Abstract
To interpret LC-MS/MS data in proteomics, most popular protein identification algorithms primarily use predicted fragment m/z values to assign peptide sequences to fragmentation spectra. The intensity information is often undervalued, since it is not as easy to predict and incorporate into algorithms. Nevertheless, the use of intensity to assist peptide identification is an attractive prospect and can potentially improve the confidence of matches and generate more identifications. Based on our previously reported study of fragmentation intensity patterns, we developed a protein identification algorithm, SeQuence IDentfication (SQID), which makes use of the coarse intensity from a statistical analysis. The scoring scheme was validated by comparing with Sequest and X!Tandem using three datasets, and the results indicate an improvement in the number of identified peptides, including unique peptides that are not identified by Sequest or X!Tandem. The software and source code are available under the GNU GPL license at: http://quiz2.chem.arizona.edu/wysocki/bioinformatics.htm.
Keywords: protein identification algorithm, intensity, tandem mass spectrometry, database search
Introduction
Tandem mass spectrometry is widely used in proteomic studies because of its ability to identify large numbers of peptides from complex mixtures. In a typical LC-MS/MS experiment, the digested peptides are separated by one or two stages of liquid chromatography and ionized by electrospray ionization. The intact mass of each peptide is measured by mass spectrometry, then the peptide is mass-selected and fragmented to produce MS/MS spectra. These spectra are processed by protein identification algorithms to determine peptide sequences, which, in turn, infer protein sequence. Due to the large number of spectra generated in modern proteomic experiments, protein identification algorithms are a necessity.
Most commonly used algorithms are designed for sequence identification from fragmentation spectra produced by collision induced dissociation (CID), in which peptide precursor ions collide with inert gas molecules and dissociate. CID typically results in fragmentation along the peptide backbone at the amide bonds, producing predominantly N-terminal b and C-terminal y ions. Other ion types, including neutral water and ammonia losses and side chain cleavages are also possible. Because the masses of the product ions are predictable, the sequence of the original peptide can be reconstructed from the MS/MS spectrum by matching experimental fragment ion masses with theoretical ones. For a long time, m/z has been the main information used by popular algorithms, including Sequest 1, X!Tandem 2 and Mascot 3, to assign peptide sequences to fragmentation spectra. The process consists of searching a protein database or translated nucleotide database by m/z for possible peptide candidates, comparing each experimental spectrum with a large number of constructed theoretical spectra or peak lists that correspond to candidate peptide sequences, and assigning a score to each candidate sequence based on the similarity between the theoretical and experimental spectra or on the probability that their match is not random. The strength of the match is finally evaluated according to the top score and the score difference between the top and other candidates.4
One limitation of the process described above is that all the major ions of a given series in a theoretical spectrum are assumed to have the same intensity regardless of the properties of the peptide; intensity information contained in an experimental spectrum is essentially abandoned. Though in many cases the m/z information alone is enough to provide reliable identification, intensity can potentially improve the confidence and generate more identifications because it is also highly dependent on the sequence of the peptide and the amino acid residue compositions. Preferential cleavage, for example, is expected at the N-terminus of proline in the presence of a mobile proton or the C-terminus of aspartic acid when no mobile proton is available.5–7 Nevertheless, intensity is still seldom given much weight in algorithms because of the limited ability to predict and quantify the chemical rules of peptide fragmentation. Various factors, including peptide length, charge state, amino acid content, and charge location, can complicate the process of gas phase peptide dissociation and make the resulting peak intensities difficult to predict and interpret. 8
Clearly, the use of intensity to improve peptide identification provides an attractive prospect and efforts have been made by different groups.9–16 Elias and coworkers, for example, used a probabilistic decision tree - specifically, a treelike feather extracting graph, which requires the members of each branch to have similar properties - to model the probability of observing certain peak intensities in a mass spectrum so as to improve peptide identification.13 Another algorithm, MASPIC, developed by Narasimhan et al., considered the possibility of random intensity matches as an alternative to using m/z only, based on the assumption that a random match is more likely to correspond to low intensity peaks since these peaks are more common in tandem mass spectra.14 Zhang reported a kinetic model for prediction of low-energy CID spectra from sequences, assuming that the intensity of a fragment ion is determined by the dissociation pathway and the rate of the dissociation.9,12 Another intensity model which considers more peptide features and fragmentation rules was developed by Zhou.16 Intensity is emphasized in these algorithms not because that it is more critical than m/z, but because it can provide additional information that can assist with the peptide identification.
The goal of the work presented here is to develop a simple, fast database search algorithm that incorporates rough intensity information to assist peptide identification. In our previously reported study of fragmentation intensity patterns, we introduced a routine to mine a large number of spectra with known sequences for fragment ion intensity based on pairwise amino acid (AA) cleavage patterns, and the relative peak intensity for each AA pair was recorded. Because the probability that a data peak of a specific intensity corresponds to any given AA pair cleavage is directly proportional to the probability of that AA pair cleavage resulting in a peak of that intensity, we can evaluate whether the intensity for a certain AA pair in an experimental spectrum is consistent with statistical values. We applied this approach to our SQID algorithm described in this paper. As with other algorithms, the SQID score depends on the presence or absence of ion series peaks at the expected m/z, but is also heavily affected by intensity information to increase the evidence for sequence identification. This is analogous to the manual process of verifying peptide identifications by looking for known fragmentation motifs (e.g. looking for enhanced cleavage at the N-terminus of proline), but with the objectivity of using statistical information gathered in the data-mining process.
Methods
Algorithm Design
SQID is designed for identification of peptides from ion trap tandem mass spectra in LC-MS/MS experiments but with the ability to extend to spectra acquired using different instruments or dissociation methods (e.g. ETD, ECD) in the future, as long as appropriate training datasets are available. It is written in C language and has been tested in Windows XP and Windows 7 operation systems. The software is available with source code under GNU GPL license at: http://quiz2.chem.arizona.edu/wysocki/bioinformatics.htm. SQID contains a one-time training stage to generate intensity tables that are used in scoring. In the training stage, spectra with known sequence are used to generate the pairwise intensity statistical lookup tables, which quantify the probability to observe a strong peak given a certain amino acid pair. The tables from the training stage are stored in the algorithm and do not need to be re-generated. The scoring process makes use of information from the experimental spectrum and intensity tables to evaluate a match. The algorithm design is described below.
