Abstract
The development of small molecule chemical probes or therapeutics that target RNA remains a significant challenge despite the great interest in such compounds. The most significant barrier to compound development is a lack of knowledge of the chemical and RNA motif spaces that interact specifically. Herein, we describe a bioactive small molecule probe that targets expanded r(CGG) repeats, or r(CGG)exp , that causes Fragile X-associated Tremor Ataxia Syndrome (FXTAS). The compound was identified by using information on the chemotypes and RNA motifs that interact. Specifically, 9-hydroxy-5,11-dimethyl-2-(2-(piperidin-1-yl)ethyl)-6H-pyrido[4,3-b]carbazol-2-ium, binds the 5’CGG/3’GGC motifs in r(CGG)exp and disrupts a toxic r(CGG)exp -protein complex in vitro. Structure-activity relationships (SAR) studies determined that the alkylated pyridyl and phenolic side chains are important chemotypes that drive molecular recognition to r(CGG)exp . Importantly, the compound is efficacious in FXTAS model cellular systems as evidenced by its ability to improve FXTAS-associated pre-mRNA splicing defects and to reduce the size and number of r(CGG)exp -protein aggregates. This approach may establish a general strategy to identify lead ligands that target RNA while also providing a chemical probe to dissect the varied mechanisms by which r(CGG)exp promotes toxicity.
INTRODUCTION
RNA plays diverse and important roles in biological processes (1). Aberrant RNA function causes many severe diseases (2). For example, microRNA dysregulation can contribute to cancer (3) and single nucleotide mutations in mRNAs cause beta-thalassemia and inherited breast cancer (4). RNA trinucleotide repeat expansions cause or contribute to various neurological disorders (5) including Fragile X-associated Tremor Ataxia Syndrome (FXTAS), myotonic dystrophy type 1 (DM1), and Huntington’s disease (HD) (6) and may contribute to Fragile X Syndrome (FXS) (7).
Although RNA transcripts with expanded repeats cause the diseases mentioned above, the physiological response to the repeats, and thus the causes of disease, are quite different. Differences are mainly due to the location of the expanded repeats in a given mRNA. For example, HD is caused by an expansion of r(CAG) in the coding region of huntingtin mRNA. In the most well established mechanism of HD, disease is caused when expanded r(CAG) repeats are translated into a toxic polyQ version of huntingtin (8). Thus, HD is caused by a gain-of-function at the protein level. In FXS, >200 copies of d(CGG) in the 5’ untranslated region (UTR) of the fragile X mental retardation 1 (FMR1) gene causes disease by initiating local methylation, transcriptional silencing, and loss of fragile x mental retardation protein (FMRP) (9). Thus, FXS is caused by a loss-of-function. Lastly, FXTAS and DM1 are caused when expanded repeats present in UTR’s sequester proteins that are involved in pre-mRNA splicing regulation (10, 11). Sequestration of these proteins causes the aberrant splicing of a variety of pre-mRNAs, leading to the expression of defective proteins. Thus, FXTAS and DM1 are caused by an RNA gain-of-function.
Despite the contribution of expanded RNA repeats to diseases, there are few compounds that target these RNAs in particular and non-ribosomal RNAs in general. Our group recently reported two approaches to design small molecules (12, 13) and modularly assembled compounds (14) that bind RNA and modulate its function in vivo. In particular, we have used information about RNA motif-small molecule interactions (15–17) and chemical similarity searching (18–21) to design bioactive ligands that target r(CUG)exp and r(CAG)exp , which cause DM1 and HD, respectively (12–14). These approaches were thus applied to develop bioactive small molecules that target r(CGG)exp .
All RNA trinucleotide repeats fold into a hairpin with periodically repeating 1x1 nucleotide internal loops in the stem (10). We therefore probed an RNA-focused small molecule library enriched with chemotypes that bind RNA 1x1 nucleotide internal loops, such as the 1x1 nucleotide GG internal loop in r(CGG)exp (13). It was previously that Hoechst 33258 (22) and a related derivative (16) bind 1x1 nucleotide GG internal loops. Therefore, one of the chemotypes used as a basis for our small molecule library was Hoechst.
Since FXTAS is caused by sequestration of proteins that regulate pre-mRNA splicing, a high throughput protein-displacement assay was used to screen for inhibitors. From this library, a designer small molecule, 9-hydroxy-5,11-dimethyl-2-(2-(piperidin-1-yl)ethyl)-6H-pyrido[4,3-b]carbazol-2-ium, was identified. The compound binds tightly to 1x1 nucleotide GG loops and is efficacious in cell culture models of FXTAS. Specifically, it improves pre-mRNA splicing defects and reduces the size and number of r(CGG)exp nuclear foci. Thus, this compound may serve as a chemical probe to understand how r(CGG)exp causes FXS and may contribute to FXTAS. Collectively, these studies suggest that small molecules targeting traditionally recalcitrant RNA targets can be developed.
