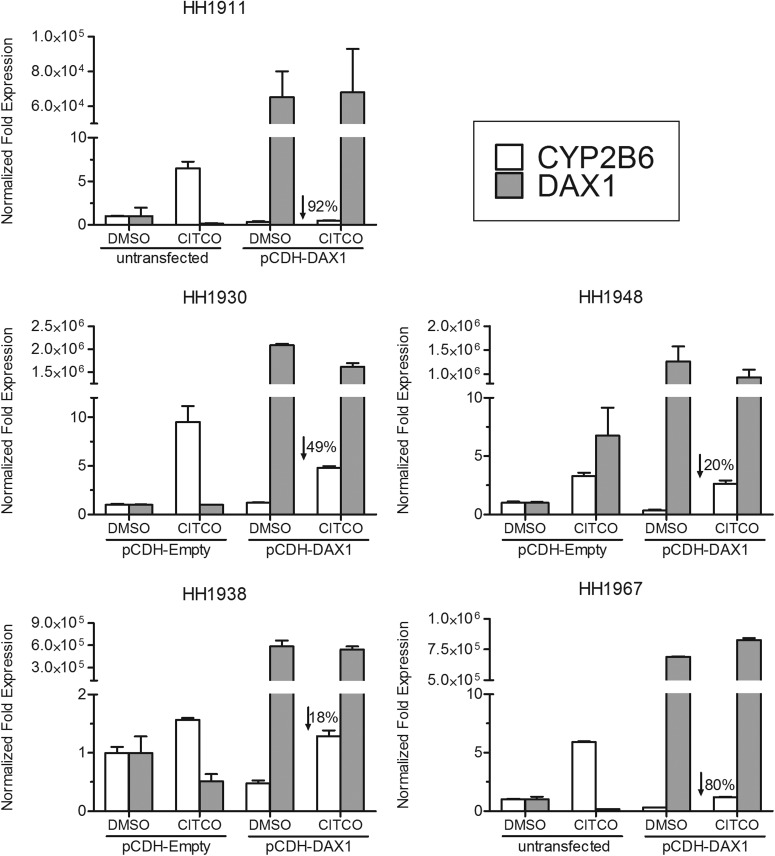
Fig. 6.
Effect of DAX-1 on CAR-mediated induction of CYP2B6 by CITCO in primary human hepatocytes. Human primary hepatocytes were transfected with DAX-1, empty vector and then treated with CITCO or DMSO. Real-time polymerase chain reaction was performed on cDNA samples isolated from the cells to quantify DAX-1, CYP2B6, and GAPDH (as an internal reference). Values were expressed as fold induction relative to untransfected or empty vector-transfected, DMSO-treated cells and represent the average of two replicate samples. Percentages indicate the decrease in CYP2B6 induction relative to untransfected or empty vector-transfected, CITCO-treated cells.
Coimmunoprecipitation of DAX-1 with CAR1.
Additional evidence of a direct interaction between CAR1 and DAX-1 was obtained with immunoprecipitation experiments conducted using lysates from COS-1 cells that were cotransfected with 3XFLAG-CAR1 and pCMV6-DAX1 and treated with model CAR direct and indirect activators as well as the inverse agonist ANDRO. Anti-FLAG beads were used for immunoprecipitation, and the precipitated proteins were subjected to Western blot analysis with anti-DAX1 antibody. As shown in Fig. 5, B and C, the highest levels of DAX-1 were detected from the cells that were treated with CITCO, with lower levels detected in the other treatment groups. These results agree with the AlphaScreen results suggesting that DAX-1 preferentially binds with CAR1 in the ligand-bound state.
Overexpression of DAX-1 in Primary Human Hepatocytes.