Step 1: Collect pairwise cleavage intensities
The dataset used for training contains 138033 unique D. melanogaster (version: drosophila-7-14-2008-it) and S. cerevesiae (version: yeast-5-04-2009-it) ion trap spectra extracted from the National Institute of Standards and Technology (NIST) Libraries of Peptide Tandem Mass Spectra (http://peptide.nist.gov/). 17 It is a set of spectra with known sequences and consists of singly-, doubly-, and triply-charged tryptic peptides ranging from 5 to 56 amino acid residues in length. It contains unmodified peptides as well as peptides with carbamidomethylation of cysteine or oxidation of methionine. Currently we do not treat these modified residues (C+57, M+16) as unique amino acids and their cleavage intensities are combined with those of corresponding unmodified residues (C, M). For each training spectrum, the mass of each expected b and y ion was calculated based on the assigned peptide sequence. Ions outside of the ion trap mass range (high mass cutoff = 2000; low mass cutoff = (precursor m/z)*0.28) were not included (the low and high mass cutoffs can be adjusted as necessary to match the instrument type). The peak intensity of each b and y ion was scaled to the most abundant peak of its own series. The intensity information was sorted by ion type and by the amino acid residue pair cleavage responsible for the fragment ions. Using all training spectra, a histogram was generated containing the relative peak intensities for every expected peak sorted by amino acid pair. When the expected peak was not present, a zero value was included.
Step 2: Calculate probability of strong fragment ions for each AA pair
The relative abundance information for each amino acid pair was separated into three bins: no abundance (intensity = 0), weak (>0 –33%) or strong (>33% – 100%). The ranges defined as weak and strong intensity were empirically determined and the intensity strength for a certain amino acid pair is roughly proportional to the probability of observing a strong peak from that amino acid pair. The probability of having a strong peak (Pr) is defined as the number of strong peaks divided by the total number of expected peaks for the amino acid pair cleavage:
Pr = (number of strong peaks) / (total number of expected peaks)
For instance, the AP pair has a y ion Pr of 0.57, meaning that there is a 57% probability of seeing a cleavage between A and P with a strong y ion peak (>33%). In contrast, the PA pair has a Pr of 0.03, which means that there is only 3% probability of seeing a strong y ion peak for cleavage of the PA pair. In general, these values are in agreement with empirical knowledge and provide a quantitative basis for rough peak intensity prediction given a peptide sequence. Part of the pairwise cleavage intensity probabilities are shown in Table 1. The full table is available in supporting information.
Table 1.
Pairwise cleavage intensity probability table for selected amino acid pairs, based on spectra of 138033 singly, doubly, and triply charged unique peptide sequences.
| Amino acid pair | Pr of y ion | Pr of b ion |
|---|---|---|
| AA | 0.23 | 0.11 |
| AC | 0.29 | 0.10 |
| AD | 0.20 | 0.06 |
| AE | 0.17 | 0.07 |
| AP | 0.57 | 0.28 |
| AQ | 0.16 | 0.09 |
| DN | 0.31 | 0.14 |
| DP | 0.70 | 0.41 |
| PN | 0.02 | 0.01 |
| PP | 0.08 | 0.01 |
| YW | 0.35 | 0.13 |
| YY | 0.31 | 0.11 |
Step 3: Scoring experimental spectra
Experimental spectra are assigned peptide sequences by scoring a list of candidate peptide sequences against each spectrum. Each experimental spectrum is modified by eliminating precursor ions, water and ammonia loss products from precursor ions (mass tolerance is the same as fragment tolerance), and isotopes (SQID uses a simple deisotoping algorithm for ion trap data: if the two peaks differ by 1 +/− 0.25 and the intensity of the first peak is greater than that of the second one, the second peak is considered to be an isotope peak and removed. The main purpose of deisotoping is to ensure that isotopes of high abundance peaks will not be accidentally selected as top peaks in intensity score calculation). The top 80 of the most abundant peaks from the simplified spectrum are kept for scoring. For each spectrum, a list of candidate peptides (with mass within user-defined tolerance of the precursor mass of the experimental spectrum) is generated from a user-defined FASTA protein database. Each candidate sequence is scored by the following method:
Calculate the masses of expected fragment ions from the candidate peptide sequence (same high and low mass cutoffs as in training). In the present work only b and y ions are considered along with H2O and NH3 losses from b and y ions. Doubly charged fragments are considered in the circumstance that the precursor ion is triply charged and the mass of the fragment is greater than 900. The value of 900 was empirically determined based on the fact that we seldom see doubly-charged ions at m/z < 450.
Count the number of matched peaks in the experimental spectrum corresponding to the masses of the expected ions for the candidate sequence, within a user defined fragment threshold. If an expected water loss or ammonia loss product is observed, the total number of matched peaks is increased by 0.5. The number of matched fragments is used as a preliminary score and only the top 200 candidates are retained.
Count the number of consecutive ion pairs for a match. For instance, if y5 and y6 ions are found, it is counted as a consecutive ion pair. Though in many cases consecutive ion pairs increases almost linearly with the number of matched ions, we show later that including them can provide better discrimination than using the number of matched ions alone.
For the K most abundant peaks in an experimental spectrum (K depends on the mass of peptide, and equals the integer portion of [2+mass/330]), the Pr of amino acid pairs that result in these peaks are summed and the sum is used as the intensity score: . The intensity score is affected by two factors: how many top peaks are matched and how well the corresponding intensity matches. Because the Pr of amino acid pairs range from 0.01 to 0.72, both factors could play an importance role depending on the sequence.
The final SQID score is calculated as:
| (Equation 1) |
, where m is the number of matched peaks, n is the number of consecutive ions pairs, Pr is the probability for a certain AA pair to have strong peaks, and K is the number of most intense peaks used to calculate the intensity score. In the scoring function, (m+n) measures the number of matched peaks and numbers of consecutive ion pairs, and increased m and n will increase the confidence of a match; the term (1+∑Pr)/(1+0.155K) measures whether the observed intensity (the numerator) is better than the expected value (the denominator). We expect that the average Pr of the top K peaks is greater than 0.155, the average of all Pr values in the statistical table. A more detailed discussion of the scoring function can be found in a latter section of this report. The specific form of the score function was empirically determined in a trial-and-error manner to reach optimized performance using the Pacific Northwest National Laboratories (PNNL) dataset (the first testing dataset), and then applied to other datasets without any changes. Besides the SQID score, a delta score is used to give further discrimination. Calculation of a delta score in SQID is the same as that in Sequest: the difference of top score and second score was divided by the top score, which shows the percentage difference of the second score to the top score.
The matched peptide sequences and final scores can be reported either as a single tab delimited file or as separate text files. The results can be reported in .OUT format (mimicking Sequest output) for importing into Scaffold 18 to compare with other algorithms, as was done in the present work. Work is in progress to allow future versions of Scaffold to include a separate SQID input.