RESULTS & DISCUSSION
FXTAS is caused by a pathogenic mechanism in which there is a gain-of-function by an expanded r(CGG) repeat, or r(CGG)exp (10). Like other expanded RNA trinucleotide repeating transcripts, r(CGG)exp folds into a hairpin structure with regularly repeating 1x1 nucleotide internal loops, or 5’CGG/3’GGC motifs (Figure 1) (23). FXTAS patients are carriers of pre-mutation alleles (55–200 repeats) and have increased FMR1 mRNA levels and normal or moderately low FMRP protein expression levels (24, 25). Evidence for RNA gain-of-function comes from animal models and cell-based assays. For example, insertion of untranslated r(CGG)exp (of the length that cause FXTAS) into mice and Drosophila cause deleterious effects like those observed in humans that have FXTAS (26, 27). In particular, it has been shown that there is genetic interaction between r(CGG)exp and Purα mediates neurodegeneration (28). In cell-based models, r(CGG)exp forms nuclear inclusions, and the size of inclusions scales with the length of the repeat and the age of death from the disease (29, 30).
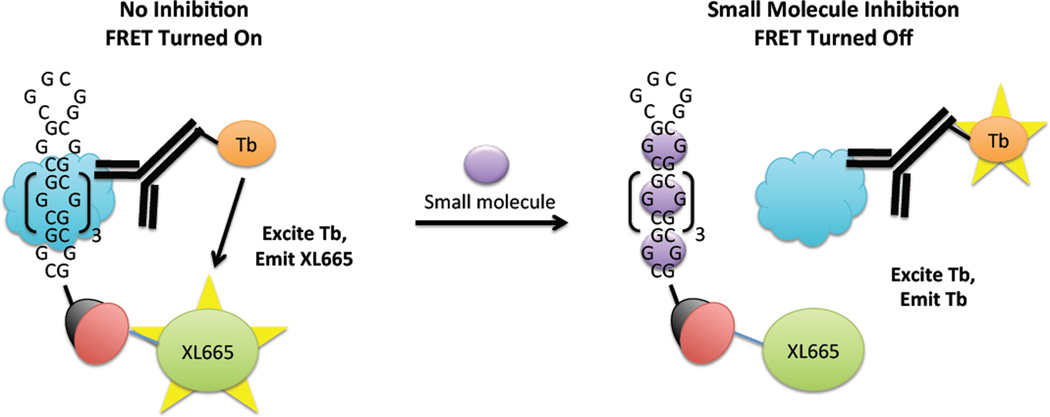
Figure 1.
Schematic of the protein displacement assay that was used to identify small molecule inhibitors of the r(CGG)12-DGCR8Δ interaction and to determine their potencies. The r(CGG)12 oligonucleotide is labeled with a 5’-biotin while DGCR8Δ (blue cloud) contains a histidine (His) tag. Left, in the absence of inhibitor, DGCR8Δ binds to r(CGG)12. Binding is quantified by using two antibodies that form a FRET pair—an anti-His antibody labeled with Tb that binds to DGCR8Δ and streptavidin labeled with XL665 that binds to r(CGG)12. The two fluorophores are within close enough proximity to form a FRET pair. Tb is excited at 345 nm; the resulting emission (~545 nm) excites XL665, which emits at 665 nm. Right, in the presence of inhibitor, the r(CGG)12-DGCR8Δ interaction is disrupted, and the two fluorophores are not within close enough proximity to form a FRET pair. Therefore, emission is only observed at 545 nm (due to Tb). XL665 emission is not observed.
A more detailed mechanism for the RNA gain-of-function has recently been elucidated from studies of patient-derived cell lines. In studies by the Charlet group, it was shown that r(CGG)exp sequesters and inactivates the Src-Associated substrate during mitosis of 68 kDa protein (Sam68) (10). Sam68 is a nuclear RNA-binding protein involved in alternative splicing regulation (32), and the sequestration of Sam68 by r(CGG)exp leads to the pre-mRNA splicing defects observed in FXTAS patients (10). The RNA-protein complex is a scaffold for the assembly of other proteins such as muscleblind-like 1 protein (MBNL1) and heterogeneous nuclear ribonucleoprotein (hnRNP). Additional data indicate that Sam68 does not bind r(CGG)exp directly, rather the interaction is mediated by another protein. The Charlet group has recently determined that this protein is DGCR8 (Sellier, C. (IGBMC, University of Strasbourg), Freyermuth, F. (IGBMC, University of Strasbourg), Tabet, R. (IGBMC, University of Strasbourg), Tran, T. (The Scripps Research Institute), He, F. (University of Michigan), Ruffenach, F. (IGBMC, University of Strasbourg), Alunni, V. (IGBMC, University of Strasbourg), Moine, H. (IGBMC, University of Strasbourg), Thibault, C. (IGBMC, University of Strasbourg), Page, A. (IGBMC, University of Strasbourg), Tassone, F. (University of California, Davis), Willemsen, R., (Erasmus MC, Rotterdam) Disney, M.D. (The Scripps Research Institute), Todd, P.K. (University of Michigan), Hagerman, P. (University of California, Davis), and Charlet-Berguerand, N. (IGBMC, University of Strasbourg); unpublished data and reference 31). Taken together, targeting r(CGG)exp with a small molecule to inhibit protein binding is an attractive treatment for FXTAS. We therefore screened a library enriched in small molecules that are biased, or focused, for binding RNA to identify lead ligands that bind r(CGG)exp.
