Abstract
Background
Information regarding changes in organismal status is transmitted to the stem cell regulatory machinery by a limited number of signal transduction pathways. Consequently, these pathways derive their functional specificity through interactions with stem cell intrinsic master regulators, notably transcription factors. Identifying the molecular underpinnings of these interactions is critical to understanding stem cell function.
Scope of review
This review focuses on studies in Drosophila that identify the gene regulatory basis for interactions between three different signal transduction pathways and an intrinsic master transcriptional regulator in the context of hematopoietic stem-like cell fate choice. Specifically, the interface between the GATA:FOG regulatory complex and the JAK/STAT, BMP, and Hedgehog pathways is examined.
Major conclusions
The GATA:FOG complex coordinates information transmitted by at least three different signal transduction pathways as a means to control stem-like cell fate choice. This illustrates emerging principles concerning regulation of stem cell function and describes a gene regulatory link between changes in organismal status and stem cell response.
General significance
The Drosophila model system offers a powerful approach to identify the molecular basis of how stem cells receive, interpret, and then respond to changes in organismal status.
Keywords: GATA, Friend of GATA, JAK/STAT, Bone Morphogenic Protein, Hedgehog, Drosophila, Hematopoiesis
1. Introduction
Stem cells have the dual capacity to self-renew and differentiate, thereby replenishing the stem cell pool and producing the entire spectrum of cells that form a given tissue. These characteristics underlie the ability of stem cells to maintain tissue homeostasis throughout the life of an organism by replacing lost or damaged tissue and/or mounting a response to environmental assaults. This involves communicating changes in the status of the organism (organismal status), such as immune challenge and nutritional deprivation or wounding, to the often sequestered stem cell compartment [1–4]. Signal transduction pathways serve in this capacity by transmitting remote information to the stem cell regulatory machinery in order to maintain homeostasis and initiate the appropriate cellular response. Interestingly, this information is thought to be communicated by relatively few signal transduction pathways, which also function across different stem cell systems [5]. Additionally, within a given system, these pathways have been shown to promote both stem cell self-renewal and differentiation [6–9]. Signal transduction pathways appear to derive their functional specificity by interacting with intrinsic stem cell master regulators, notably transcription factors [5]. Additional levels of functional specificity are achieved by the convergence of multiple signal transduction pathways that interface with one or more intrinsic master regulators. Ultimately, this convergence alters the stem cell-specific gene regulatory landscape and thereby determines cell fate choice [5;10].
The blood organ is a dynamic system that produces a number of different cell types that respond to a wide range of changes, including immune challenge and aging [1–4;11–18]. Consequently, hematopoiesis is an excellent system to investigate how signal transduction pathways interface with specific master regulators to control cell fate choice. Although Drosophila has a rudimentary hematopoietic system, powerful genetics coupled with a short generation time makes this an ideal model to investigate the molecular basis for regulatory strategies that link these changes in organismal status with cell fate choice. Importantly, because these studies are conducted in vivo, the response to changing conditions is governed by the regulatory complexity imposed by the whole organism [19].
Recent studies in the fly suggest that the GATA transcription factor when bound to the co-regulator Friend of GATA (GATA:FOG complex) is a master regulator, which controls hematopoietic cell fate choice through interactions with the following three signal transduction pathways: 1) Janus kinase/Signal Transducer and Activator of Transcription (JAK/STAT); 2) Bone Morphogenic Protein (BMP); and 3) Hedgehog (Hh) [20–22]. Specifically, the GATA:FOG complex functions to maintain multilineage developmental potential (multipotency) and block differentiation of blood cell progenitors (stem-like cells). Importantly, changes in the relative levels of GATA to FOG also alter fate choice [21]. GATA:FOG complex formation is regulated by the JAK/STAT and BMP signal transduction pathways, which may be important mechanisms that mediate the response to changing environmental conditions [20;21]. Of equal importance are the downstream effectors of the GATA:FOG complex. GATA singularly, and when bound to FOG, regulates the Hedgehog expression domain [22]. Tight regulation of Hedgehog is required to maintain the stem-like cell population [23]. Collectively, these observations suggest that the GATA:FOG complex serves as a nexus that coordinates information carried by three different pathways to regulate cell fate choice. Overall, these findings may provide a conceptual framework for studies designed to investigate how information is received, interpreted, and acted upon by stem cell systems. This review presents the findings that identified the molecular basis for the interaction between these signal transduction pathways and the GATA:FOG complex. Additionally, a discussion of how these interactions may control cell fate choice during steady-state hematopoiesis and in response to immune challenge is presented.
2. Drosophila hematopoietic system
2.1. The Drosophila hemocytes and hematopoietic organ
Drosophila blood cell progenitors have been described as stem-like cells because they share key characteristics with mammalian hematopoetic stem cells (HSCs), including quiescence, multipotency, and niche-dependence [23–25]. Drosophila stem-like cells give rise to all three of the mature blood cell types: 1) plasmatocytes are operational macrophages that mediate phagocytosis of bacterial pathogens and apoptotic bodies; 2) crystal cells are named for their crystalline inclusion bodies, and are involved in wound healing; and 3) lamellocytes are normally rare blood cells that are produced in large numbers in response to various types of immune challenge [20;21;23;24;26–38].
Drosophila hematopoiesis takes place during two spatially and temporally distinct periods or waves, which is similar to the pattern seen in vertebrate blood systems. The first wave takes place in the embryonic head mesoderm, whereas the second wave takes place in a specialized organ known as the lymph gland [39]. An elegant study using lineage analyses of transplanted cells demonstrated that the blood cells of the head mesoderm and the cells of the primordial lymph gland arise from two different anlagen. Furthermore, this approach was instrumental in demonstrating that blood cells from both the first wave (head mesoderm) and second wave (lymph gland) persist throughout the adult stage of the fly [40].
During embryogenesis, the lymph gland is specified from the cardiogenic mesoderm and develops from hemangioblast-like cells that have the potential to become either heart (dorsal vessel) or blood cells. The embryonic lymph gland is a bilateral organ containing one pair of primary lobes that flank the heart [41]. The primary lobes contain two distinct cells types, comprising approximately 20 hematopoietic stem-like cells and a cluster of five or six non-hematopoietic cells that sit at the posterior base and give rise to the Posterior Signaling Center (PSC). The PSC functions as the stem cell niche [23;24;42].
In Drosophila development, the embryonic stage is followed by three larval instars. During the larval instars, the lymph gland cells proliferate, increasing in number by approximately 100-fold. By the early third larval instar, additional paired secondary lymph gland lobes have formed posterior to the primary lobes [35]. The lymph gland reaches full maturity by the middle of the third larval instar [24]. The primary lobes contain stem-like cells, precursors, and terminally differentiated blood cells [23;24;35;43]. At this stage, the primary lobe is organized into three regions or zones with distinct hematopoietic functions (Figure 1). The first is the PSC or niche, which maintains stem-like cell quiescence and multipotency through several signaling pathways [23;24;44;45]. The second or medullary zone contains the stem-like cells. During the process of differentiation, these cells migrate to the third region called the cortical zone. Here, they continue to develop and give rise to all three blood cell types [23;24;35;44].
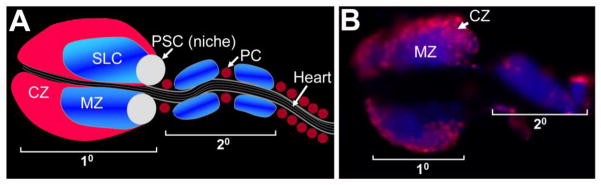
Figure 1. The Drosophila hematopoietic lymph gland.
(A) Schematic of the mature third larval instar lymph gland showing primary (10) and secondary (20) lobes. The relative positions of the three domains within the primary lobe are shown, specifically cortical zone (CZ), medullary zone (MZ), and stem cell niche (PSC; Posterior Signaling Center). Stem-like cells (SLC) reside in the MZ. Pericardial nephrocytes (PC) are insects renal cells that filter the blood and reside in two rows that flank the insect heart. (B) Lymph gland showing plasmatocytes in the CZ stained with the specific marker P1 (red). The medial region of the lymph gland contains densely packed, unstained MZ cells. The lymph gland is counterstained with Dapi (blue) and the 10 and 20 lobes are marked.