Performance test
Datasets
Three ion trap datasets were used to test the performance of SQID:
PNNL dataset: contains 28311 spectra (25% singly charged, 62% doubly charged and 13% triply charged) from unmodified Deinococcus radiodurans and Shewanella oneidensis peptides collected on a Thermo LCQ ion trap mass spectrometer. 7, 19, 20 When these spectra were collected, FT-ICR was used simultaneously for accurate mass measurements. Each LCQ spectrum was then analyzed by the Sequest search engine with D. radiodurans and S. oneidensis protein databases to assign a sequence. Preliminary identifications of peptides with a minimum cross-correlation score of 1.5 (Xcorr ≥ 1.5) were validated by measurements of Accurate Mass Tags (AMTs) from FT-ICR (mass measurement accuracy < 10 ppm). Though these spectra are of high quality, the error rate of the initial assigned sequences is unclear considering the low Xcorr threshold used. As a result, in current work we use an alternative strategy (see next section) to evaluate the confidence of matches instead of using those initially assigned sequences. The dataset is available at http://quiz2.chem.arizona.edu/wysocki/bioinformatics.htm. Spectra were identified against Deinococcus radiodurans and Shewanella oneidensis database (7984 entries).
18 Protein Mixture dataset: This dataset contains 37044 spectra collected by the Keller group, Institute for Systems Biology, Seattle, from a mixture of 18 purified proteins using a Thermo Finnigan ESI-ITMS 21. The dataset was collected with 22 LC-MS/MS runs, and only the most abundant peak in each full scan was selected for fragmentation, followed by 3 minutes of dynamic exclusion. The dataset is available at http://regis-web.systemsbiology.net/PublicDatasets/omics_dataset/. Spectra were searched against a reverse version of Deinococcus radiodurans and Shewanella oneidensis database (7984 entries) plus the 18 protein mixture and common contaminants (trypsin, human keratin, protein standards for MS calibration such as bovine serum albumin and angiotensin, etc).
Yeast Dataset: This dataset of 54799 spectra from a MudPIT experiment of yeast-extract was collected by the students (group A) of Dr. Andrew Link during the 2006 Cold Spring Harbor Laboratory Proteomics course, on a Thermo LTQ ion trap mass spectrometer. 22 These MudPIT data include six SCX/RP LC separations and the top five most abundant peaks in each full scan were selected for fragmentation. The raw file is available at: http://www.mc.vanderbilt.edu/root/vumc.php?site=msrc/bioinformatics&doc=21164. Spectra were identified against a yeast database (14590 entries) extracted from the NCBI non-redundant database (ftp.ncbi.nih.gov/blast/db/fasta/). All sequences with “yeast” or “Saccharomyces cerevisiae” in the description line were included.
Search parameters and false discovery rate determination
The three datasets above were converted to .DTA file format using Bioworks (Version 3.2). Sequest (Version 28, rev.12) and X!Tandem (Version Tornado 2008.02.01.3) were run simultaneously with SQID to evaluate the performance of SQID. Sequest was chosen because it uses a similar scoring that involves no expectation value calculation; X!Tandem was chosen because it is open source and based on expectation values. All algorithms were used with a parent mass tolerance of 1.5 Da and a fragment mass tolerance of 0.5 Da, and a maximum of two missed tryptic cleavage sites. Refinement for X!Tandem was disabled, and the maximum valid E-value for reporting was set to 10000. PNNL and 18 protein mixture datasets were searched with semi-tryptic cleavage (tryptic required at one terminus only) and without chemical modifications. The yeast dataset was searched with full tryptic cleavage (both termini) and with variable modification of C+57 (carbamidomethylation) and M+16 (oxidation). These modified amino acids are treated as C and M in the SQID intensity score calculation.
For the PNNL dataset and the yeast dataset, the false discovery rate (FDR) was determined using a target-decoy database search strategy. The database mentioned above was appended with a reverse database using “decoy.pl” program from Matrix Science (http://www.matrixscience.com/help/decoy_help.html#WHAT). At a certain score threshold, the spectra matched to target sequences were labeled “Target” and the ones matched to decoy sequences were labeled “Decoy”. The false discovery rate (FDR) was calculated as: FDR = (2×Decoy) / (Target + Decoy). 23 FDR is further expressed as a q-value, 24 which is the minimum FDR threshold at which a given match is considered positive. For easier understanding, the q-value can be regard as a measurement of FDR.
For the 18 protein mixture dataset, an identification was assumed to be “True” in the circumstance that the top hit belongs to any of those 18 proteins or common contaminants. At a score threshold which allows “x” spectra (among which “y” of them are true) to pass, FDR was simply calculated as: FDR = 1-y/x.
Results and Discussion
Calculation of intensity score
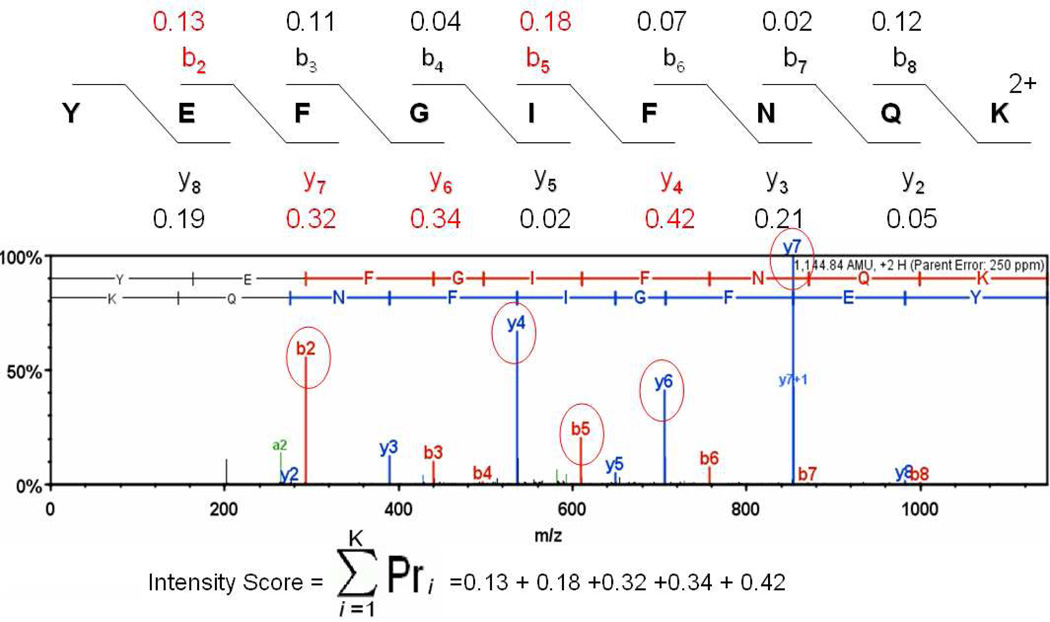
Intensity information is incorporated into SQID by using statistical intensity tables. Figure 1 is an example of how the intensity score is calculated. For the experimental spectrum with precursor MH+ 1144.8, the top five peaks are used for intensity scoring. Given the candidate sequence YEFGIFNQK 2+, SQID will first determine that the top five peaks (integer portion of [2+(1144.8/330)]) are matching to two b ions (b2, b5) and three y ions (y4, y6, y7), which correspond to EF and IF pairs for b ions and EF, FG and IF pairs for y ions. By looking up the intensity table, the probabilities to have strong (>33%) peaks (Pr) for each ion pair are 0.13 (EF pair, b2 ion), 0.18 (IF pair, b5 ion), 0.32 (EF pair, y7 ion), 0.34 (FG pair, y6 ion) and 0.42 (IF pair, y4 ion). The sum of the above values returns the intensity score. From the graph, it can be clearly seen that the Pr of the top five peaks (shown in red) are among the largest compared with Pr for other peaks (in black), which means that the most abundant peaks in the spectrum are also expected to be statistically strong based on the training set. In general, a higher intensity score indicates that the statistical fragmentation trends are reflected in the match so the confidence of the identification is increased.