Probing an RNA-Focused Small Molecule Library to Identify Inhibitors of the r(CGG)-DGCR8Δ Complex
In order to construct a library of small molecules that is enriched in compounds that have the potential to recognize RNA 1x1 nucleotide internal loops like the ones that are displayed in r(CGG)exp (Figure 1), previously reported chemical similarity searches were employed (12, 13). Those searches identified compounds that are similar to the bis-benzimidazole Hoechst 33258, 4',6-diamidino-2-phenylindole (DAPI), and pentamidine. This RNA-focused collection of small molecules contained two small molecules that improve defects that are associated with r(CAG)exp and r(CUG)exp in cell culture models of HD and DM1, respectively (12, 13). Thus, Hoechst-, pentamidine-, and DAPI-like compounds were screened to identify inhibitors of the r(CGG)-DGCR8Δ protein complex.
Screening was completed using a time-resolved FRET assay that has been previously described for identifying inhibitors of the r(CUG)exp-MBNL1 and r(CAG)exp-MBNL1 complexes (Figure 1) (12, 13). Briefly, a 5’-biotinylated RNA oligonucleotide containing 12 r(CGG) repeats is incubated with His6-tagged DGCR8Δ. The ligand of interest is then added, followed by addition of two antibodies that recognize the RNA (streptavidin-XL665) or DGCR8Δ (Tb labeled anti-His6). If the compound does not displace DGCR8Δ, then Tb and XL-665 are within close enough proximity to form a FRET pair. If, however, the ligand displaces the protein, then no FRET is observed.
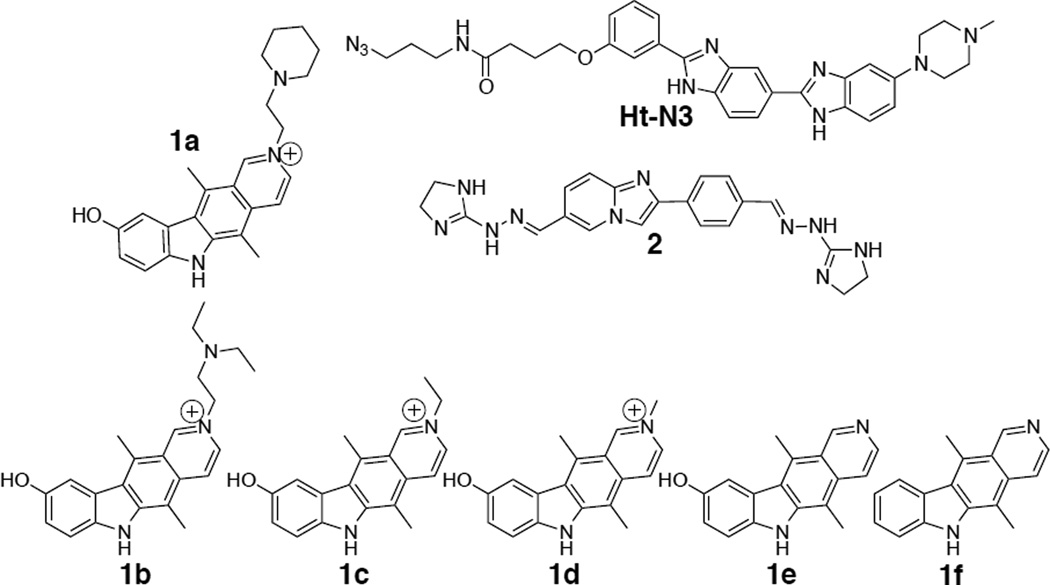
From this screen, three compounds (Figure 2) were identified that disrupt the r(CGG)exp-DGCR8Δ complex in the low to mid micromolar range. (Either no or very slight inhibition was observed for all other compounds at 100 µM). They include compounds 1a, 2, and Ht-N3 (Figure 2). Interestingly, all three compounds were derived from the Hoechst or bis-benzimidazole query. Dose-response curves show that 1a and Ht-N3 disrupt the r(CGG)12-DGCR8Δ complex with IC50’s of 12 and 33 µM, respectively. Compound 2, however, only disrupts ≈25% of the r(CGG)12-DGCR8Δ complex at 100 µM.
Figure 2.
The structures of the small molecules identified from an RNA-focused library that inhibit the r(CGG)12-DGCR8Δ interaction and derivatives of the most potent compound (1a). 1b – 1f were used to construct structure-activity relationships and define the active pharmacophore. Inhibition is markedly decreased for derivatives 1e and 1f (Table 1).
Molecular Recognition of r(CGG)exp by 1a
To further investigate the chemotypes in compound 1a that allow effective recognition of r(CGG)exp and inhibition of the r(CGG)12-DGCR8Δ complex, a series of derivatives were studied (Figure 2). These compounds probe the role of: (i) the identity of the alkylated pyridyl side chain; (ii) the phenolic side chain; and, (iii) the positive charge. The IC50’s for inhibition of protein binding for 1b, 1c, and 1d are similar to that of 1a (5–12 µM). The IC50 of compounds 1e is ~25 µM while 1f has no effect on protein binding at 25 µM. Table 1 summarizes the IC50’s and the percentage of protein displaced from r(CGG)12 at 25 µM of each compound. Taken together, the presence of a positive charge due to the alkylated pryidyl side chain and the presence of the exocyclic hydroxyl group are required for compound potency.
Table 1.