2.2 The Drosophila stem-like cells
Mammalian HSCs are characterized using functional assays that assess the capacity to continuously regenerate all blood cell types. This involves transplantation of heterogeneous populations of cells into irradiated animals and assaying for repopulation of all the blood lineages [46;47]. This method is considered the gold standard for identifying HSCs. Repopulation assays are currently not feasible for studies using Drosophila. Instead, investigators rely on lineage tracing studies and the persistence of marked clones to identify putative stem cell populations [48;49]. Using this approach, one study provided evidence for Drosophila HSCs within the embryo and first larval instar lymph gland. However, it was not possible to definitively identify HSCs in the second and third larval instar using this approach. The interval between clone induction in the second larval instar and analyses in the third larval instar is too short to distinguish between transient clones representing dividing precursors and persistent clones representing self-renewing stem cells [49]. In another study using this method, the authors concluded that all lymph gland cells become committed precursors by the end of the first larval instar [48]. However, heat shock was used to induce clones in both of these studies [48;49]. Heat shock changes the gene expression landscape, which could alter progenitor cell potential and may account for some of the differences between these two studies. On the other hand, lineage tracing studies performed without the use of heat shock showed that second larval instar progenitors can give rise to all the mature blood cell types [35]. This suggests that when the lymph gland reaches maturity in the mid-third instar, the medullary zone contains a heterogeneous population of cells, which most likely ranges from bona fide stem cells to more advanced progenitors. Indeed, a recent study demonstrated for the first time the heterogeneity of the medullary zone population. This study showed that two distinct markers, Domeless-Gal4 (Dome) and ZCL2897 (ZCL), are differentially expressed within this population. Three different cell types were observed - Domehi ZCLlo, Domehi ZCLhi, and Domelo ZCLlo [50]. These results represent an important first step towards characterization of the medullary zone cell population. Nevertheless, this undifferentiated population of cells gives rise to all three blood cell types and, as such, is multipotent [35;48]. In addition to multipotency, medullary zone cells are also quiescent and niche dependent [23;24;35]. Thus, based on these stem cell characteristics, the cells of the medullary zone have been described as stem-like [25].
2.3 The utility of the Drosophila hematopoietic system
The hematopoietic system in the fly is ideally suited to rapidly identify gene regulatory strategies that control cell fate choice during steady-state conditions and in response to changes in organismal status. Many of the factors that regulate hematopoiesis are evolutionarily conserved [19;39]. The lymph gland is readily accessible, and the niche, stem-like cells, and differentiating cells are compartmentalized and easily identifiable (Figure 1). The zonal arrangement of the lymph gland facilitates studies designed to map the origin of signal transduction pathways involved in specific cellular processes [23;24;44;45]. A short generation time and powerful genetic methods insure rapid identification of interactions between hematopoietic regulators in vivo. Simplicity and limited genetic redundancy facilitate the identification of gene functions that are often obscured in mammalian model systems [39]. The genetic control of the stem-like cell response to changes in organismal status is routinely assessed in the context of the regulatory complexity imposed by the whole organism [24;30;31]. Finally, rapid in vivo cis-regulatory analyses can be used to establish direct target/regulator pairs and thereby provide confirmation of gene interactions that can only be inferred from in silico analyses [21;22;51–53].
3. The GATA:FOG complex maintains hematopoietic stem-like cell multipotency
3.1. The GATA:FOG complex
GATA transcription factors activate gene expression and are named for the consensus WGATAR recognition sequence. These factors regulate a variety of biological processes and control the development of a number of tissues across taxa, ranging from fungi to plants and from invertebrates to vertebrates. There are six mammalian GATA factors, and GATA-1, −2 and −3 regulate hematopoiesis [39;54–57]. GATA-2 functions at the apex of hematopoiesis by maintaining the HSC population [57–59]. All three GATA factors function later in hematopoiesis to control lineage commitment and differentiation of specific blood cell types [39;54;56;57;60;61]. Drosophila has five GATA factors, and Serpent (Srp; dGATAb) was first identified over 30 years ago as part of the seminal work by Nüsslein-Volhard and Wiechaus, which resulted in the discovery of a number of key developmental regulators [62]. Srp functions in a variety of developmental processes [39;56]. The name Serpent derives from the phenotype of srp null mutant embryos, which have a slightly twisted snake-like appearance [62]. Srp acts analogously to GATA-2 in that it is required to maintain the stem-like cell pool [41]. Srp also acts later in hematopoiesis to direct blood lineage commitment and differentiation [32;51;52;63–66]. A recent report suggests that a second GATA factor, Pannier (Pnr, dGATAa), directs development of the plasmatocyte lineage [67].
In general, GATA factors have conserved N- and C-terminal zinc-finger protein domains. The C-terminal zinc-finger binds the WGATAR recognition sequence. The N-terminal zinc-finger stabilizes DNA binding and interacts with the GATA transcriptional co-regulator, FOG [39;55;57;61;63;68–73]. In Drosophila, the srp gene is alternatively spliced to produce two isoforms. One isoform (SrpNC) contains both canonical zinc-fingers. The other isoform (SrpC) lacks the N-terminal zinc-finger and consequently does not bind the Drosophila FOG homolog, U-shaped (Ush) [63;65;74]. Like srp, ush was also identified in the screen conducted by Nüsslein-Volhard and Wieschaus [75]. Loss of Ush function results in embryos exhibiting arrested development and a shape that resembles the letter “U”.
FOG transcriptional regulators are multitype zinc-finger proteins that activate or repress GATA-directed transcription, depending on the gene regulatory context [39;56;57;60;61;70–74;76–84]. There are two mammalian FOG proteins, FOG-1 and -2, whereas Ush is the only Drosophila FOG protein (dFOG) [70;71;81–85]. During mammalian hematopoiesis, FOG-1 interacts with a cognate hematopoietic GATA factor to form the corresponding GATA:FOG complex. GATA:FOG complexes regulate the development of a number of blood lineages. These complexes promote erythrocyte and megakaryocyte differentiation [70;71;76;79;80], and block granulocyte, eosinophil, mast cell, and helper T cell differentiation [78;86–89]. This latter function is conserved between flies and mammals, as the GATA:FOG complex also blocks the differentiation of all three Drosophila blood cell types [21;32;51;52;63;74].
3.2 Drosophila FOG functions to maintain stem-like cell multipotency and block differentiation
Work from our laboratory provided evidence that dFOG is required to maintain hematopoietic stem-like cell multipotency by blocking differentiation [21]. In support of this conclusion, we showed that dFOG is expressed in stem-like cells and maintains the stem-like cell pool. The loss of one functional copy of dFOG (heterozygotes) significantly reduced the stem-like cell population and increased the number of steady-state effector cells, specifically crystal cells and plasmatocytes. Interestingly, the relative level of dFOG expression appears to control stem-like cell fate choice. While the loss of one copy leads to an increase in plasmatocyte and crystal cell differentiation, loss of greater than one copy leads to an increase in the normally rare immune effector lamellocytes [21;32]. This occurs at the expense of both plasmatocytes and crystal cells. Reduction of dFOG to less than one functional copy also severely depleted the stem-like cell pool [21]. Thus, the relative level of dFOG expression not only regulates the choice between the maintenance of multipotency and differentiation but also regulates the choice between steady-state effector cells and immune response cells.
As stated above, FOG proteins bind GATA factors to modify GATA-activated gene expression. During lymph gland hematopoiesis, dFOG most likely interacts with SrpNC to form the GATA:FOG complex, which maintains stem-like cell multipotency and controls lineage choice. Conversely, a reduction in the level of dFOG may free SrpNC to act with lineage-specific factors to drive commitment and differentiation. Support for this model comes from the following observations: First, Srp and dFOG are co-expressed in stem-like cells and a subset of differentiating cells [21;64;74]. Second, dFOG is downregulated in crystal cells and plasmatocytes and is not expressed in lamellocytes [21]. Finally, dFOG may antagonize SrpNC function during lamellocyte differentiation [32].