Figure 1.
The calculation of intensity score in SQID. The bottom is a labeled experimental spectrum when matching it to the candidate sequence YEFGIFNQK2+. The most abundant peaks used for the intensity score calculation are circled. The numbers above b ions and below y ions are the probabilities of observing strong peaks with Pr values extracted from the intensity table.
Effect of individual components in the SQID score function
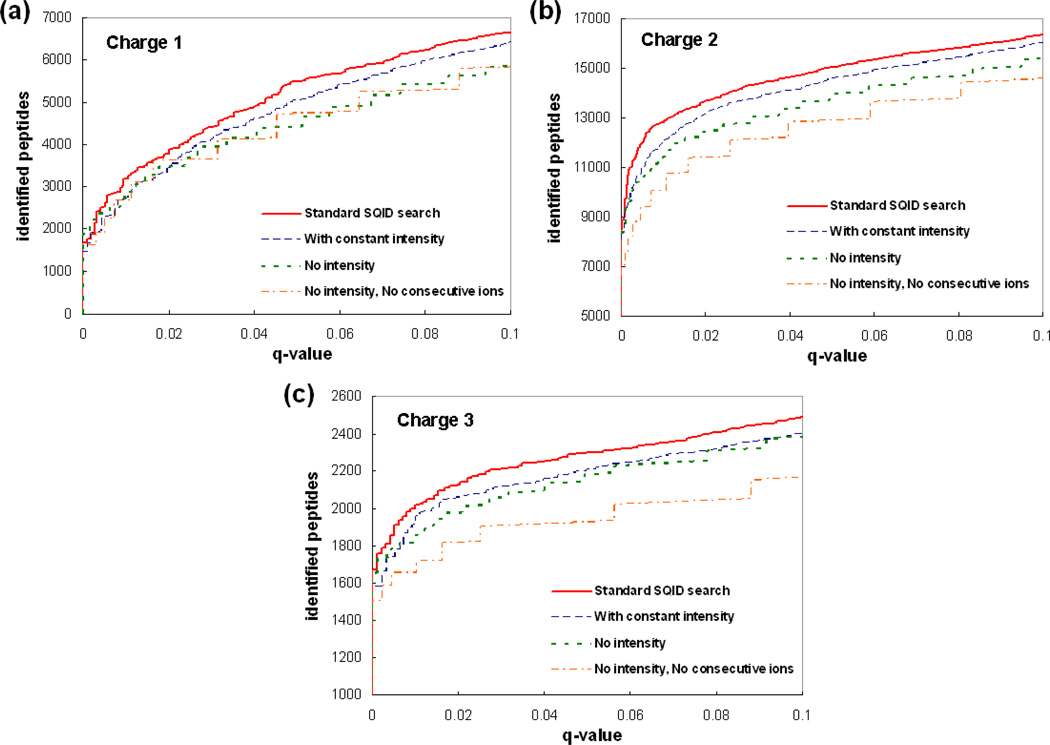
In addition to the number of matched ions used in most algorithms, the SQID score function (Equation 1) involves two features to improve peptide identification: consecutive ion series n and intensity (1+∑Pr)/(1+0.155K). To evaluate their contributions, four searches were conducted using the PNNL dataset: 1) A “Standard SQID search” using equation 1, with Pr values adopted from probability table (variable intensity). 2) “With constant intensity” for each ion type, where the Pr value is 0.22 for y ions and 0.09 for b ions. These are the average Pr values for each ion type. 3) “No intensity”: both and K equal zero, which completely removes the effect of intensity and ion type. The score function equals m+n. 4) “No intensity, no consecutive ion series”. The score equals the number of matched ions m. For each search, the results were ranked by the top scores from high to low, then FDR and q-values were determined as described earlier. By plotting q-value versus the number of peptide hits, Figure 2 showed that more peptides were identified when adding consecutive ion pairs as well as the intensity related terms to the scoring function. From the plots, it should be noticed that by using the number of matched ions alone (m), a significant number of peptides can be identified (orange dot-dashed lines). This illustrates that m/z is powerful information for peptide identification. By adding consecutive ion series (score function is now m+n), the performance increases as charge state increases (green doted lines). This may be explained by the fact that higher charged peptides normally have longer sequences and more theoretical peaks, which will increase the chance of finding consecutive ion pairs. At 0.05 q-value cutoff, the performance improved 8% for doubly charged spectra. Adding an intensity term with a constant intensity (blue dashed line) will give a score bonus when a theoretical peak is matched to a high abundance peak, with a higher bonus for y ions and lower bonus for b ions. This step gave an additional 6% (based on “no intensity search”) for doubly charged spectra at 0.05 q-value cutoff. Lastly, the standard SQID score (red solid line), which gives a statistically determined score bonus when a theoretical peak is matched to a high abundance peak, improved the overall performance by another 4% (based on “constant intensity”) for doubly charged spectra at 0.05 q-value cutoff. The actual performance boost differed for different charge states and q-value cutoffs, e.g. at 0.005 q-value cutoff (2+), “Standard SQID search” outperformed “constant intensity” by 13.4%. If the differences between “No intensity” (green dotted line) and “Standard SQID search” (red solid line) were counted as the contribution of intensity incorporation, normally the value was in the range of 5 to 15 %.
Figure 2.
Plot of q-value versus number of identified peptides showing the effect of individual components in the SQID score function for a) singly b) doubly c) triply charged peptides. More peptides were identified when adding consecutive ion pairs as well as the intensity related terms to the scoring function.
Comparison of algorithms
A completely objective comparison of algorithms is always difficult because each algorithm uses different spectrum preprocessing methods, different scoring schemes and different score reporting. In the spectrum preprocessing step, Sequest preprocesses the spectrum by keeping the top 200 peaks and separates the spectrum into ten bins for normalization. SQID keeps the top 80 peaks after removing parent related peaks and obvious non-monoisotopic peaks. X!Tandem simply keeps the 50 most abundant peaks by default. In term of score report, X!Tandem reports “E-value” and a much less important hyperscore, while SQID and Sequest report the main scores as well as delta scores. In this work, the default spectra preprocessing methods were used, and only the main scores, Xcorr, SQID score and E-value were used for a relatively fair comparison. It is important to note that these parameters can potentially affect the search results demonstrated below.