CITCO induces CYP2B6 in human liver via activation of CAR (Maglich et al., 2003). To assess the impact of DAX-1 on CAR-mediated CYP2B6 induction ex vivo, primary human hepatocytes from five individual donors were transfected with DAX-1, treated with CITCO, and then assessed for CYP2B6 mRNA expression. The donor characteristics are shown in Table 3. On the basis of the visualization of GFP, the transfection efficiency was estimated as ranging from approximately 5 to 20% in the different hepatocyte donor preparations (E. M. Laurenzana and C. J. Omiecinski, unpublished observations). However, it should be noted that the pCDH-dCMV vector used in the transfections contains dual promoters, with GFP driven by a separate promoter than DAX-1, and thus the relative efficiencies of expression of these proteins may be different. In this respect, the Ct values obtained for DAX-1 expression clearly demonstrated DAX-1 overexpression in the cultured cells. Real-time polymerase chain reaction analyses of the respective hepatocyte cDNAs showed that DAX-1 expression attenuated CAR-mediated induction of CYP2B6 by CITCO in all of the donors, with the extent of CYP2B6 inhibition ranging from 18 to 92% across individual hepatocyte cultures.
TABLE 3.
Human hepatocyte donor information
| Donor ID | Sex | Age | Source | Disease or Cause of Death | Chemotherapy |
|---|---|---|---|---|---|
| 1911 | M | 14 | Surgical Resection | Unknown | Unknown |
| 1930 | M | 60 | Surgical Resection | Metastatic adenocarcinoma | Yes |
| 1938 | F | 49 | Surgical Resection | Metastatic colon cancer | Yes |
| 1948 | F | 60 | Surgical Resection | Metastatic cancer origin unknown | Unknown |
| 1967 | M | 25 | Donor | Drug overdose | No |
ID, identification; M, male; F, female.
Effect of CAR Activation on DAX-1 Expression in Primary Human Hepatocytes.
To assess whether CAR activation induces hepatic DAX-1 expression, untransfected primary human hepatocytes were treated with prototypical CAR (PB and CITCO) or CAR2 (DEHP) activators for 24 h and assessed quantitatively for levels of DAX-1 mRNA expression. Four separate hepatocyte donors were tested, but in no case was DAX-1 induction detected by these treatments (E. M. Laurenzana and C. J. Omiecinski, unpublished observations).
Discussion
Screening a panel of peptides containing different LXXLL motifs (or NR boxes) led to the identification of DAX-1 as a potential CAR-interacting protein and repressor of CAR transcriptional activity. DAX-1 has been most clearly characterized for its role in human development, because mutations in the gene are associated with both X-linked congenital adrenal hypoplasia and hypogonadotropic hypogonadism (Zanaria et al., 1994; Tabarin et al., 2000). More recently, a role for DAX-1 as a regulator of liver physiology has emerged, with its reported activity as a corepressor of hepatocyte nuclear factor 4α (HNF4α) and liver X receptor (LXR) resulting in negative regulation of gluconeogenic pathways (Nedumaran et al., 2009) and lipogenesis, respectively (Nedumaran et al., 2010). Although DAX-1 exhibited the highest degree of homology with the detected CAR-interacting peptide, we also examined SHP, because they belong to the same NR subfamily (Ehrlund and Treuter, 2012) and, like CAR, SHP is expressed in the liver and was demonstrated previously to interact with mouse CAR (Bae et al., 2004; Park et al., 2004). Furthermore, DAX-1 and SHP both lack a traditional NR DBD, contain LXXLL NR interaction motifs otherwise found in nuclear coactivators, and act as transcriptional corepressors of ligand-activated NRs (Ehrlund and Treuter, 2012). Despite these similarities, our results demonstrate that DAX-1 is a much more potent repressor of human CAR1 activity than is SHP.