The potencies of 1a - 1f for disruption of the r(CGG)12-DGCR8Δ complex and the corresponding affinities for an RNA containing one 5’CGG/3’GGC motif. The potencies of the compounds are reported as IC50s as determined from the TR-FRET assay.
| 1a | 1b | 1c | 1d | 1e | 1f | |
|---|---|---|---|---|---|---|
| Percentage displacement at 25 µM |
85 ± 1 | 91 ± 5 | 96 ± 9 | 87 ± 5 | 46 ± 5 | 0 |
| IC50, µM | 13 ± 0.4 | 8 ± 0.3 | 13 ± 0.2 | 7 ± 0.2 | ~25 | NDa |
| Kd, nM | 76 ± 4 | 38 ± 1 | 69 ± 5 | 50 ± 18 | NMb | NMb |
ND denotes that no determination could be made.
NM denotes that no measurement was made.
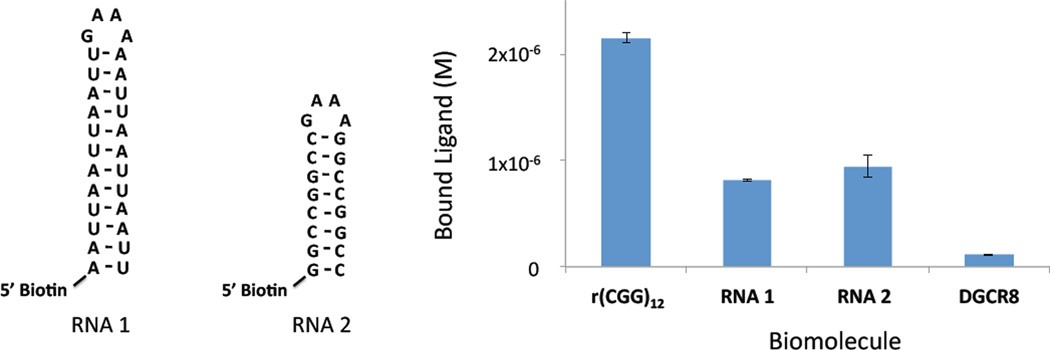
In the protein displacement assay, inhibition occurs if the small molecule binds the protein or the RNA. Therefore, we used competition dialysis (33) to assess the selectivity of 1a. A series of RNA targets, including two base paired RNAs, r(CGG)12 (a mimic of r(CGG)exp used in the displacement assay, Figure 1), and DGCR8Δ were used (Figure 3). The results of these studies show that 1a binds tightly to r(CGG)12 while very little binding is observed to DGCR8Δ. Although some binding is observed to fully paired RNAs, less than half of the amount of ligand that partitioned into r(CGG)12 partitioned into these samples. Thus, 1a binds preferentially to r(CGG)12 over the other targets tested. The binding affinities of 1a - 1d for an RNA with a 1x1 nucleotide GG Internal loop motif were also determined. The measured Kd’s are similar for all four compounds and range from ~40 – 75 nM (Table 1), as expected based on their similar potencies.
Figure 3.
Results of competition dialysis experiments used to assess the specificity of 1a for r(CGG)12. Top, the secondary structures of two fully paired RNAs used in competition dialysis. Bottom, plot of the amount of ligand bound to various RNAs and DGCR8Δ.
Biological Activity of 1a in Model Cellular Systems of FXTAS
In order to assess the bioactivity of 1a, a model cellular system of FXTAS was used (10). Previously, it has been shown that pre-mRNA splicing defects are observed in survival of motor neuron 2 (SMN2) and B-cell lymphoma x (Bcl-x) mRNAs when cells express r(CGG)exp (10). These pre-mRNA splicing defects are due to sequestration of Sam68 by r(CGG)exp; Sam68 directly regulates the alternative splicing of SMN2 and Bcl-x (10). Specifically, exon 7 of the SMN2 mRNA is included too frequently in FXTAS model systems; ~70% of SMN2 mRNA contains exon 7 when r(CGG)exp is expressed while exon 7 is included in only ~30% of SMN2 mRNA in cells that do not express r(CGG)exp (Figure 4, top). Likewise, there are two isoforms of Bcl-x mRNA, Bcl-xL and Bcl-xS. In FXTAS cellular model systems, 60% of the Bcl-x mRNA is the Bcl-xL isoform. In healthy cells, only 40% of the mRNA is the Bcl-xL isoform (Figure 4, bottom).