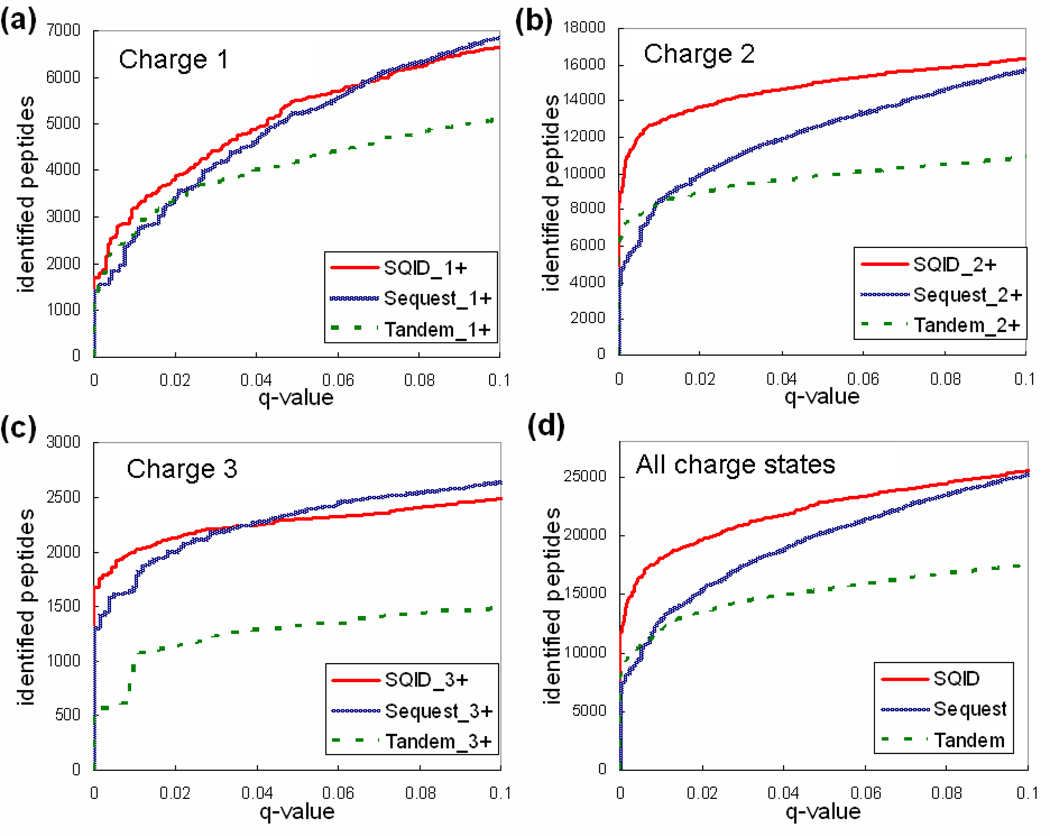
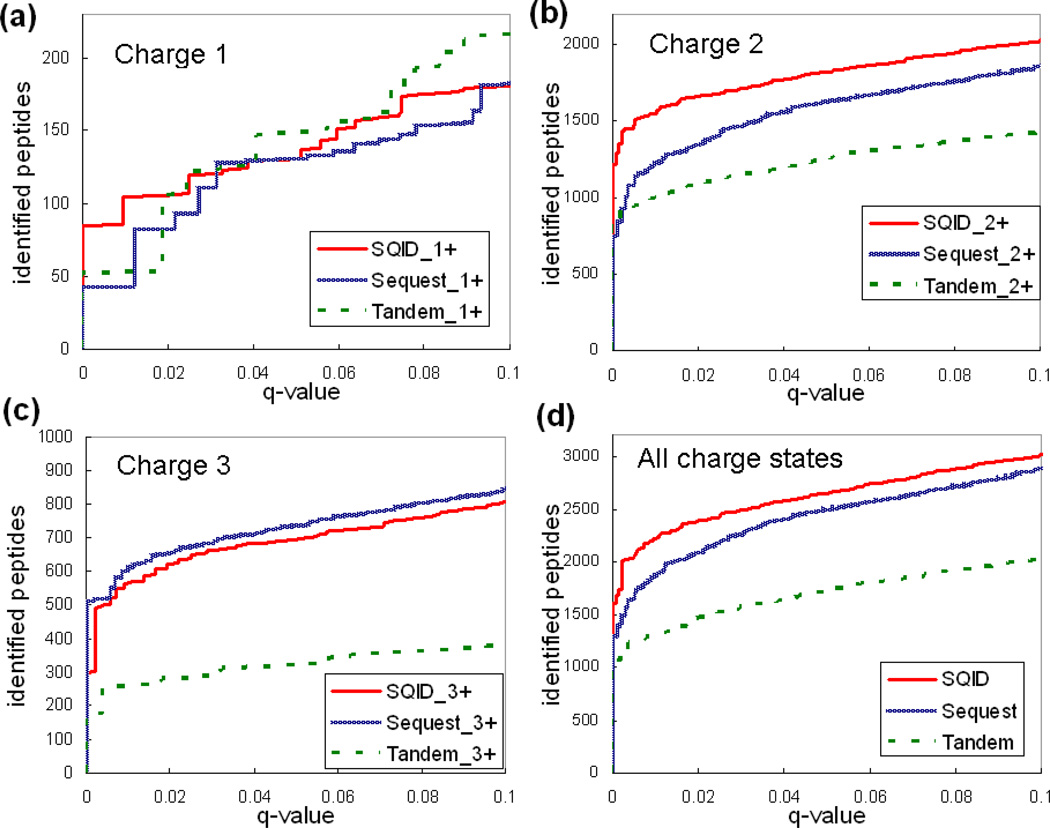
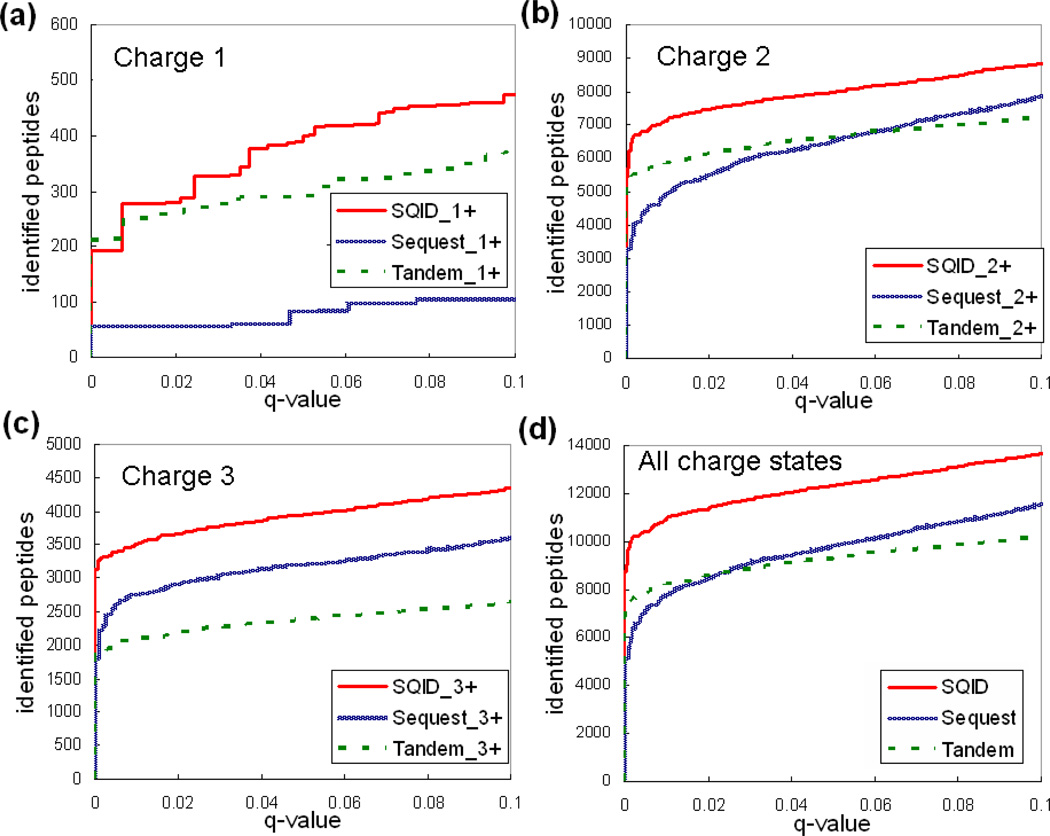
SQID was compared with Sequest and X!Tandem using the PNNL, 18 protein mixture, and yeast datasets. The main score for each algorithm, SQID score, Xcorr and E-value, was sorted for filtering and q-value determination. Figures 3, 4 and 5 compared the search results of SQID, Sequest and Tandem for each dataset, at each charge state. Tables 2, 3 and 4 list the unique peptide (no duplicated sequences) overlap table for these datasets at 0.05 q-value cutoffs. The performance of SQID varied for different datasets, charge states and q-values. For the PNNL dataset, it can be seen that SQID yielded similar performance with Sequest for singly and triply charged peptides, but had much more identification for doubly charged peptides, especially at low q-value cutoffs. At a q-value of 0.05, a total of 22135 unique peptides were identified by SQID, compared with 19678 by Sequest and 14878 by X!Tandem (12% and 48% more identification). The 18 protein mixture dataset showed a smaller difference between SQID and Sequest at all charge states, but X!Tandem still lagged behind. At 0.05 q-value cutoff 292, 273 and 241 unique peptides were identified by SQID, Sequest and X!Tandem, respectively. For the yeast dataset, SQID exhibited strong performances for all charge states in a wide confidence range. At q-value cutoff 0.05, the number of unique peptides lead Sequest or X!Tandem by 25% (4355 for SQID, 3319 for Sequest and 3501 for X!Tandem). It was also noted that compared with X!Tandem, Sequest showed a reduced performance for this dataset. This may be due to the fact that the spectra are relatively noisy, and Sequest relies primarily on the number of matched ions and keeps more peaks in spectrum preprocessing. For all three datasets, SQID identified a significant number of unique peptides that were not identified by either Sequest or X!Tandem, and the overlap regions between SQID and Sequest or SQID and X!Tandem were normally larger than the region between Sequest and X!Tandem (Tables 2, 3 and 4). SQID also showed a better discrimination power at lower q-value cutoffs, which can be seen from the figures below.