Similar to SHP (Lee et al., 2000), DAX-1 likely functions through two different protein interaction mechanisms for the repression of NR activity. The first is based on the four NR boxes in the DAX-1 sequence. The three NR boxes localized in its N-terminal region are analogous to the LXXLL motifs commonly found in NR coactivators. A fourth sequence, PCFQVLP, is thought to mimic the LXXLL motifs and forms the interface for the interaction with the AF-2 domain of LRH-1 (Sablin et al., 2008). It is interesting to note that SHP also contains a similar PSFCHLP sequence, although no functional relevance has been demonstrated (Ehrlund and Treuter, 2012). These four DAX-1 NR boxes allow for competition with coactivators. A second mechanism mediating DAX-1 repression lies with a C-terminal transcription silencing domain (TSD). The TSD allows for the recruitment of transcriptional corepressors such as NCoR (Crawford et al., 1998), SMRT (Agoulnik et al., 2003), and Alien (Altincicek et al., 2000).
We further tested CAR1 and its splice variants, CAR2 and CAR3, for differences in their interactions with DAX-1. For CAR1, CAR2, and CAR3, mutation of the first two NR boxes (M1 and M2, Fig. 1) had no effect on DAX-1 repression of CAR activity, whereas mutation of the third NR box and the atypical PCFVQLP NR box (M3 and PCF, Fig. 1) inhibited DAX-1's repression of CAR activity by approximately 50 to 75%. The results also suggested that the third NR box interacted more strongly with CAR2 and CAR3 than with wild-type CAR1. These differences may stem from conformational changes in the CAR variant proteins resulting from amino acid insertions in their sequences (Auerbach et al., 2003). Nevertheless, the third LXXLL and the PCFQVLP NR boxes of DAX-1 were the most critical in mediating the functional interaction with CAR. It is interesting to note that deletion of DAX-1's TSD restored CAR1 activity to control levels, whereas the comparative activities of CAR2 and CAR3 were only restored to approximately 65% of control. Therefore, these results suggest that: 1) the third LXXLL and the PCFQVLP NR boxes of DAX-1 are the principal effectors of coactivator competition and interaction with CAR and 2) DAX-1 competition with coactivators may be more for CAR2 and CAR3 than CAR1.
Initial mammalian two-hybrid studies failed to detect a direct interaction between CAR1 and DAX-1. Explanations for this result may implicate another “bridging” protein required for the interaction and/or that the intrinsic repressor effects from the TSD of DAX-1 are blocking activation of the GAL4 reporter in these assays. The latter possibility appears likely, because the basal activity generated with pm-CAR1 and VP16-empty was repressed in the pm-CAR1:VP16-DAX-1 assay (Fig. 4A, left). To address whether TSD was masking the detection of the CAR1-DAX-1 interaction, a DAX-1-TSD construct was used in the mammalian two-hybrid assays. The basal activity of pm-CAR1 was partially restored with VP16-DAX-1-TSD. We further tested DAX-1 and DAX-1-TSD in a mammalian two-hybrid competition assay with CAR1 and SRC1. Although intact DAX-1 inhibited the CAR1-SRC1 interaction 50 to 75% depending on the vector orientation, DAX-1-TSD was not as efficient. In summary, these results support the concept that competition with SRC1 and the intrinsic transcription repressor domain are both important mechanisms in DAX-1 repression of CAR transcriptional activity. However, the results did not clarify whether a direct interaction existed between CAR1 and DAX-1.
In these respects, both AlphaScreen and coimmunoprecipitation experiments clearly indicated a direct interaction between these proteins. Furthermore, in both assays the CAR1-DAX-1 direct interaction was enhanced by the presence of the ligand CITCO, which is consistent with known functions of DAX-1 acting as a repressor of ligand-activated receptors. In general, corepressors bind to NRs in the absence of ligand (Perissi et al., 1999). Although CAR1 is “constitutively active” in vitro, binding of the inverse agonist ANDRO favors the recruitment of the corepressor SMRT (Dussault et al., 2002). Thus, although SMRT or other corepressors help to regulate activity of “unactivated” CAR, DAX-1 serves to repress the activity of CAR1 as well as the ligand-activated variants of the receptor, CAR2 and CAR3. In this manner, DAX-1 may provide another level through which CAR activity is fine-tuned.