Collectively, these studies strongly suggest that the GATA:FOG complex maintains stem-like cell multipotency by blocking differentiation. They also provide a compelling case for the position that changes in the relative levels of GATA to FOG can alter stem-like cell fate choice. This may be an important regulatory mechanism controlled by the JAK/STAT and BMP signal transduction pathways in response to immune challenge. The evidence supporting this hypothesis is presented in Sections 4 and 5.
4. JAK/STAT signaling promotes formation of the GATA:FOG complex to maintain stem-like cell multipotency
4.1. The JAK/STAT pathway
In mammals, the core components of the JAK/STAT signal transduction pathway include a wide range of extracellular ligands and transmembrane receptors, including four JAKs and seven STAT transcription factors [6;90–92]. In contrast, the fly contains a simplified version of the pathway with only three cytokine-like ligands, namely Unpaired (Upd)-1, -2, and 3, and one receptor, Domeless, which is homologous to the interleukin receptors. Additionally, the pathway has only one JAK (Hopscotch, Hop) and one STAT (STAT92E). Nonetheless, this streamlined JAK/STAT pathway is sufficient to regulate the myriad of processes that coordinate the life cycle of the fly [6;90].
The JAK/STAT pathway is activated when an extracellular ligand binds to the transmembrane receptor. This results in the phosphorylation of the receptor/JAK complex by a receptor-associated JAK tyrosine kinase, which creates a STAT docking site. STATs bind to this site and are phosphorylated and form either homo- or hetero-dimers. STAT dimers translocate to the nucleus where they bind a palindromic DNA sequence and activate gene transcription [6;90;91].
The JAK/STAT pathway regulates a number of processes, including embryonic development, hematopoiesis, and the immune response. Additionally, pathway dysregulation is implicated in various cancers [6;90;91;93]. During mammalian hematopoiesis, STATs promote HSC self-renewal as well as lymphoid and erythroid differentiation [7–9]. JAK/STAT function is remarkably conserved between flies and mammals. In Drosophila, the pathway maintains hematopoietic stem-like cell multipotency and regulates blood cell differentiation and cellular immunity [6;16;24;26;31;67;94;95]. Similar to mammalian systems, dysregulation of the pathway leads to tumor formation that is associated with reduced viability [6;90;94].
4.2. JAK/STAT maintains stem-like cell multipotency
The JAK/STAT pathway maintains stem-like cell multipotency and is downregulated in response to immune challenge [24;95]. JAK/STAT signaling is most likely active in the medullary zone stem-like cells. This is strongly supported by the fact that three different bona fide STAT target reporter genes, domeless-Gal4, domeMESO, and 10X-STAT-GFP, are expressed in the medullary zone stem-like cells and downregulated in the differentiating cells of the cortical zone [21;24;35;45;96]. However, evidence for JAK/STAT signaling within a given cell type would be strengthened by using in situ hybridization or immunofluorescence to detect endogenous STAT transcript or protein. In the stem-like cells, iJAK/STAT signaling appears to be maintained by the stem cell niche in so far as loss of niche function leads to loss of JAK/STAT signaling [24]. The cytokine-like ligand, Upd-3, is expressed in both the niche and stem-like cells. However, studies using tissue-specific RNAi knockdown showed that loss of Upd-3 expression in the niche had no effect on JAK/STAT activity in stem-like cells. In contrast, when Upd-3 expression was knocked down in stem-like cells, JAK/STAT activity was dramatically reduced [95]. Thus, an unknown factor from the niche may activate Upd-3 expression in the medullary zone stem-like cells to maintain JAK/STAT signaling [24;95].
Functional analyses were used to demonstrate that STAT maintains stem-like cell multipotency and block differentiation. During wasp parasitization, JAK/STAT signaling must be downregulated to initiate the cellular immune response, which culminates in the production of lamellocytes [95]. This is accomplished by altering the expression of two different pathway components. One component is Upd-3, which when downregulated in response to parasitization leads to reduced expression of the Domeless receptor [95]. In this regard, studies have shown that although binding of Upd ligands to Domeless activates JAK/STAT signaling, the domeless gene is also a downstream target of JAK/STAT signaling [96]. The second component, a newly identified dominant negative regulator of JAK/STAT signaling named Latran, is upregulated in response to wasp parasitization [95]. Latran is expressed exclusively in stem-like cells and forms inactive heterodimers with Domeless that downregulate the JAK/STAT pathway. Thus, wasp parasitization both reduces Domeless expression and increases Latran expression. This results in an increased number of inactive heterodimers that block JAK/STAT signaling. Consequently, in Latran loss-of-function mutants, JAK/STAT signaling remains active during wasp parasitization, which severely dampens the cellular immune response [95]. Collectively, these data suggest that JAK/STAT signaling maintains stem-like cell multipotency by blocking differentiation.
It is important to note that downregulation of JAK/STAT is necessary but not sufficient to drive lamellocyte differentiation in response to immune challenge [95]. This indicates that while loss of JAK/STAT activity within the medullary zone primes stem-like cells for differentiation, it alone does not lead to terminal differentiation. This important observation may explain the seemingly contradictory results obtained by groups analyzing STAT loss-of-function in cells that are contained within a larger population of STAT heterozygous cells (STAT-null clones). In two different studies, the authors reported that STAT-null clones did not express differentiation markers and thus failed to differentiate in the absence of STAT activation [45;67]. Additionally, one group observed that neither loss of domeless nor loss of hop function resulted in increased plasmatocyte differentiation [45]. These findings can readily be reconciled in a model that considers the duality of STAT function, which is discussed in more detail in Section 4.3. In this model, STAT functions in the medullary zone to prevent stem-like cells from entering the differentiation pathway (Figure 2A). However, STAT is also required later during blood cell development within the cortical zone to promote terminal differentiation (Figure 2B). Indeed, as discussed above, maintaining JAK/STAT signaling blocked differentiation in response to wasp parasitization [95], whereas STAT is also required to promote the terminal differentiation of both lamellocytes and plasmatocytes [31;67]. Additional support for the notion that STAT blocks differentiation could be obtained by showing that stem-like cell-specific marker expression is reduced in STAT-null clones. Most importantly, this model of the duality of STAT function in the fly is consistent with STAT function in mammalian systems, which is discussed in Section 4.3.
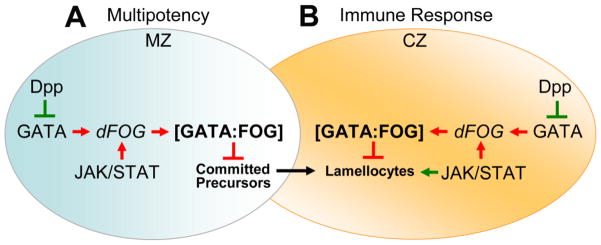
Figure 2. Antagonism between Dpp and JAK/STAT signaling regulates GATA:FOG complex formation to control cell fate choice.
(A) The GATA factor and JAK/STAT signaling co-activate dFOG gene expression. dFOG binds to SrpNC to form the GATA:FOG complex, which promotes stem-like cell multipotency and blocks differentiation. Dpp represses GATA expression and downregulates dFOG expression and complex formation. This limits the size of the stem-like cell pool. JAK/STAT signaling is initially downregulated during immune challenge (not shown). This promotes stem-like cell progression to the committed precursor stage by limiting GATA:FOG formation. (B) Later, JAK/STAT signaling is required for lamellocyte differentiation in response to immune challenge. However, JAK/STAT signaling continues to promote formation of the GATA:FOG complex. In order to counter this repressive effect, Dpp limits formation of the GATA:FOG complex to promote lamellocyte differentiation. Multipotent medullary zone (MZ) stem-like cells are depicted in blue. Differentiating cells of the cortical zone (CZ) are depicted in tan. Red arrows and blocked lines indicate pathways that maintain multipotency and block differentiation; green arrows and blocked lines indicate pathways that promote differentiation. dFOG is depicted in italics to indicate that gene expression is upregulated by the GATA factor Srp and JAK/STAT signaling.