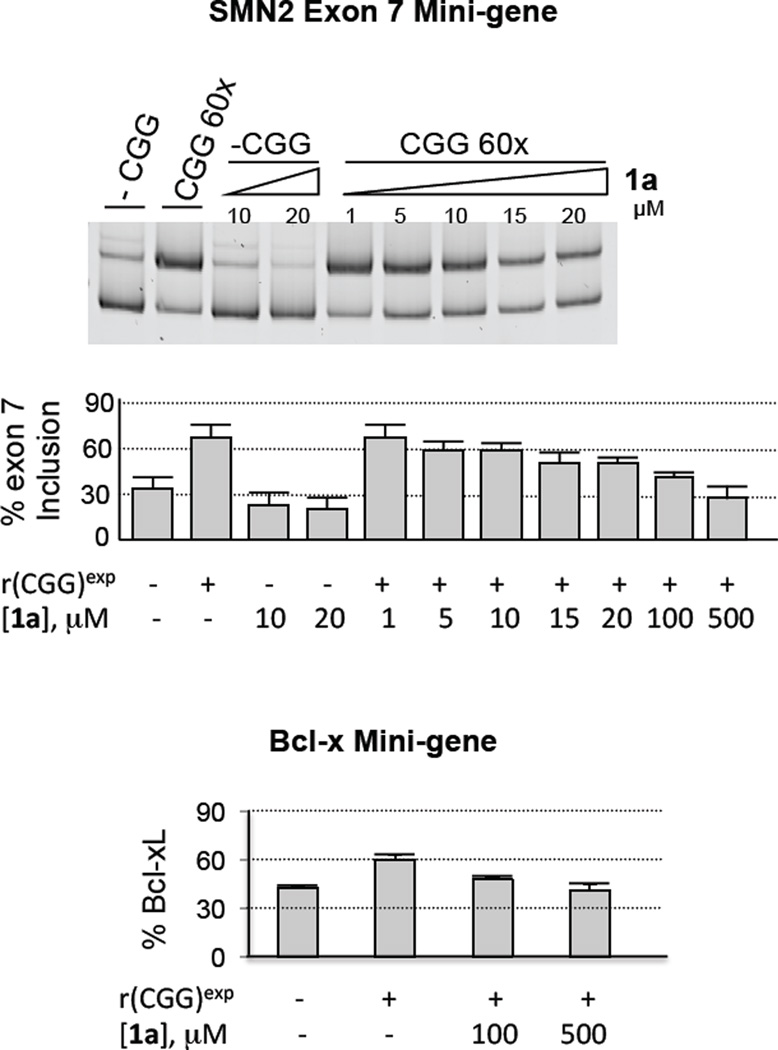
Figure 4.
In vivo efficacy of 1a against FXTAS as assessed by improvement in pre-mRNA splicing defects. Briefly, COS7 cells were transfected with an SMN2 or Bcl-x mini-gene in the presence or absence of a mini-gene that express 60 r(CGG) repeats (CGG 60X). The cells were then treated with 1a. Total RNA was harvested, and the alternative splicing of the SMN2 exon 7 or Bcl-x exon 2 was determined by RT-PCR. A, 1a improves SMN2 pre-mRNA splicing defects. B, 1a improves Bcl-x pre-mRNA splicing defects.
When cells that express r(CGG)60 are treated with 1a, improvement in SMN2 and Bcl-x pre-mRNA splicing defects are observed (Figure 4). For example, improvement of SMN2 splicing defects can be observed when cells are treated with as little as 20 µM of 1a. SMN2 mis-splicing is further improved at higher concentrations: treatment with 100 µM 1a improves pre-mRNA splicing levels to approximately 70% of wild type (absence of r(CGG)exp) while treatment with 500 µM restores pre-mRNA splicing to levels wild type (Figure 4). 1a also improves dysregulation of Bcl-x splicing. Statistically significant improvement is observed when cells are treated with 100 µM of 1a while restoration of wild type splicing patterns are observed at 500 µM (Figure 4). No statistically significant effect on SMN2 or Bcl-x splicing was observed when cells that do not express r(CGG)60 are treated with 1a. This suggests that the improvement of pre-mRNA splicing defects is due to 1a displacing proteins from r(CGG)60.
Control experiments were also completed to determine the specificity of 1a; that is if it affects the splicing of RNAs not controlled by Sam68. In these experiments, a PLEKHH2 (17) or cardiac troponin T (cTNT) (34) mini-gene was used, as their alternative splicing is not regulated by Sam68. The addition of 1a (500 µM) did not affect PLEKHH2 or cTNT alternative splicing (Figures S-4 & S-5). Thus, the effect of 1a on pre-mRNA splicing appears to be specific to the splicing of pre-mRNAs regulated by Sam68.
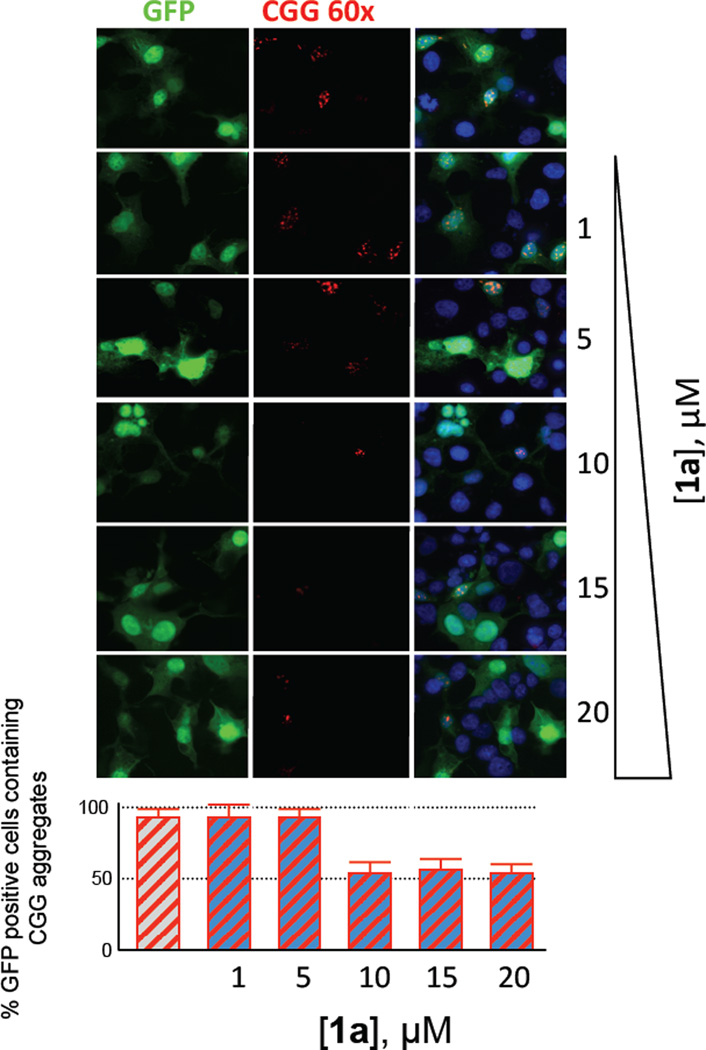
Another phenotype of cells expressing r(CGG)exp is the formation of nuclear foci. Additional studies were therefore completed by using a fluorescence in situ hybridization (FISH) assay to determine if 1a decreased the number or size of foci. As can be observed in Figure 5, 1a reduces the size and the number of foci. Collectively, the improvement in the formation of foci and in the dysregulation of pre-mRNA alternative splicing show that 1a binds r(CGG)exp in cellular systems and displaces bound proteins that are then free to complete their normal physiological functions.