Figure 3.
A comparison of SQID, Sequest and X!Tandem by plotting q-value (a measure of FDR) versus identified peptide-spectrum match for the PNNL dataset. (a) Singly charged peptides. (b) Doubly charged peptides. (c) Triple charged peptides. (d) A combination of all charge states.
Figure 4.
A comparison of SQID, Sequest and X!Tandem by plotting q-value (a measure of FDR) versus identified peptide-spectrum match for the 18 protein mixture dataset. (a) Singly charged peptides. (b) Doubly charged peptides. (c) Triple charged peptides. (d) A combination of all charge states.
Figure 5.
A comparison of SQID, Sequest and X!Tandem by plotting q-value (a measure of FDR) versus identified peptide-spectrum match for the yeast dataset. (a) Singly charged peptides. (b) Doubly charged peptides. (c) Triple charged peptides. (d) A combination of all charge states.
Table 2.
Unique peptide overlap table for PNNL dataset at 0.05 q-value cutoff. A total of 22135 unique peptides are identified by SQID, compared with 19678 by Sequest and 14878 by X!Tandem.
| ID set | Charge | Total | ||
|---|---|---|---|---|
| 1+ | 2+ | 3+ | ||
| SQID, Sequest, Tandem overlap | 2886 | 7777 | 860 | 11523 |
| SQID, Sequest overlap | 1146 | 3695 | 983 | 5824 |
| SQID, Tandem overlap | 680 | 1198 | 95 | 1973 |
| Sequest, Tandem overlap | 227 | 88 | 89 | 404 |
| SQID only | 625 | 1889 | 301 | 2815 |
| Sequest only | 832 | 730 | 365 | 1927 |
| Tandem only | 259 | 479 | 240 | 978 |
Table 3.
Unique peptide overlap table for 18 protein mixture dataset at 0.05 q-value cutoff. A total of 292 unique peptides are identified by SQID, compared with 273 by Sequest and 241 by X!Tandem.
| ID set | Charge | Total | ||
|---|---|---|---|---|
| 1+ | 2+ | 3+ | ||
| SQID, Sequest, Tandem overlap | 18 | 139 | 47 | 204 |
| SQID, Sequest overlap | 3 | 24 | 16 | 43 |
| SQID, Tandem overlap | 4 | 9 | 1 | 14 |
| Sequest, Tandem overlap | 2 | 2 | 7 | 11 |
| SQID only | 0 | 12 | 19 | 31 |
| Sequest only | 3 | 3 | 9 | 15 |
| Tandem only | 8 | 2 | 2 | 12 |
Table 4.