A recent report provided evidence for an additional mechanism whereby STAT maintains stem-like cell multipotency. In this model, STAT is upregulated in the differentiating cells of the cortical zone by the Drosophila version of the Platelet Derived Growth Factor (PDGF; Pvr) signal transduction pathway. STAT then activates Adenosine deaminase growth factor A (Adgf-A), which signals from the cortical zone to the stem-like cells of the medullary zone to maintain the multipotent state. However, JAK and Domeless do not appear to participate in this alternate Pvr/STAT/Adgf-A pathway. Thus, STAT upregulation in this context results from non-canonical signaling [45].
4.3. The duality of JAK/STAT signaling during Drosophila hematopoiesis
In addition to maintaining stem-like cell multipotency, JAK/STAT signaling is also required for terminal differentiation of lamellocytes and plasmatocytes [31;67]. Studies of a constitutively active form of JAK (HopscotchTumorous-lethal; HopTum-l) were the first to suggest that the pathway was required for differentiation of lamellocytes in response to immune challenge. Specifically, these studies showed that hopTum-l mutants have dramatically increased numbers of lamellocytes [26;94;97;98]. The role of the pathway as a positive effector of the immune response was later confirmed by studies showing that loss-of-function hop mutants failed to produce lamellocytes in response to immune challenge [31]. The body of work on JAK/STAT regulation of Drosophila hematopoiesis, when taken together, indicates that the pathway is downregulated to permit stem-like cells to enter the differentiation pathway in response to immune challenge, and is required later in the process for the terminal differentiation of lamellocytes (Figure 2). This type of functional duality is not unusual and is conserved across taxa. Specifically, mammalian STAT5 is required for HSC self-renewal as well as lymphoid and erythroid differentiation [7–9]. These observations demonstrate that JAK/STAT signaling regulates hematopoiesis in a highly context dependent manner and supports the assertion that interactions with cell-specific master regulators are determinants of JAK/STAT function. In particular, our work suggests that JAK/STAT signaling promotes GATA:FOG complex formation as a means to maintain stem-like cell multipotency [21].
4.4. JAK/STAT upregulates dFOG gene expression to maintain stem-like cell multipotency
Several initial observations implicated dFOG as a downstream target of JAK/STAT signaling. First, constitutively activated JAK/STAT leads to increased dFOG expression in hopTum-l mutants [21;32;99]. In this regard, dFOG-positive cells were visible throughout the primary lobes of hopTum-l mutant lymph glands [21;32]. Furthermore, circulating hemocytes from hopTum-l mutant larvae had elevated levels of dFOG transcript [99]. Second, the phenotypes of dFOG and STAT heterozygous mutants are remarkably similar, showing increased plasmatocyte and crystal cell production and decreased numbers of stem-like cells [21;24]. Finally, our laboratory identified a dFOG hematopoietic cis-regulatory element (dFOG CRM) that contains a consensus STAT binding site [21;51]. Using genetic and in vivo cis-regulatory analyses, our studies showed that STAT function is required for dFOG gene expression in stem-like cells. Loss of one copy of STAT resulted in loss of both dFOG protein expression and dFOG CRM activity. Furthermore, mutating the STAT binding site dramatically reduced CRM activity. Finally, while HopTum-l was able to activate the wild-type dFOG CRM, it could not activate the dFOG CRM containing the mutant version of the STAT binding site. Collectively, these data showed that STAT upregulates dFOG in stem-like cells [21]. Subsequently, dFOG most likely binds SrpNC to form the GATA:FOG complex that maintains stem-like cell multipotency and blocks differentiation (Figure 2A). Overall, the findings presented here may represent a conserved regulatory strategy that controls the response of stem/progenitor cells to changing environmental conditions such as immune challenge.
Although JAK/STAT signaling promotes the terminal differentiation of lamellocytes, it also appears to upregulate the lamellocyte repressor, dFOG, under these conditions. This conclusion is supported by studies showing that dFOG expression is upregulated by constitutively active JAK/STAT signaling [21;32;99]. While this may be an important negative feedback mechanism that controls lamellocyte number, it could also compromise the efficacy of the cellular immune response. BMP signaling may counter JAK/STAT activation of dFOG expression by antagonizing GATA:FOG complex formation (Figure 2B). This may insure the production of lamellocytes in response to immune challenge. Support for this model is presented in Section 5.
5. BMP signaling represses GATA:FOG complex formation to promote stem-like cell differentiation
5.1. The BMP signaling pathway
BMP is a member of the transforming growth factor-β (TGF-β) superfamily. These are structurally-related cytokines that bind transmembrane receptors to initiate a conserved intracellular signaling cascade [100–103]. The mouse genome contains 33 genes that encode members of the TGF-β superfamily. In contrast, the Drosophila genome contains seven superfamily members [104]. The superfamily is further subdivided into TGF-βs, activins, inhibins, and BMPs. Drosophila Decapentaplegic (Dpp) is structurally and functionally related to BMP. Indeed, expression of human BMPs in flies can rescue loss of Dpp function, and Dpp can induce bone formation in mammalian cell culture [104].
In addition to TGF-β superfamily ligands, the core components of the pathway include transmembrane receptors and intracellular signal mediators. There are two types of single pass serine/threonine kinase receptors, designated type I and type II. Seven type I and five type II receptors have been reported in mammalian systems, whereas three type I and two type II receptors have been identified in flies [102;105]. The intracellular mediators are known as Smad proteins. Smads were first identified in worms and flies and derive their name from a combination of the C. elegans homolog, SMA, and the Drosophila homolog, MAD, Mothers Against Dpp. Smads fall into three functional categories designated receptor-regulated (R-Smads), common mediator (Co-Smads), and inhibitor (I-Smads).
The canonical TGF-β signaling pathway is activated when a TGF-β ligand dimer binds to the type II receptor, which then recruits the type I receptor. This facilitates phoshorylation of the type I receptor by the constitutively active type II receptor kinase. The type I receptor then phosphorylates R-Smads, enabling them to associate with the Co-Smads. This produces a heteromeric complex that translocates to the nucleus and activates gene expression through the Smad binding elements (SBE) [100–105]. Typically this involves cooperative interactions with other transcriptional regulators [5;10;100;103].
TGF-β signaling regulates a wide variety of functions, including embryonic development, cellular proliferation, apoptosis, differentiation, and migration. During hematopoiesis, various superfamily members regulate processes that range from the initial specification of embryonic HSCs to the terminal differentiation of adult blood lineages [5;100;103]. During Drosophila hematopoiesis, Dpp limits the number of stem-like cells in the early embryonic lymph gland as well as those that reside in the mature third larval instar lymph gland. Additionally, Dpp is required for lamellocyte differentiation in response to immune challenge [20].
5.2. Dpp limits stem-like cell number during early lymph gland development
Dpp is a critical regulator of tissue development throughout the life cycle of the fly. During Drosophila embryogenesis, Dpp signaling regulates many developmental processes [106]. Among these are three rounds of Dpp signaling from the dorsal ectoderm that influence cell fate within an increasingly restricted spatial domain [20;107–109]. The second and third rounds signal to the mesoderm to modulate differentiation of cardio-hematopoietic precursors [20;108;109]. Specifically, the second round of Dpp signaling is required to specify the cardiogenic mesoderm, from which the lymph gland is derived [20;41;107–109]. In contrast, the third round of signaling limits the stem-like cell pool by repressing Srp expression in the embryonic lymph gland [20]. In this regard, it was previously shown that Srp is required for maintenance of the embryonic lymph gland stem-like cells [41]. Dpp blocks Srp expression by downregulating one of its upstream activators, Zfh1 (Zinc-finger homeodomain 1). Zfh1 is a transcriptional regulator that shares homology with murine zinc-finger E-box binding homeobox 1 (Zeb1) [20].
5.3. Dpp limits the expression of dFOG and Srp (dGATAb) to promote stem-like cell differentiation in the mature lymph gland
In addition to the embryonic lymph gland, Dpp limits the number of stem-like cells in the mature third larval instar lymph gland [27]. This has been shown to involve two different mechanisms. One mechanism involves autocrine Dpp signaling that limits the number of cells within the PSC. This in turn limits the number of stem-like cells [110]. In contrast, the first mechanism to be discovered does not involve PSC signaling. However, this PSC-independent mechanism does impact GATA:FOG complex formation and will therefore be examined in more detail in this section [20].