Figure 5.
1a decreases r(CGG)exp-protein aggregates as assessed by fluorescence in situ hybridization (FISH). Briefly, COS7 cells were co-transfected with a plasmid encoding 60 r(CGG) repeats and a plasmid encoding GFP. Cells were then treated with 1a and probed with 5’(CCG)8-Cy3 DNA oligonucleotide probe. Only cells that are GFP positive were analyzed for the presence of nuclear foci. Top, confocal microscopy images of cells treated with different concentrations of 1a. For all panels: left, GFP fluorescence (indicates transfected cells); middle, Cy3 fluorescence (indicates r(CGG)exp); right: overlay of GFP, Cy3, and DAPI (indicates nuclei) fluorescence images. Bottom, plot of the number of r(CGG) aggregates as a function of the concentration of 1a.
Comparison to Other Small Molecules that Target RNA
A few bioactive small molecules have been shown to bind to expanded triplet repeats in vivo and to improve associated defects (12–14, 17). For example, a bis-benzimidazole (13), pentamidine (17), and modularly assembled bis-benzimidazoles (14) target the r(CUG)exp that causes DM1. Each improves pre-mRNA splicing defects. In general, modularly assembled ligands that target multiple 5’CUG/3’GUC motifs in r(CUG)exp simultaneously are the more potent inhibitors. For example, a monomeric bis-benzimidazole (H1) improves pre-mRNA splicing defects in DM1 model systems to wild type levels when 2000 µM of compound is used. A dimeric modularly assembled compound that displays two copies of a bis-benzimidazole, 2H-4, improves pre-mRNA splicing levels back to wild type when cells are treated with 50 µM compound. This represents a greater than 40-fold enhancement in bioactivity provided by a modular assembly approach even though the assembled compounds are of higher molecular weight and not classically “drug-like.” The improved bioactivity of the modularly assembled compound could be due to the increased surface area occupied by the compound, residence time on the RNA target, and the affinity and selectivity of modularly assembled ligands for r(CUG)exp (14, 16).
In order to synthesize second-generation modularly assembled compounds that target r(CGG)exp, a site that can be used to conjugate 1a-like compounds onto an assembly scaffold must be identified. Fortuitously, our SAR studies showed that the side chain that emerges from the pyridyl group can be altered since it does not affect potency. Thus, this site is ideal for the addition of reactive groups that can be anchored onto an assembly scaffold.
Implications
The identification of a bioactive small molecule that targets r(CGG)exp not only provides lead compounds that could become therapies for FXTAS, but also for other disorders that are mediated by r(CGG)exp. Notably, this includes Fragile X Syndrome (FXS), the most common single gene cause of autism (35). FXS is caused by hypermethylation of the FMR1 locus, leading to transcriptional silencing and loss of FMRP protein (9). There is some evidence that an RNAi-like mechanism may also contribute in which r(CGG)exp is cleaved into small RNAs that enable transcriptional silencing (7). If r(CGG)exp does indeed contribute to FXS, then a small molecule that targets r(CGG)exp could help differentiate modes of toxicity and/or disease mechanisms.
Lastly, the ability of a small molecule to target r(CGG)exp in cellular models of FXTAS and reverse a pre-mRNA splicing defect provides further evidence for an RNA gain-of-function mechanism. Since this study is another example of a small molecule that targets a non-ribosomal RNA that causes disease, it provides further evidence that small molecules can be developed to target non-coding regions in RNA even though these targets have been thought to be recalcitrant to small molecule intervention.
MATERIALS & METHODS
Small Molecules
All small molecules were procured from the National Cancer Institute (NCI), Sigma Aldrich, or The Scripps Research Institute. The purities of the compounds used in additional studies (IC50’s, affinities, etc.) were determined by HPLC, and their masses were confirmed by ESI mass spectrometry. All compounds were ≥95% pure. These data are available in the Supporting Information (Figure S-6 and Table S-2).