In vivo, CAR is a hepatic “xenosensor” that upon chemical activation translocates to the nucleus where it facilitates the transcription of genes encoding xenobiotic metabolism and transport (Timsit and Negishi, 2007). CYP2B6 is a prototypical phase I gene activated by CAR in human liver (Honkakoski et al., 2003). The current studies demonstrated that DAX-1 can mediate CAR transcriptional activity in primary human hepatocytes, with the overexpression of DAX-1 resulting in decreased CAR-mediated induction of CYP2B6 in CITCO-treated cells. Given the interindividual differences noted in inducer responsiveness among humans (Dekeyser et al., 2009), the degree of CYP2B6 induction in CITCO-treated cultures was variable among donors, as was the extent of DAX-1 repression. In general, greater levels of CYP2B6 repression were observed in donors that were most responsive to CITCO induction. Differential xenobiotic responsiveness among humans likely results from a complex interplay of genetics, previous chemical exposures, and perhaps differences in the expression profiles of the xenobiotic receptors that mediate these responses. Because CAR activation in the liver mediates the induction of xenobiotic metabolic enzymes and transporters, DAX-1 repression may contribute to the etiology of adverse drug reactions in select individuals.
Although the physiological implications of DAX-1 repression of hepatic CAR activity are currently unclear, DAX-1 appears to negatively regulate hepatic gluconeogenesis and lipogenesis in mice (Nedumaran et al., 2009, 2010). These reports demonstrated that hepatic DAX-1 expression is modulated by insulin and nutritional status and that DAX-1 repressed the transcriptional activity of HNF4α, a transcription factor known to positively regulate gluconeogenic enzymes (Nedumaran et al., 2009). Furthermore, adenoviral-mediated overexpression of DAX-1 in mice fed a high-fat diet significantly reduced fasting blood-glucose levels. In another study, DAX-1 was shown to repress hepatic LXRα transcriptional activity, resulting in decreased in expression of SREBP-1c, a transcription factor that mediates the expression of lipogenic enzymes, as well as decreased liver triglyceride levels (Nedumaran et al., 2010). It is interesting to note that CAR activation decreases lipogenesis in mouse models fed a high-fat diet, and different, but perhaps interrelated, mechanisms have been proposed for this effect. CAR activation induces the expression of Insig-1, which in turn represses the activation of SREBP-1c, resulting in decreased expression of lipogenic enzymes (Roth et al., 2008). Another study outlined an indirect mechanism whereby CAR-mediated induction of SULT2B1b results in the inactivation of LXRα oxysterol agonists, in turn causing decreased activation of SREBP-1c by LXRα (Dong et al., 2009). Furthermore, CAR activation also has been shown to suppress the expression of gluconeogenic enzymes in mouse models, resulting in improved antidiabetic effects (Dong et al., 2009). Given these observations, DAX-1 antagonism of CAR activity in the liver would be predicted to have opposing effects compared with DAX-1 antagonism of hepatic HNF4α and LXRα. Thus, it is conceivable that DAX-1 binds to CAR or HNF4α and LXRα preferentially, based on nutritional status and perhaps the ligand bound to the receptor. Future studies are required to sort out the complex NR-mediated regulatory mechanisms controlling hepatic lipogenesis and gluconeogenesis.
Given that DAX-1 and SHP share structural and functional similarities (Ehrlund and Treuter, 2012) and that hepatic SHP expression is induced by bile acid-activated farnesoid X receptor, ultimately down-regulating bile acid and fatty acid synthesis (Lee et al., 2007), we investigated whether CAR activation might induce DAX-1 expression in primary human hepatocytes. After 24 h of treatment with CAR agonists, no induction of DAX-1 expression was observed (E. M. Laurenzana and C. J. Omiecinski, unpublished observations); however, it is intriguing to speculate that DAX-1 may be induced by a downstream metabolic product to mediate CAR activity.