Dpp is essential for embryonic development. However, the use of a homozygous mutant that can survive to adulthood, dppd6, enabled identification of a PSC-independent mechanism that limits the number of stem-like cells in the mature lymph gland. The dppd6 mutation results from the removal of specific dpp cis-regulatory elements [107;108]. Odd-skipped (Odd) is a marker for stem-like cells and the number of Odd-expressing stem-like cells increased in dppd6 mutants [20;27]. This was accompanied by a significant reduction in terminally differentiated plasmatocytes. Furthermore, both dFOG and Srp expression increased in dppd6 mutants. In fact, this increase may underlie the observed expansion of the stem-like cell pool [20;27]. In this model, Dpp signaling limits formation of the GATA:FOG complex to promote stem-like cell differentiation, whereas with loss of signaling, increased formation of GATA:FOG complex limits stem-like cell differentiation and expands the size of this population (Figure 2A). In addition, the cellular immune response may be regulated by this strategy. Lamellocytes differentiate in response to Salmonella typhimurium challenge. Furthermore, lamellocyte differentiation requires Dpp signaling as these cells were rarely observed in Salmonella infected dppd6 mutants. Thus, Dpp may promote lamellocyte differentiation in response to immune challenge by limiting the formation of the GATA:FOG complex in a progenitor population (Figure 2B), similar to the role of Dpp in stem-like cells [20].
Taken together, these studies detailing the interactions between the GATA:FOG complex and the JAK/STAT and Dpp signaling pathways suggest the following dynamic model of hematopoiesis. In the fly lymph gland, JAK/STAT signaling upregulates dFOG, which results in increased formation of the GATA:FOG complex and maintenance of stem-like cell multipotency. Subsequently, during immune challenge, JAK/STAT signaling is downregulated. The resulting downregulation of dFOG allows the stem-like cells to enter the differentiation pathway (Figure 2A). Later, JAK/STAT is required again to promote lamellocyte differentiation. However, JAK/STAT signaling continues to upregulate the lamellocyte repressor, dFOG. To counter the effect of JAK/STAT, Dpp signaling antagonizes the expression of Srp and its target, dFOG, to promote lamellocyte differentiation (Figure 2B). Thus, the combined positive and negative regulatory inputs from these signal transduction pathways control dFOG expression and, thereby, the choice between maintenance of multipotency and the response to immune challenge (Figures 2 and 3). As a result, analyses of Dpp and JAK/STAT regulation of GATA:FOG complex formation can serve as a framework for studies that investigate the molecular basis for convergence of multiple pathways and how this regulates cell fate choice during steady-state hematopoiesis and in response to immune challenge.
Figure 3. The GATA:FOG complex links three signal transduction pathways that control stem-like cell fate choice.
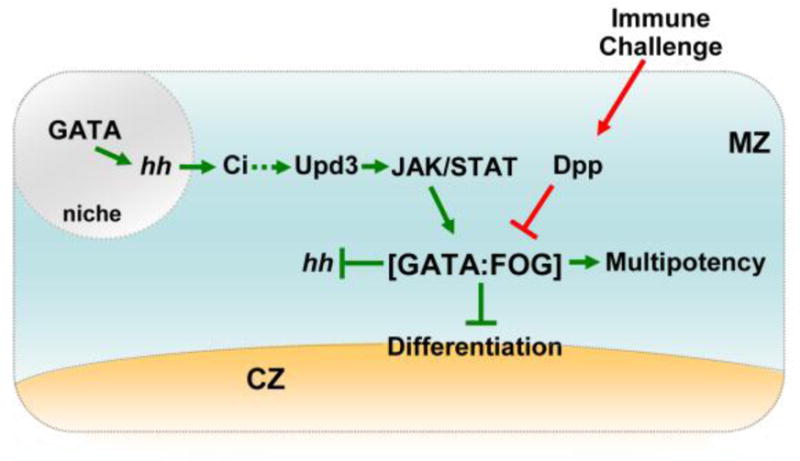
The GATA:FOG complex is a master regulator that promotes medullary zone (MZ) stem-like cell multipotency and blocks differentiation. Cell fate choice is modulated by GATA:FOG complex formation through opposing signals from the JAK/STAT and Dpp pathways. Thus, the complex serves as a nexus that responds to signals from the following sources: 1) the niche through JAK/STAT signaling; and 2) immune challenge through Dpp signaling. Hh signaling is activated in the niche by GATA and maintains stem-like cell multipotency. This may involve upregulating JAK/STAT signaling. Additionally, the GATA:FOG complex limits Hh expression in the MZ. Overall, tight regulation of Hh is necessary to maintain the appropriate number of MZ stem-like cells. The niche is depicted in grey, MZ stem-like cells are depicted in blue, and differentiating cells of the cortical zone (CZ) are depicted in tan. The green arrows and blocked lines indicate pathways that promote stem-like cell multipotency. Red arrows indicate pathways that promote differentiation. The hatched arrow indicates a possible link between Hh and JAK/STAT signaling. The hh gene is depicted in italics to indicate that gene expression is regulated by GATA and the GATA:FOG complex.
6. The GATA:FOG complex regulates hedgehog gene expression
The GATA:FOG complex can be thought of as a gene network switch that is modulated by JAK/STAT and Dpp signaling. Additional critical components of this network are the downstream targets of the complex that control multipotency. Importantly, this includes regulating the function of a third signal transduction pathway. In this regard, a recent report showed that the GATA:FOG complex regulates the expression of the gene that encodes Hedgehog (Hh), which is a diffusible ligand that transmits signals from the niche to the stem-like cells of the medullary zone [22]. Thus, the GATA:FOG complex is a central regulator that connects and modulates the function of three different signal transduction pathways (Figure 3).
6.1. The Hedgehog signaling pathway
Hh is a secreted ligand that activates a conserved signal transduction pathway, which regulates development, cell proliferation, migration, and differentiation during tissue and organ formation [111–113]. The Hh ligand is produced and secreted by a single cell type, but has been shown to travel across as many as 10 cell lengths to activate gene expression in a target cell population [114]. The hh gene was another important developmental regulator discovered in the Nüsslein-Volhard and Wieschaus screen [115]. The name Hedgehog describes the original phenotype of the mutation that disrupts the larval body plan, resulting in a duplication of bristle-like cuticular processes (denticles). This gives the appearance of a continuous lawn of denticles that resembles a hedgehog [111;112].
Drosophila has a single Hh ligand, whereas mammals have three ligands, Sonic Hh (Shh), Indian Hh (Ihh) and Desert Hh (Dhh). Additional Hh pathway components include a ligand binding receptor, signal transducers, and downstream transcriptional effectors. Hh binds Patched (Ptc), which is a 12-pass transmembrane receptor. Ptc is the sole receptor in the fly, whereas mammals have two Patched receptors, Ptch1 and 2. Smoothen (Smo) is a 7-pass transmembrane signal transducer. Flies and mammals each have one Smo transducer. In Drosophila, the downstream transcriptional effector is Cubitus interruptus (Ci), a zinc-finger transcription factor that translates the signal into specific changes in the gene expression output. There are three mammalian homologs of the Drosophila Ci protein, Glioma-associated oncogene homolog (Gli) 1, 2 and 3 [111–113].
Pathway activation begins with the post-translational modification of Hh to produce an active signaling molecule [112;113]. Subsequent binding of Hh to Ptc activates the signal transduction pathway by actually repressing Ptc activity. In the absence of Hh binding, Ptc suppresses Smo activity, whereas Hh binding blocks the ability of Ptc to repress Smo [112;113;116]. Activated Smo blocks the proteolytic cleavage of Ci and, as a result, full length Ci translocates to the nucleus to activate specific target genes [112;113;116]. In the absence of Hh signaling, proteolytic cleaveage of Ci produces a truncated protein that acts as a transcriptional repressor [112;113]. A more detailed description of the factors and processes that mediate Hh signaling can be found in several excellent review articles [111–113;116]. The role of Hh signaling in vertebrate hematopoiesis is both controversial and poorly understood [112;116]. However, Hh signaling appears to support the survival of chronic myeloid leukemic (CML) stem cells. As a result, the pathway is under investigation as a potential therapeutic target [116]. In Drosophila, Hh signaling is a critical regulatory link between the niche and the stem-like cells [23].