Unique peptide overlap table for yeast dataset at 0.05 q-value cutoff. A total of 4355 unique peptides are identified by SQID, compared with 3319 by Sequest and 3501 by X!Tandem.
| ID set | Charge | Total | ||
|---|---|---|---|---|
| 1+ | 2+ | 3+ | ||
| SQID, Sequest, Tandem_overlap | 42 | 1595 | 724 | 2361 |
| SQID, Sequest overlap | 3 | 227 | 209 | 439 |
| SQID, Tandem overlap | 83 | 349 | 83 | 515 |
| Sequest, Tandem overlap | 0 | 137 | 94 | 231 |
| SQID unique | 57 | 580 | 403 | 1040 |
| Sequest unique | 1 | 166 | 121 | 288 |
| Tandem unique | 15 | 246 | 133 | 394 |
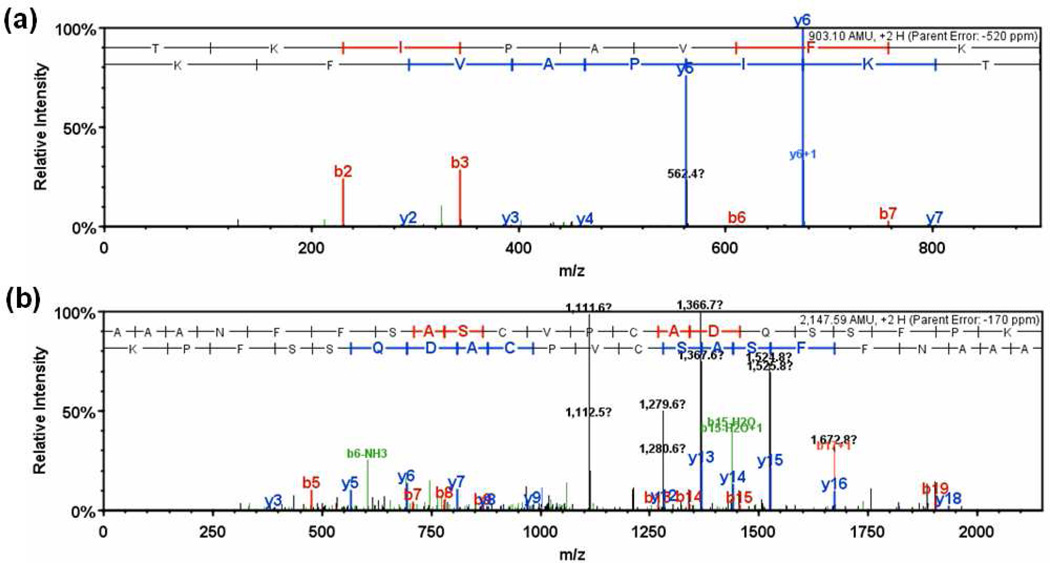
The performance difference between SQID and other algorithms can be contributed to many aspects including spectrum quality, spectrum preprocessing, the incorporation of intensity, consecutive ion series, etc., and it is very difficult to individually quantify each of these terms. Here we show as examples two spectra where intensity scoring played important roles. Figure 6a is an example peptide (TKIPAVFK 2+, from one of the 18 proteins) that was identified by SQID but missed by both Sequest and X!Tandem. The spectrum contains two dominant cleavages (IP, KI) and the other fragments are very small. SQID identified this peptide with 8.5 fragments, 5 consecutive ion series, and a very high intensity score 1.44 (for the top 4 peaks). The final SQID score was 20.33, which was 100% confident (q-value equals 0 at 20.33). This peptide ranked 8th in Sequest with a Xcorr 1.61; X!Tandem missed this identification, and got a wrong hit with E-value 4.7. In contrast, Figure 6b is a peptide (AAANFFSASCVPCADQSSFPK 2+, from one of the 18 proteins) that was identified by Sequest and X!Tandem but missed by SQID. Though there were a high number of fragment matches, the most abundant peaks essentially can not be matched to any fragments, which gave an intensity score 0. This greatly reduced the final SQID score (the peptide scored 5.35, ranked 6th). Sequest identified this peptide as a top hit with Xcorr 3.73 (100% confident) and X!Tandem identified it with E-value 0.0084 (99% confident).
Figure 6.
Example spectra that are a) identified by SQID but missed by Sequest and X!Tandem (TKIPAVFK 2+), b) identified by Sequest and X!Tandem but missed by SQID (AAANFFSASCVPCADQSSFPK 2+).
Further comparison with Sequest
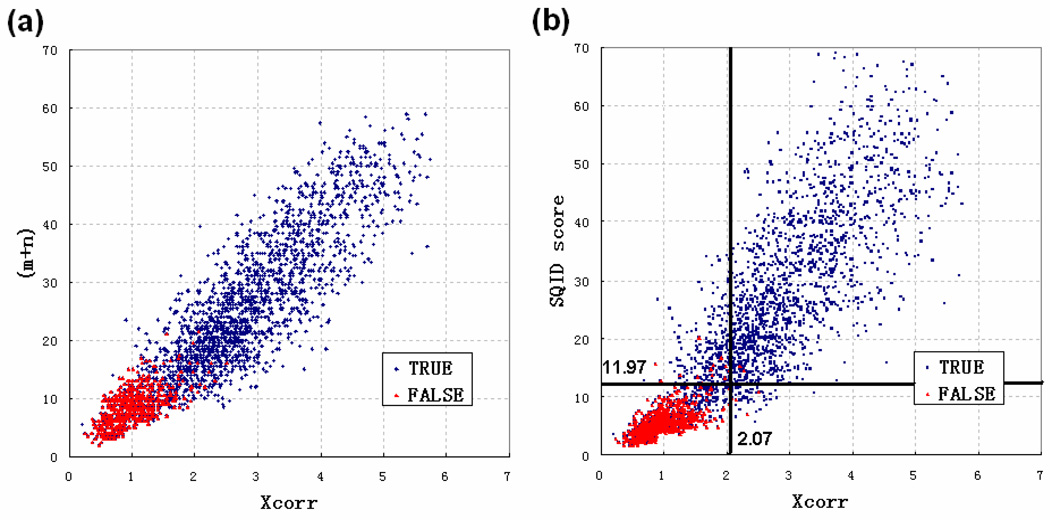
Because neither SQID nor Sequest are probability based, it is more informative to compare their scores, especially when they are matching the same peptide sequence to the same spectra. We extracted the doubly charged spectra that reached the same peptide identifications by SQID and Sequest in the 18 protein mixture dataset. Of the 18496 spectra of doubly charged peptides, only 2571 (13.9%) identifications were overlapping by SQID and Sequest, among which 1912 (74.4% of 2571) belonged to the 18 proteins or contaminations (True) and the remaining 659 belonged to reverse proteins (False). Figure 6a shows the plot of Xcorr with (m+n), the SQID score without the intensity part, and Figure 6b plots Xcorr with the entire SQID score (with the intensity part incorporated). It can be seen that (m+n) is almost proportional to Xcorr for both true and false identifications. However, the full SQID score increases much slower than Xcorr for false identifications in Figure 7b, and a better separation between true and false is achieved. The two bold lines are the corresponding Xcorr and SQID score thresholds (experimentally determined from Figure 4b) for 0.05 q-value cutoffs. At this confidence level, the peptides in the upper-right corner (Xcorr>= 2.07, SQID score>= 11.97) can be identified by both algorithms; the peptides in the upper-left corner (Xcorr < 2.07, SQID score>= 11.97) will be identified only by SQID and the peptides in the lower-right corner will be identified only by Sequest. Therefore, the plot suggests that combining different algorithms is potentially the most beneficial approach for maximizing the number of confident hits.