Another tissue in which DAX-1-mediated repression of CAR activity may be important is the adrenal gland. DAX-1 is highly expressed in the adrenal gland, and mutations in DAX-1 result in adrenal hypoplasia congenita (for a review, see Lalli and Sassone-Corsi, 2003; Niakan and McCabe, 2005). Relatively high levels of CAR1 and CAR variants also are detected in the adrenal gland (Savkur et al., 2003; Arnold et al., 2004; Lamba et al., 2004), although a physiologic role in adrenal function has not been identified. However, a glucocorticoid response element has been identified in the CAR promoter sequence, and studies in human hepatocytes demonstrated that activated GR induces CAR expression (Pascussi et al., 2000, 2003). Furthermore, DAX-1 is known to repress ligand-activated GR transactivation (Zhou et al., 2008). Thus, it seems likely that interactions of DAX-1, CAR, and GR in the adrenal gland may influence glucocorticoid homeostasis; however, additional studies are necessary to explore these interactions.
This study demonstrates for the first time that DAX-1 functions as a corepressor for activated human CAR and its splice variants CAR2 and CAR3. These effects are likely mediated through competition with coactivators, such as SRC1, and via the intrinsic TSD of DAX-1, which is responsible for the recruitment of corepressors. Although the physiological implications of the CAR-DAX-1 interaction are yet to be determined, DAX-1-mediated repression of CAR transcriptional activity represents an additional level through which CAR activity may be precisely regulated in liver hepatocytes.
Supplementary Material
Acknowledgments
We thank Denise Coslo for technical assistance with the primary human hepatocytes.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This study was supported by a grant from the National Institutes of Health National Institute of General Medicine [Grant GM066411].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- NR
- nuclear receptor
- AF
- activation function
- ANDRO
- 5α-androstan-3α-ol
- CAR
- constitutive androstane receptor
- CITCO
- 6-(4-chlorophenyl)imidazo[2,1-b]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime
- CMV
- cytomegalovirus
- DAX-1
- dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1
- DBD
- DNA-binding domain
- DMSO
- dimethyl sulfoxide
- CAS
- Chemical Abstract Service
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HNF4α
- hepatocyte nuclear factor 4α
- LBD
- ligand-binding domain
- LXR
- liver X receptor
- PB
- phenobarbital
- PBREM
- phenobarbital response enhancer module
- RID
- receptor interaction domain
- RXR
- retinoid X receptor
- SHP
- small heterodimer protein
- SRC-1
- steroid receptor coactivator 1
- TSD
- transcription silencing domain
- VP16
- virus protein 16
- XREM
- xenobiotic response enhancer module
- NFDM
- nonfat dried milk
- HRP
- horseradish peroxidase
- TTBS
- Tween Tris-buffered saline.
Authorship Contributions
Participated in research design: Laurenzana, Chen, and Omiecinski.
Conducted experiments: Laurenzana, Chen, Kannuswamy, Sell, and Li.
Contributed new reagents or analytic tools: Strom and Li.
Wrote or contributed to the writing of the manuscript: Laurenzana and Omiecinski.