6.2. Hedgehog signaling from the niche blocks stem-like cell differentiation
In the Drosophila hematopoietic lymph gland, the PSC functions as a stem cell niche (see Section 2) [23;24;42]. In this role, the PSC regulates medullary zone stem-like cells, and loss of the PSC leads to a reduction of this cell population with a concomitant increase in terminally differentiated plasmatocytes and crystal cells [23;24]. Regulation of stem-like cell fate choice is mediated through the Hh signaling pathway [23]. This was demonstrated by showing that Hh was expressed in the PSC, and that loss of Hh resulted in increased numbers of differentiated plasmatocytes and crystal cells. The roles of two additional members of the canonical Hh signaling pathway, Ptc and Ci, have also been examined. Ptc and Ci were shown to be expressed in stem-like cells. Additionally, loss of Ci function produced increased numbers of differentiated cells, which is similar to the loss-of-function hh mutant phenotype [23]. Collectively, these results indicate that the canonical Hh pathway provides a regulatory basis for niche control of stem-like cell fate choice (Figure 3).
6.3. The GATA:FOG complex regulates hedgehog gene expression in the Drosophila hematopoietic organ
The finding that Hh is a key link between the niche and the stem-like cell population provided an opportunity to probe deeper into the underlying mechanisms that control cell fate choice. This question was addressed by first characterizing the CRM that specifically directs hh gene expression in the PSC. This enabled the search and subsequent rapid identification of transacting factors that control hh gene expression in vivo. Using this approach, the GATA factor Srp, the GATA:FOG complex, and Suppressor of Hairless [Su(H)] were shown to be direct regulators of hh gene expression [22].
The hh CRM is a 190 bp fragment that contains GATA and Su(H) binding sites. The CRM is located within the 1st intron of the hh gene and mutational analyses showed that the GATA sites were required for activity in the PSC. Subsequent genetic analyses confirmed that Srp was required for hh gene expression in the PSC [22]. As a result, loss of Srp function in the PSC should mimic loss of hh expression and thus lead to increased numbers of differentiated plasmatocytes and crystal cells. Indeed, Srp acts upstream of Hh to limit niche-directed stem-like cell differentiation (Figure 3). Furthermore, loss of Srp function in the PSC, and by extension loss of Hh signaling, resulted in loss of medullary zone stem-like cells [22]. This latter result extended previous work by providing compelling evidence that Hh not only blocks differentiation but also maintains the stem-like cell pool [22;23]. In addition to Srp, in vivo cis-regulatory and genetic analyses were used to show that Su(H) actually blocked hh expression in the medullary zone stem-like cells [22].
Srp is expressed throughout the hematopoietic cells of the lymph gland and the non-hematopoietic cells of the PSC [64]. This raised the question as to what limits hh gene expression to the PSC. The most likely explanation is that SrpNC interacts with dFOG to produce a repressor complex that blocks hh gene expression in the hematopoietic cells (Figure 3). Consistent with this notion, dFOG is expressed in the hematopoietic cells of the lymph gland but not in the non-hematopoietic cells of the PSC [27;35]. Accordingly, misexpression of dFOG in the PSC blocked hh expression, which provided further evidence for this hypothesis. Conversely, loss of dFOG expression resulted in de novo expression of both the hh CRM and endogenous protein in the hematopoietic lymph gland cells. Thus, while Srp activates hh expression in the non-hematopoietic cells of the PSC, the GATA:FOG complex in concert with Su(H) blocks expression in the hematopoietic cells [22].
The PSC produces filopodia that are thought to facilitate the delivery of Hh ligand to cells removed from immediate contact with the niche [23;24]. Although Srp is not required for the specification of the PSC, it is required for proper differentiation. Loss of Srp expression in the PSC resulted in loss of filopodia. Thus, Srp promotes Hh signaling through the following two mechanisms: 1) direct upregulation of hh gene expression; and 2) production of filopodia that facilitate delivery of Hh signal to the target cell population. Conversely, the GATA:FOG complex limits the hh expression domain thereby providing an additional regulatory tier that modulates niche-directed control of stem-like cell fate choice [22]. Thus, the GATA:FOG complex coordinates the input and output of information transmitted by three different signal transduction pathways as a means to regulate stem-like cell fate choice (Figure 3).
7. Summary and Future Directions
In summary, the GATA:FOG complex maintains stem-like cell multipotency, whereas downregulation of the complex promotes differentiation [21]. GATA:FOG complex formation is regulated by the antagonistic activity of JAK/STAT and Dpp signaling [20;21]. JAK/STAT signaling promotes complex formation through upregulation of dFOG expression, whereas Dpp signaling limits complex formation through downregulation of srp expression and loss of dFOG expression [20;21]. Additionally, the GATA:FOG complex regulates Hh signaling through hh gene expression [22]. Thus, the GATA:FOG complex coordinates information transmitted by three different signal transduction pathways as a means to control stem-like cell fate choice (Figure 3). These findings illustrate three emerging principles of how signal transduction pathways regulate stem cell function. First, these observations support the notion that the specific function of a given signal transduction pathway is highly context dependent and, as a result, reliant on the ability to act with key master regulators [5]. Second, these results explain how signal transduction pathways act antagonistically to control master regulator function and thereby determine cell fate choice. Finally, this body of work describes a gene regulatory link between changes in organismal status and stem cell response. Consequently, these studies in the fly can serve as a framework to investigate how the stem cell receives, interprets, and then responds to changes in organismal status. Going forward, it will be important to determine the extent to which the GATA:FOG complex functions as a central hub that integrates signal transduction pathways to control hematopoietic stem-like cell fate choice. Specifically, does the complex interface with other major developmental pathways, such as Wingless, Notch, and Toll? Furthermore, is the GATA:FOG complex a master regulator of the stem-like cell response to stress-induced signaling? In particular, while the complex appears to regulate the response to immune challenge, does it also mediate the response to nutrient deprivation or increased levels of reactive oxygen species (ROS)? These latter conditions have recently been shown to promote stem-like cell differentiation [34;117]. Finally, aside from hh, what are the downstream targets of the GATA:FOG complex that maintain stem-like cell multipotency and block differentiation? Given the importance of GATA:FOG complex in mammalian hematopoiesis, such studies in the fly will undoubtedly provide valuable information about how this complex functions in mammalian model systems. Overall, the Drosophila model system offers a powerful approach to identify the molecular underpinnings of how stem cells sense changing conditions and subsequently respond to preserve tissue homeostasis.
Highlights.
We discuss interactions between the GATA:FOG complex and the JAK/STAT, BMP and Hedgehog pathways.
These interactions control stem-like cell fate choice during Drosophila hematopoiesis.
This discussion illustrates emerging principles regarding regulation of stem cell function.
This discussion also describes a gene regulatory link between changes in organismal status and stem-like cell response.