Figure 7.
A plot of Xcorr versus a) m+n (numbers of matched peaks and numbers of consecutive pairs) and b) SQID score for 2571 peptide-spectrum matches extracted from the 18 protein mixture dataset. Every data point is scored by Sequest and SQID using the same experimental spectrum and the same peptide sequence. The blue spots are true identifications and red spots are false identifications.
Conclusions
In general, SQID shows improved performance compared with popular algorithms as shown by results for three different datasets, with a large number of unique identifications. Combining SQID with other algorithms will thus be potentially beneficial, such as increasing the number of peptide hits and the confidence of identifications. SQID also has the potential to be applied to electron transfer dissociation (ETD) spectra as long as corresponding intensity tables are elucidated. By analyzing over 10000 high resolution ETD spectra from the Coon group, the University of Wisconsin-Madison, we found that the peak intensities in ETD spectra are also highly dependent on the amino acid composition, e.g. amino acid pairs containing basic residues tend to have enhanced cleavage, while pairs containing hydrophobic residues have weaker intensities. This study could help intensity prediction in ETD, and at the same time, provide evidence to clarify the controversial dissociation mechanisms. As a new algorithm, SQID still requires further optimization to improve the overall performance. Future efforts will include creating intensity histograms for other instrument types (e.g. Q-TOF, Orbitrap) and proteases (e.g. GluC, Aspn, chymotrypsin) to enable SQID searches on these data, incorporating different intensity histograms corresponding to specific sequence motifs, combining SQID score and delta score to give a single discrimination score, and developing programs that directly modify scores from other search engines.
Supplementary Material
Acknowledgements
The authors appreciate NIST, PNNL, Dr. Andrew Keller and Dr. Andrew Link for collecting and sharing the corresponding datasets for training and testing SQID. We also thank Dr. David Tabb for helpful discussions and suggestions. The study is funded by NIH Grant 2R01GM051387 to V.W.
Footnotes
Supporting Information Available:
Four supplementary tables, seven supplementary figures, and the intensity histogram for tryptic ion trap data that support the article are provided. This information is available free of charge via the Internet at http://pubs.acs.org. The SQID program and source code are under GNU GPL license and can be downloaded at http://quiz2.chem.arizona.edu/wysocki/bioinformatics.htm. The search results from SQID, Sequest and X!Tandem are available from the authors upon request.
References
- 1.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Craig R, Beavis RC. Tandem: matching proteins with mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 3.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Sadygov RG, Cociorva D, Yates JR. Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat. Methods. 2004;1(13):195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- 5.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. Mobile and localized protons: a framework for understanding peptide dissociation. Journal of Mass Spectrometry. 2000;35(12):1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH. Statistical characterization of the charge state and residue dependence of low-energy CID peptide dissociation patterns. Anal. Chem. 2005;77(18):5800–5813. doi: 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Tseng GC, Yuan S, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH. A data-mining scheme for identifying peptide structural motifs responsible for different MS/MS fragmentation intensity patterns. Proteome Res. 2008;7(1):70–79. doi: 10.1021/pr070106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrometry Rev. 2005;24(4):508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z. Prediction of low-energy collision-induced dissociation spectra of peptides with three or more charges. Anal. Chem. 2004;76(14):3908–3922. doi: 10.1021/ac049951b. [DOI] [PubMed] [Google Scholar]
- 10.Gibbons FD, Elias JE, Gygi SP, Roth FP. SILVER helps assign peptides to tandem mass spectra using intensity-based scoring. J Am Soc Mass Spectrom. 2004;15(6):910–912. doi: 10.1016/j.jasms.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Havilio M, Haddad Y, Smilansky Z. Intensity-based statistical scorer for tandem mass spectrometry. Anal. Chem. 2003;75(3):435–444. doi: 10.1021/ac0258913. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z. Prediction of low-energy collision-induced dissociation spectra of peptides. Anal. Chem. 2004;76(14):3908–3922. doi: 10.1021/ac049951b. [DOI] [PubMed] [Google Scholar]
- 13.Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nature Biotechnol. 2004;22(2):214–219. doi: 10.1038/nbt930. [DOI] [PubMed] [Google Scholar]
- 14.Narasimhan C, Tabb DL, Verberkmoes NC, Thompson MR, Hettich RL, Uberbacher EC. MASPIC: intensity-based tandem mass spectrometry scoring scheme that improves peptide identification at high confidence. Anal Chem. 2005;77(23):7581–7593. doi: 10.1021/ac0501745. [DOI] [PubMed] [Google Scholar]
- 15.Sun S, Meyer-Arendt K, Eichelberger B. Improved validation of peptide MS/MS assignments using spectral intensity prediction. Molecular and Cellular Proteomics. 2007;6(1):1–17. doi: 10.1074/mcp.M600320-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Zhou C, Bowler LD, Feng J. A machine learning approach to explore the spectra intensity pattern of peptides using tandem mass spectrometry data. BMC Bioinformatics. 2008;9:325. doi: 10.1186/1471-2105-9-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein SE, Pudnick PA. Gaithersburg, MD: National Institute of Standards and Technology; NIST Peptide Tandem Mass Spectral Libraries. Yeast Peptide Mass Spectral Reference Data, Yeast, ion trap, Official Build Date: May. 14, 2009. Drosophila Peptide Mass Spectral Reference Data, Drosophila, ion trap, Official Build Date: July. 14, 2008. 20899. Downloaded from http://peptide.nist.gov on April 1 2010. [Google Scholar]
- 18.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10(6):1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 19.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong K, Zhao R, Smith RD. Globalanalysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci U S A. 2002;99(17):11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolker E, Picone AF, Galperin MY, Romine MF, Higdon R, Makarova KS, Kolker N, Anderson GA, Qiu X, Auberry KJ, Babnigg G, Beliaev AS, Edlefsen P, Elias DA, Gorby YA, Holzman T, Klappenbach JA, Konstantinidis KT, Land ML, Lipton MS, McCue L, Monroe M, Pasa-Tolic L, Pinchuk G, Purvine S, Serres MH, Tsapin S, Zakrajsek BA, Zhu W, Zhou J, Larimer FW, Lawrence CE, Riley M, Collart FR, Yates JR, Smith RD, Giometti CS, Nealson KH, Fredrickson JK, Tiedje JM. Global profiling of Shewanella oneidensis MR-1: expression of hypothetical genes and improved functional annotations. Proc Natl Acad Sci U S A. 2005;102(6):2099–2104. doi: 10.1073/pnas.0409111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller A, Purvine S, Nesvizhskii AI, Stolyar S, Goodlett DR, Kolker E. Experimental protein mixture for validating tandem mass spectral analysis. OMICS. 2002;6(2):207–212. doi: 10.1089/153623102760092805. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. Proteome Res. 2007;6(9):3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. Proteome Res. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci U. S. A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.