Oligonucleotide Preparation and Purification
The RNAs used in the protein displacement assay (5'-biotin-(CGG)12) and competition dialysis were purchased from Dharmacon. The ACE protecting groups were cleaved using Dharmacon’s deprotection buffer (100 mM acetic acid, adjusted to pH 3.8 with TEMED) by incubating at 60 °C for 2 h. The samples were lyophilized, resuspended in water, and desalted using a PD-10 gel filtration column (GE Healthcare). The concentrations were determined by absorbance at 90 °C using a Beckman Coulter DU800 UV-Vis spectrophotometer equipped with a Peltier temperature controller unit. Extinction coefficients (at 260 nm) were calculated using the HyTher server (36, 37), which uses nearest neighbors parameters (38).
DGCR8Δ Expression and Purification
His-tagged DGCR8Δ was expressed in Escherichia coli BL21 cells via induction with 1 mM IPTG for 4 h. Cells were lysed in 50 mL of Lysis Buffer (50 mM Tris-Cl pH 8.0, 150 mM NaCl, 2 mM 2-mercaptoethanol, 10 mM imidazole, 0.1% (v/v) Igepal, 2 mg mL−1 lysozyme, and 1 mM PMSF) for 30 min on ice. DNase I was then added to a final concentration of 1 U mL−1, and cells were sonicated (60% power for 9 x 10 s). The DGCR8Δ protein was purified via FPLC (Akta Explore, GE Healthcare) using a HiTrap Ni-column (GE Healthcare), followed by a cation exchange column (HiTrap SP FF, GE Healthcare) and a Superdex 75 size exclusion column. The protein was concentrated and dialyzed in a Vivaspin 15 centrifugal concentrator (Sartorius Stedim Biotech) into Storage Buffer (10 mM Tris-Cl pH 7.6, 200 mM NaCl, 1 mM EDTA, and 5 mM DTT, and 30% Glycerol) and stored at −20 °C.
Determination of Compound Potency via a Protein Displacement Assay
The protein displacement assay used to identify inhibitors of the r(CGG)12-DGCR8Δ complex is based on PubChem BioAssay AID 2675 (Figure 1), which utilizes time resolved (TR)-FRET between antibodies that bind the RNA and the protein. The assay was conducted in 1X TR-FRET Assay Buffer (20 mM HEPES pH 7.5, 110 mM KCl, 110 mM NaCl, 0.1% (w/v) BSA, 2 mM MgCl2, 2 mM CaCl2, 0.05% Tween-20, and 5 mM DTT) with 5 µM yeast extract bulk tRNA (Roche Diagnostics), 160 nM RNA, 154.5 nM His-tagged DGCR8Δ, 40 nM Streptavidin-XL665 (HTRF, Cisbio Bioassays) and 4.4 ng µl−1 Anti-His6-Tb (HTRF, Cisbio Bioassays).
The RNA was folded by incubation at 60 °C for 5 min in 1X Folding Buffer (20 mM HEPES, pH 7.5, 110 mM KCl, and 110 mM NaCl) followed by slow cooling to room temperature. Then, DGCR8Δ and the other buffer components specified above were added to the folded RNA. After incubating for 15 min at room temperature, 9 µL of the mixture was transferred to a microcentrifuge tube containing 1 µL ligand at varying concentrations. A 9 µL aliquot of this final mixture was transferred to a well of a 384-well white plate (Greiner) and incubated for 1 h at room temperature. To exclude ligands that perturb F545/F665, a 9 µL control solution containing antibodies and different ligand concentrations in 1X TR-FRET Assay Buffer but no RNA or protein was also transferred to the plate.
The time resolved fluorescence at 545 nm and 665 nm was measured using a SpectraMax M5 plate reader (Molecular Devices, Inc.) with excitation wavelength of 345 nm, cut-off at 420 nm, 200 µs delay, and 1500 µs integration time. The ratio of fluorescence intensities at 545 nm and 665 nm (F545/F665) for a series of ligand dilutions were fit to equation 1:
| (eq. 1) |
where y is the percentage of DGCR8Δ displacement, B is the percentage of DGCR8Δ displacement in the absence of ligand (0%), A is the maximum percentage displacement of DGCR8Δ (typically 100%), and the IC50 is the concentration of ligand where half of the protein is displaced from the RNA.
Competition Dialysis
Competition dialysis was completed as previously described (33). Briefly, 2 µM RNA or protein was transferred into Slide-a-Lyzer MINI dialysis units with a molecular weight cut-off of 2,000 (Thermo Scientific), and the units were placed into a solution of 0.7 µM ligand. Two blank units containing only buffer were used to monitor equilibration by checking the absorbance at the peak wavelength. After the blank units reached equilibrium, sodium dodecyl sulfate (SDS) was added to a final concentration of 1%, and the absorbance was measured. This absorbance was used to determine total ligand concentration (Ct). The concentration of the dialysate (free ligand concentration, Cf) was determined analogously. The bound ligand concentration (Cb) was then determined using equation 2:
| (eq. 2) |
where Cb, Ct, and Cf are concentrations of bound, total, and free ligand, respectively.