Acknowledgments
This work was supported by Public Health Service grant DK072229 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 4.Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol. 2010;10:201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- 5.Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, Li P, Durand EM, Mosimann C, Heffner GC, Daley GQ, Paulson RF, Young RA, Zon LI. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory L, Came PJ, Brown S. Stem cell regulation by JAK/STAT signaling in Drosophila. Semin Cell Dev Biol. 2008;19:407–413. doi: 10.1016/j.semcdb.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Li G, Wang Z, Zhang Y, Kang Z, Haviernikova E, Cui Y, Hennighausen L, Moriggl R, Wang D, Tse W, Bunting KD. STAT5 requires the N-domain to maintain hematopoietic stem cell repopulating function and appropriate lymphoid-myeloid lineage output. Exp Hematol. 2007;35:1684–1694. doi: 10.1016/j.exphem.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato Y, Iwama A, Tadokoro Y, Shimoda K, Minoguchi M, Akira S, Tanaka M, Miyajima A, Kitamura T, Nakauchi H. Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J Exp Med. 2005;202:169–179. doi: 10.1084/jem.20042541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wierenga AT, Vellenga E, Schuringa JJ. Maximal STAT5-induced proliferation and self-renewal at intermediate STAT5 activity levels. Mol Cell Biol. 2008;28:6668–6680. doi: 10.1128/MCB.01025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WY, Wilson NK, Landry JR, Wood AD, Kolb-Kokocinski A, Green AR, Tannahill D, Lacaud G, Kouskoff V, Gottgens B. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 12.Feng CG, Weksberg DC, Taylor GA, Sher A, Goodell MA. The p47 GTPase Lrg-47 (Irgm1) links host defense and hematopoietic stem cell proliferation. Cell Stem Cell. 2008;2:83–89. doi: 10.1016/j.stem.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passegue E, Ernst P. IFN-alpha wakes up sleeping hematopoietic stem cells. Nat Med. 2009;15:612–613. doi: 10.1038/nm0609-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 15.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, Lio P, Macdonald HR, Trumpp A. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 16.Bodian DL, Leung S, Chiu H, Govind S. Cytokines in Drosophila hematopoiesis and cellular immunity. Prog Mol Subcell Biol. 2004;34:27–46. doi: 10.1007/978-3-642-18670-7_2. [DOI] [PubMed] [Google Scholar]
- 17.Govind S. Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 2008;15:29–43. doi: 10.1111/j.1744-7917.2008.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MJ, Kalamarz ME, Paddibhatla I, Small C, Rajwani R, Govind S. Virulence factors and strategies of Leptopilina spp.: selective responses in Drosophila hosts. AdvParasitol. 2009;70:123–145. doi: 10.1016/S0065-308X(09)70005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans CJ, Hartenstein V, Banerjee U. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev Cell. 2003;5:673–690. doi: 10.1016/s1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- 20.Frandsen JL, Gunn B, Muratoglu S, Fossett N, Newfeld SJ. Salmonella pathogenesis reveals that BMP signaling regulates blood cell homeostasis and immune responses in Drosophila. Proc Natl Acad Sci US A. 2008;105:14952–14957. doi: 10.1073/pnas.0808208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H, Wu X, Fossett N. Upregulation of the Drosophila Friend of GATA gene U-shaped by JAK/STAT signaling maintains lymph gland prohemocyte potency. Mol Cell Biol. 2009;29:6086–6096. doi: 10.1128/MCB.00244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokusumi Y, Tokusumi T, Stoller-Conrad J, Schulz RA. Serpent, suppressor of hairless and U-shaped are crucial regulators of hedgehog niche expression and prohemocyte maintenance during Drosophila larval hematopoiesis. Development. 2010;137:3561–3568. doi: 10.1242/dev.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446:325–328. doi: 10.1038/nature05650. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Agosto JA, Mikkola HK, Hartenstein V, Banerjee U. The hematopoietic stem cell and its niche: a comparative view. Genes Dev. 2007;21:3044–3060. doi: 10.1101/gad.1602607. [DOI] [PubMed] [Google Scholar]
- 26.Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- 27.Gao H, Wu X, Fossett N. Odd-skipped maintains prohemocyte potency and blocks blood cell development in Drosophila. Genesis. 2011;49:105–116. doi: 10.1002/dvg.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honti V, Csordas G, Markus R, Kurucz E, Jankovics F, Ando I. Cell lineage tracing reveals the plasticity of the hemocyte lineages and of the hematopoietic compartments in Drosophila melanogaster. Mol Immunol. 2010;47:1997–2004. doi: 10.1016/j.molimm.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Markus R, Laurinyecz B, Kurucz E, Honti V, Bajusz I, Sipos B, Somogyi K, Kronhamn J, Hultmark D, Ando I. Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. Proc Natl Acad Sci US A. 2009;106:4805–4809. doi: 10.1073/pnas.0801766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorrentino RP, Carton Y, Govind S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev Biol. 2002;243:65–80. doi: 10.1006/dbio.2001.0542. [DOI] [PubMed] [Google Scholar]
- 31.Sorrentino RP, Melk JP, Govind S. Genetic analysis of contributions of dorsal group and JAK-Stat92E pathway genes to larval hemocyte concentration and the egg encapsulation response in Drosophila. Genetics. 2004;166:1343–1356. doi: 10.1534/genetics.166.3.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorrentino RP, Tokusumi T, Schulz RA. The Friend of GATA protein U-shaped functions as a hematopoietic tumor suppressor in Drosophila. DevBiol. 2007;311:311–323. doi: 10.1016/j.ydbio.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Williams MJ. Drosophila hemopoiesis and cellular immunity. J Immunol. 2007;178:4711–4716. doi: 10.4049/jimmunol.178.8.4711. [DOI] [PubMed] [Google Scholar]
- 34.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132:2521–2533. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- 36.Chiu H, Ring BC, Sorrentino RP, Kalamarz M, Garza D, Govind S. dUbc9 negatively regulates the Toll-NF-kappa B pathways in larval hematopoiesis and drosomycin activation in Drosophila. Dev Biol. 2005;288:60–72. doi: 10.1016/j.ydbio.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Paddibhatla I, Lee MJ, Kalamarz ME, Ferrarese R, Govind S. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS Pathog. 2010;6:e1001234. doi: 10.1371/journal.ppat.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu P, Pan PC, Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- 39.Fossett N, Schulz RA. Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation. 2001;69:83–90. doi: 10.1046/j.1432-0436.2001.690202.x. [DOI] [PubMed] [Google Scholar]
- 40.Holz A, Bossinger B, Strasser T, Janning W, Klapper R. The two origins of hemocytes in Drosophila. Development. 2003;130:4955–4962. doi: 10.1242/dev.00702. [DOI] [PubMed] [Google Scholar]
- 41.Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat Genet. 2004;36:1019–1023. doi: 10.1038/ng1404. [DOI] [PubMed] [Google Scholar]
- 42.Crozatier M, Ubeda JM, Vincent A, Meister M. Cellular immune response to parasitization in Drosophila requires the EBF orthologue collier. PLoS Biol. 2004;2:E196. doi: 10.1371/journal.pbio.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 44.Sinenko SA, Mandal L, Martinez-Agosto JA, Banerjee U. Dual role of wingless signaling in stem-like hematopoietic precursor maintenance in Drosophila. Dev Cell. 2009;16:756–763. doi: 10.1016/j.devcel.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mondal BC, Mukherjee T, Mandal L, Evans CJ, Sinenko SA, Martinez-Agosto JA, Banerjee U. Interaction between differentiating cell- and niche-derived signals in hematopoietic progenitor maintenance. Cell. 2011;147:1589–1600. doi: 10.1016/j.cell.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuzaki Y, Kinjo K, Mulligan RC, Okano H. Unexpectedly efficient homing capacity of purified murine hematopoietic stem cells. Immunity. 2004;20:87–93. doi: 10.1016/s1074-7613(03)00354-6. [DOI] [PubMed] [Google Scholar]
- 47.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 48.Krzemien J, Oyallon J, Crozatier M, Vincent A. Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Dev Biol. 2010;346:310–319. doi: 10.1016/j.ydbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Minakhina S, Steward R. Hematopoietic stem cells in Drosophila. Development. 2010;137:27–31. doi: 10.1242/dev.043943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalamarz M, Paddibhatla I, Nadar C, Govind S. Sumoylation is tumor-suppressive and confers proliferative quiescence to hematopoietic progenitors in Drosophila melanogaster larvae. Biology Open. 2012;1:161–172. doi: 10.1242/bio.2012043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muratoglu S, Garratt B, Hyman K, Gajewski K, Schulz RA, Fossett N. Regulation of Drosophila friend of GATA gene, u-shaped, during hematopoiesis: a direct role for serpent and lozenge. Dev Biol. 2006;296:561–579. doi: 10.1016/j.ydbio.2006.04.455. [DOI] [PubMed] [Google Scholar]