References
- Agoulnik IU, Krause WC, Bingman WE, 3rd, Rahman HT, Amrikachi M, Ayala GE, Weigel NL. (2003) Repressors of androgen and progesterone receptor action. J Biol Chem 278:31136–31148 [DOI] [PubMed] [Google Scholar]
- Altincicek B, Tenbaum SP, Dressel U, Thormeyer D, Renkawitz R, Baniahmad A. (2000) Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J Biol Chem 275:7662–7667 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda A, Pascual A. (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81:1269–1304 [DOI] [PubMed] [Google Scholar]
- Arnold KA, Eichelbaum M, Burk O. (2004) Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl Recept 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Dekeyser JG, Stoner MA, Omiecinski CJ. (2007) CAR2 displays unique ligand binding and RXRalpha heterodimerization characteristics. Drug Metab Dispos 35:428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. (2003) Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res 31:3194–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Stoner MA, Su S, Omiecinski CJ. (2005) Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3). Mol Pharmacol 68:1239–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y, Kemper JK, Kemper B. (2004) Repression of CAR-mediated transactivation of CYP2B genes by the orphan nuclear receptor, short heterodimer partner (SHP). DNA Cell Biol 23:81–91 [DOI] [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. (1994) A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol 14:1544–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Norris JD, Grøn H, Paige LA, Hamilton PT, Kenan DJ, Fowlkes D, McDonnell DP. (1999) Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol Cell Biol 19:8226–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Chen Q, Xu Y, Zhou Q, Zhu J, Zhang H, Wu Q, Xu J, Yu C. (2012) SRC-3 is required for CAR-regulated hepatocyte proliferation and drug metabolism. J Hepatol 56:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Dorn C, Sadovsky Y, Milbrandt J. (1998) Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol Cell Biol 18:2949–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKeyser JG, Laurenzana EM, Peterson EC, Chen T, Omiecinski CJ. (2011) Selective phthalate activation of naturally occurring human constitutive androstane receptor splice variants and the pregnane X receptor. Toxicol Sci 120:381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKeyser JG, Stagliano MC, Auerbach SS, Prabhu KS, Jones AD, Omiecinski CJ. (2009) Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol Pharmacol 75:1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. (1998) Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol 12:302–313 [DOI] [PubMed] [Google Scholar]
- Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, Stevens RD, Ilkayeva O, Newgard CB, Chan L, et al. (2009) Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci USA 106:18831–18836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussault I, Lin M, Hollister K, Fan M, Termini J, Sherman MA, Forman BM. (2002) A structural model of the constitutive androstane receptor defines novel interactions that mediate ligand-independent activity. Mol Cell Biol 22:5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlund A, Treuter E. (2012) Ligand-independent actions of the orphan receptors/corepressors DAX-1 and SHP in metabolism, reproduction and disease. J Steroid Biochem Mol Biol 130:169–179 [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. (1998) Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature 395:612–615 [DOI] [PubMed] [Google Scholar]
- Goyak KM, Johnson MC, Strom SC, Omiecinski CJ. (2008) Expression profiling of interindividual variability following xenobiotic exposures in primary human hepatocyte cultures. Toxicol Appl Pharmacol 231:216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. (2003) Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem 278:45062–45071 [DOI] [PubMed] [Google Scholar]
- Heery DM, Hoare S, Hussain S, Parker MG, Sheppard H. (2001) Core LXXLL motif sequences in CREB-binding protein, SRC1, and RIP140 define affinity and selectivity for steroid and retinoid receptors. J Biol Chem 276:6695–6702 [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed] [Google Scholar]
- Holter E, Kotaja N, Mäkela S, Strauss L, Kietz S, Jänne OA, Gustafsson JA, Palvimo JJ, Treuter E. (2002) Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX-1. Mol Endocrinol 16:515–528 [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Sueyoshi T, Negishi M. (2003) Drug-activated nuclear receptors CAR and PXR. Ann Med 35:172–182 [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. (1999) The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402:93–96 [DOI] [PubMed] [Google Scholar]
- Ito M, Yu R, Jameson JL. (1997) DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol 17:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Mangelsdorf DJ. (2009) Nuclear receptor regulation of stemness and stem cell differentiation. Exp Mol Med 41:525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GS, Lee GY, Nedumaran B, Park YY, Kim KT, Park SC, Lee YC, Kim JB, Choi HS. (2008) The orphan nuclear receptor DAX-1 acts as a novel transcriptional corepressor of PPARgamma. Biochem Biophys Res Commun 370:264–268 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lee SK, Na SY, Choi HS, Lee JW. (1998) Molecular cloning of xSRC-3, a novel transcription coactivator from Xenopus, that is related to AIB1, p/CIP, and TIF2. Mol Endocrinol 12:1038–1047 [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y. (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24:7931–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y, Negishi M, Kodama S. (2008) The roles of nuclear receptors CAR and PXR in hepatic energy metabolism. Drug Metab Pharmacokinet 23:8–13 [DOI] [PubMed] [Google Scholar]
- Lalli E, Sassone-Corsi P. (2003) DAX-1, an unusual orphan receptor at the crossroads of steroidogenic function and sexual differentiation. Mol Endocrinol 17:1445–1453 [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lamba V, Yasuda K, Lin YS, Assem M, Thompson E, Strom S, Schuetz E. (2004) Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J Pharmacol Exp Ther 311:811–821 [DOI] [PubMed] [Google Scholar]
- Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD. (2000) The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol 20:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Chanda D, Sim J, Park YY, Choi HS. (2007) Structure and function of the atypical orphan nuclear receptor small heterodimer partner. Int Rev Cytol 261:117–158 [DOI] [PubMed] [Google Scholar]
- Lempiäinen H, Molnár F, Macias Gonzalez M, Peräkylä M, Carlberg C. (2005) Antagonist- and inverse agonist-driven interactions of the vitamin D receptor and the constitutive androstane receptor with corepressor protein. Mol Endocrinol 19:2258–2272 [DOI] [PubMed] [Google Scholar]
- Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. (2008) The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol Pharmacol 74:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Suino K, Daugherty J, Xu HE. (2005) Structural and biochemical mechanisms for the specificity of hormone binding and coactivator assembly by mineralocorticoid receptor. Mol Cell 19:367–380 [DOI] [PubMed] [Google Scholar]
- Maglich JM, Parks DJ, Moore LB, Collins JL, Goodwin B, Billin AN, Stoltz CA, Kliewer SA, Lambert MH, Willson TM, et al. (2003) Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem 278:17277–17283 [DOI] [PubMed] [Google Scholar]
- Masson D, Qatanani M, Sberna AL, Xiao R, Pais de Barros JP, Grober J, Deckert V, Athias A, Gambert P, Lagrost L, et al. (2008) Activation of the constitutive androstane receptor decreases HDL in wild-type and human apoA-I transgenic mice. J Lipid Res 49:1682–1691 [DOI] [PubMed] [Google Scholar]
- Min G, Kemper JK, Kemper B. (2002) Glucocorticoid receptor-interacting protein 1 mediates ligand-independent nuclear translocation and activation of constitutive androstane receptor in vivo. J Biol Chem 277:26356–26363 [DOI] [PubMed] [Google Scholar]
- Muangmoonchai R, Smirlis D, Wong SC, Edwards M, Phillips IR, Shephard EA. (2001) Xenobiotic induction of cytochrome P450 2B1 (CYP2B1) is mediated by the orphan nuclear receptor constitutive androstane receptor (CAR) and requires steroid co-activator 1 (SRC-1) and the transcription factor Sp1. Biochem J 355:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedumaran B, Hong S, Xie YB, Kim YH, Seo WY, Lee MW, Lee CH, Koo SH, Choi HS. (2009) DAX-1 acts as a novel corepressor of orphan nuclear receptor HNF4alpha and negatively regulates gluconeogenic enzyme gene expression. J Biol Chem 284:27511–27523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedumaran B, Kim GS, Hong S, Yoon YS, Kim YH, Lee CH, Lee YC, Koo SH, Choi HS. (2010) Orphan nuclear receptor DAX-1 acts as a novel corepressor of liver X receptor alpha and inhibits hepatic lipogenesis. J Biol Chem 285:9221–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, McCabe ER. (2005) DAX1 origin, function, and novel role. Mol Genet Metab 86:70–83 [DOI] [PubMed] [Google Scholar]