- 52.Muratoglu S, Hough B, Mon ST, Fossett N. The GATA factor Serpent cross-regulates lozenge and u-shaped expression during Drosophila blood cell development. Dev Biol. 2007;311:636–649. doi: 10.1016/j.ydbio.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokusumi T, Sorrentino RP, Russell M, Ferrarese R, Govind S, Schulz RA. Characterization of a lamellocyte transcriptional enhancer located within the misshapen gene of Drosophila melanogaster. PLoS One. 2009;4:e6429. doi: 10.1371/journal.pone.0006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 56.Sorrentino RP, Gajewski KM, Schulz RA. GATA factors in Drosophila heart and blood cell development. Semin Cell Dev Biol. 2005;16:107–116. doi: 10.1016/j.semcdb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. NatRevGenet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 58.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 59.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 60.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 61.Morceau F, Schnekenburger M, Dicato M, Diederich M. GATA-1: friends, brothers, and coworkers. Ann NY Acad Sci. 2004;1030:537–554. doi: 10.1196/annals.1329.064. [DOI] [PubMed] [Google Scholar]
- 62.Nusslein-Volhard C, Wiechaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux’s Archives of Developmental Biology. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- 63.Fossett N, Hyman K, Gajewski K, Orkin SH, Schulz RA. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc Natl Acad Sci USA. 2003;100:11451–11456. doi: 10.1073/pnas.1635050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science. 2000;288:146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- 65.Waltzer L, Bataille L, Peyrefitte S, Haenlin M. Two isoforms of Serpent containing either one or two GATA zinc fingers have different roles in Drosophila haematopoiesis. EMBO J. 2002;21:5477–5486. doi: 10.1093/emboj/cdf545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferjoux G, Auge B, Boyer K, Haenlin M, Waltzer L. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev Biol. 2007;305:726–734. doi: 10.1016/j.ydbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 67.Minakhina S, Tan W, Steward R. JAK/STAT and the GATA factor Pannier control hemocyte maturation and differentiation in Drosophila. Dev Biol. 2011;352:308–316. doi: 10.1016/j.ydbio.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crispino JD. GATA1 in normal and malignant hematopoiesis. SeminCell Dev Biol. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 69.Fossett N, Schulz RA. Conserved cardiogenic functions of the multitype zinc-finger proteins: U-shaped and FOG-2. Trends CardiovascMed. 2001;11:185–190. doi: 10.1016/s1050-1738(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 70.Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 71.Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- 73.Fossett N, Zhang Q, Gajewski K, Choi CY, Kim Y, Schulz RA. The multitype zinc-finger protein U-shaped functions in heart cell specification in the Drosophila embryo. Proc Natl Acad Sci USA. 2000;97:7348–7353. doi: 10.1073/pnas.97.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA. The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc Natl Acad Sci USA. 2001;98:7342–7347. doi: 10.1073/pnas.131215798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nusslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux’s Archives of Developmental Biology. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 76.Cantor AB, Katz SG, Orkin SH. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol. 2002;22:4268–4279. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol. 2005;16:117–128. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 78.Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J Exp Med. 2008;205:611–624. doi: 10.1084/jem.20070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci USA. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. ProcNatl Acad Sci USA. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cubadda Y, Heitzler P, Ray RP, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao Z, Huang Z, Olivey HE, Gurbuxani S, Crispino JD, Svensson EC. FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis. EMBO J. 2009 doi: 10.1038/emboj.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kurata H, Lee HJ, McClanahan T, Coffman RL, O’Garra A, Arai N. Friend of GATA is expressed in naive Th cells and functions as a repressor of GATA-3-mediated Th2 cell development. J Immunol. 2002;168:4538–4545. doi: 10.4049/jimmunol.168.9.4538. [DOI] [PubMed] [Google Scholar]
- 88.Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 2000;14:2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou M, Ouyang W, Gong Q, Katz SG, White JM, Orkin SH, Murphy KM. Friend of GATA-1 represses GATA-3-dependent activity in CD4+ T cells. J ExpMed. 2001;194:1461–1471. doi: 10.1084/jem.194.10.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 91.Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene. 2007;26:6724–6737. doi: 10.1038/sj.onc.1210757. [DOI] [PubMed] [Google Scholar]
- 92.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 93.Egwuagu CE. STAT3 in CD4+ T helper cell differentiation and inflammatory diseases. Cytokine. 2009;47:149–156. doi: 10.1016/j.cyto.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Makki R, Meister M, Pennetier D, Ubeda JM, Braun A, Daburon V, Krzemien J, Bourbon HM, Zhou R, Vincent A, Crozatier M. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 2010;8:e1000441. doi: 10.1371/journal.pbio.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hombria JC, Brown S, Hader S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288:420–433. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 97.Hanratty WP, Dearolf CR. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol Gen Genet. 1993;238:33–37. doi: 10.1007/BF00279527. [DOI] [PubMed] [Google Scholar]
- 98.Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kwon SY, Xiao H, Glover BP, Tjian R, Wu C, Badenhorst P. The nucleosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev Biol. 2008;316:538–547. doi: 10.1016/j.ydbio.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 100.Blank U, Karlsson S. The role of Smad signaling in hematopoiesis and translational hematology. Leukemia. 2011;25:1379–1388. doi: 10.1038/leu.2011.95. [DOI] [PubMed] [Google Scholar]
- 101.Johnson AN, Newfeld SJ. The TGF-beta family: signaling pathways, developmental roles, and tumor suppressor activities. ScientificWorldJournal. 2002;2:892–925. doi: 10.1100/tsw.2002.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Santibanez JF, Quintanilla M, Bernabeu C. TGF-beta/TGF-beta receptor system and its role in physiological and pathological conditions. Clin Sci(Lond) 2011;121:233–251. doi: 10.1042/CS20110086. [DOI] [PubMed] [Google Scholar]
- 103.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 104.Kahlem P, Newfeld SJ. Informatics approaches to understanding TGFbeta pathway regulation. Development. 2009;136:3729–3740. doi: 10.1242/dev.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Newfeld SJ, Wisotzkey RG, Kumar S. Molecular evolution of a developmental pathway: phylogenetic analyses of transforming growth factor-beta family ligands, receptors and Smad signal transducers. Genetics. 1999;152:783–795. doi: 10.1093/genetics/152.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ray RP, Arora K, Nusslein-Volhard C, Gelbart WM. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development. 1991;113:35–54. doi: 10.1242/dev.113.1.35. [DOI] [PubMed] [Google Scholar]
- 107.Frasch M. Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature. 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- 108.Johnson AN, Bergman CM, Kreitman M, Newfeld SJ. Embryonic enhancers in the dpp disk region regulate a second round of Dpp signaling from the dorsal ectoderm to the mesoderm that represses Zfh-1 expression in a subset of pericardial cells. DevBiol. 2003;262:137–151. doi: 10.1016/s0012-1606(03)00350-6. [DOI] [PubMed] [Google Scholar]
- 109.Johnson AN, Burnett LA, Sellin J, Paululat A, Newfeld SJ. Defective decapentaplegic signaling results in heart overgrowth and reduced cardiac output in Drosophila. Genetics. 2007;176:1609–1624. doi: 10.1534/genetics.107.073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pennetier D, Oyallon J, Morin-Poulard I, Dejean S, Vincent A, Crozatier M. Size control of the Drosophila hematopoietic niche by bone morphogenetic protein signaling reveals parallels with mammals. Proc Natl Acad Sci US A. 2012;109:3389–3394. doi: 10.1073/pnas.1109407109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 112.Lim Y, Matsui W. Hedgehog signaling in hematopoiesis. Crit RevEukaryotGene Expr. 2010;20:129–139. doi: 10.1615/critreveukargeneexpr.v20.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 114.Chuang PT, Kornberg TB. On the range of hedgehog signaling. Curr Opin Genet Dev. 2000;10:515–522. doi: 10.1016/s0959-437x(00)00121-0. [DOI] [PubMed] [Google Scholar]
- 115.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 116.Mar BG, Amakye D, Aifantis I, Buonamici S. The controversial role of the Hedgehog pathway in normal and malignant hematopoiesis. Leukemia. 2011;25:1665–1673. doi: 10.1038/leu.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shim J, Mukherjee T, Banerjee U. Direct sensing of systemic and nutritional signals by haematopoietic progenitors in Drosophila. Nat Cell Biol. 2012;14:394–400. doi: 10.1038/ncb2453. [DOI] [PMC free article] [PubMed] [Google Scholar]