- Park YY, Kim HJ, Kim JY, Kim MY, Song KH, Cheol Park K, Yu KY, Shong M, Kim KH, Choi HS. (2004) Differential role of the loop region between helices H6 and H7 within the orphan nuclear receptors small heterodimer partner and DAX-1. Mol Endocrinol 18:1082–1095 [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Busson-Le Coniat M, Maurel P, Vilarem MJ. (2003) Transcriptional analysis of the orphan nuclear receptor constitutive androstane receptor (NR1I3) gene promoter: identification of a distal glucocorticoid response element. Mol Endocrinol 17:42–55 [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. (2000) Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: consequences on cytochrome P450 gene regulation. Mol Pharmacol 58:1441–1450 [DOI] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. (1999) Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev 13:3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Looser R, Kaufmann M, Meyer UA. (2008) Sterol regulatory element binding protein 1 interacts with pregnane X receptor and constitutive androstane receptor and represses their target genes. Pharmacogenet Genomics 18:325–337 [DOI] [PubMed] [Google Scholar]
- Sablin EP, Woods A, Krylova IN, Hwang P, Ingraham HA, Fletterick RJ. (2008) The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci USA 105:18390–18395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Wu Y, Bramlett KS, Wang M, Yao S, Perkins D, Totten M, Searfoss G, 3rd, Ryan TP, Su EW, et al. (2003) Alternative splicing within the ligand binding domain of the human constitutive androstane receptor. Mol Genet Metab 80:216–226 [DOI] [PubMed] [Google Scholar]
- Seol W, Choi HS, Moore DD. (1996) An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science 272:1336–1339 [DOI] [PubMed] [Google Scholar]
- Shiraki T, Sakai N, Kanaya E, Jingami H. (2003) Activation of orphan nuclear constitutive androstane receptor requires subnuclear targeting by peroxisome proliferator-activated receptor gamma coactivator-1 alpha. A possible link between xenobiotic response and nutritional state. J Biol Chem 278:11344–11350 [DOI] [PubMed] [Google Scholar]
- Song KH, Park YY, Park KC, Hong CY, Park JH, Shong M, Lee K, Choi HS. (2004) The atypical orphan nuclear receptor DAX-1 interacts with orphan nuclear receptor Nur77 and represses its transactivation. Mol Endocrinol 18:1929–1940 [DOI] [PubMed] [Google Scholar]
- Tabarin A, Achermann JC, Recan D, Bex V, Bertagna X, Christin-Maitre S, Ito M, Jameson JL, Bouchard P. (2000) A novel mutation in DAX1 causes delayed-onset adrenal insufficiency and incomplete hypogonadotropic hypogonadism. J Clin Invest 105:321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit YE, Negishi M. (2007) CAR and PXR: the xenobiotic-sensing receptors. Steroids 72:231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi M, Borngraeber S, Sandler B, Fletterick RJ, Webb P, Baxter JD. (2005) Conformational adaptation of nuclear receptor ligand binding domains to agonists: potential for novel approaches to ligand design. J Steroid Biochem Mol Biol 93:127–137 [DOI] [PubMed] [Google Scholar]
- Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, et al. (2004) A structural basis for constitutive activity in the human CAR/RXRalpha heterodimer. Mol Cell 16:919–928 [DOI] [PubMed] [Google Scholar]
- Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, Guo W, Lalli E, Moser C, Walker AP, McCabe ER. (1994) An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372:635–641 [DOI] [PubMed] [Google Scholar]
- Zhang H, Thomsen JS, Johansson L, Gustafsson JA, Treuter E. (2000) DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J Biol Chem 275:39855–39859 [DOI] [PubMed] [Google Scholar]
- Zhou J, Oakley RH, Cidlowski JA. (2008) DAX-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1) selectively inhibits transactivation but not transrepression mediated by the glucocorticoid receptor in a LXXLL-dependent manner. Mol Endocrinol 22:1521–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.