RNA-Binding Assays Via Dye Displacement
Dissociation constants were determined using an in-solution, fluorescence-based assay (39–47). RNA was annealed in DNA buffer (8 mM Na2HPO4, pH7.0, 185 mM NaCl, 0.1 mM EDTA, 40 µg/mL BSA) at 60 °C for 5 min and allowed to slowly cool to room temperature. The annealed RNA was then titrated into DNA buffer containing 1000 nM Hoechst 33258. Fluorescence signal was recorded using a Bio-Tek FLX-800 plate reader, which was equipped with excitation filter at 360/40 nm and emission filter at 460/40 nm. The change in fluorescence intensity as a function of RNA concentration was fit to the following equation (eq. 3): (39, 48)
| (eq. 3) |
where I is the observed fluorescence intensity, I0 is the fluorescence intensity in the absence of RNA, Δε is the difference between the fluorescence intensity in the absence of RNA and in the presence of infinite RNA concentration and is in units of M−1, [Ht]0 is the concentration of Hoechst 33258, [RNA]0 is the concentration of the selected internal loop or control RNA, and Kt is the dissociation constant.
Ligands 1a - 1d were then added to compete for binding to the RNA (1 µM) in presence of Hoechst 33258 (1 µM). Reduction in fluorescence of Hoechst 33258 was measured using a Bio-Tek FLX-800 plate reader as a function of ligand concentration (1a – 1d) and was fit to the following equation (eq. 4): (43)
| (eq. 4) |
where Θ is the fraction bound of Hoechst 33258, Kt is the dissociation constant for Hoechst 33258, Kd is the dissociation constant of the competing ligand, [Ht]0 is the total concentration of the Hoechst 33258, [Ct]0 is the total concentration of the competing ligand, A is the fraction bound of Hoechst 33258 at infinite concentration of the competing ligand, and [RNA]0 is the total concentration of RNA.
Improvement of Splicing Defects in a Cell Culture Model Using RT-PCR
In order to determine if 1a improves FXTAS-associated splicing defects in vivo, a cell culture model system was used. Briefly, COS7 cells were grown as monolayers in 24- or 96-well plates in growth medium (1X DMEM, 10% FBS, and 1X GlutaMax (Invitrogen)). After the cells reached 90–95% confluency, they were transfected using Lipofectamine 2000 reagent (Invitrogen) or FugenHD (Roche) per the manufacturer’s standard protocol. Equal amounts of a plasmid expressing a 60 CGG repeats and a mini-gene of interest (SMN2 or Bcl-x) were used. Approximately 5 h post-transfection, the transfection cocktail was removed and replaced with growth medium containing 1a. After 16–24 h, the cells were lysed in the plate, and total RNA was harvested with a Qiagen RNAEasy kit or a GenElute kit (Sigma). An on-column DNA digestion was completed per the manufacturer’s recommended protocol.
A sample of RNA was subjected to reverse transcription-polymerase chain reaction (RT-PCR) using 5 units of AMV Reverse Transcriptase from Life Sciences or Superscript II (Invitrogen). Approximately 300 ng were reverse transcribed, and 150 ng were subjected to PCR. RT-PCR products were observed after 25–30 cycles of: 95 °C for 1 min; 55 °C for 1 min; 72 °C for 2 min and a final extension at 72 °C for 10 min. The products were separated by polyacrylamide or agarose gel electrophoresis, stained, and imaged using a Typhoon phosphorimager. The splicing isoforms were quantified using QuantityOne software (BioRad). Table S-2 lists the RT-PCR primers used for each mini-gene construct.
Two sets of control experiments were completed: (i) COS7 cells were co-transfected with a control plasmid that does not contain CGG repeats and the SMN2 or Bcl-x mini-gene as described above; and, (ii) COS7 cells were co-transfected with the mini-gene that expresses 60 r(CGG) repeats and a mini-gene that encodes a pre-mRNA whose splicing is not controlled by Sam68 (PLEKHH2 or cTNT) (49).
Disruption of Nuclear Foci Using Fluorescence In Situ Hybridization (FISH)
FISH experiments were completed as previously described.(10) Briefly, COS7 cells were plated onto glass coverslips and co-transfected with plasmids encoding for r(CGG)60 and GFP. The cells were fixed in 4% paraformaldehyde in PBS (pH 7.4) for 15 min and washed three times with PBS. Then, they were permeabilized with 0.5% Triton X-100 in PBS. Prior to addition of the FISH probe, the cells were pre-hybridized in a 2X SSC buffer containing 40% formamide and 10 mg/mL BSA for 30 min. The coverslips were hybridized for 2 h in 2X SSC buffer supplemented with 40% formamide, 2 mM vanadyl ribonucleoside, 60 µg mL−1 tRNA, 30 µg mL−1 BSA, and 0.75 µg (CCG)8-Cy3 DNA oligonucleotide probe. The cells were washed twice in 2X SSC containing 50% formamide and then twice in 2X SSC. Following FISH, the coverslips were incubated for 10 min in 2X SSC containing 1 µg mL−1 DAPI and rinsed twice in 2X SSC. The coverslips were then mounted in Pro-Long media and examined using either a simple fluorescence microscope (Leica) or a Leica DM4000 B confocal microscope.
Supplementary Material
ACKNOWLEDGMENT
We thank R. Parkesh for technical assistance. This work was funded by the National Institutes of Health (3R01GM079235-02S1 and 1R01GM079235-01A2 to MDD), INSERM AVENIR funding (to NCB), ANRGENOPAT grant P007942 (to NCB) and by The Scripps Research Institute. MDD is a Camille & Henry Dreyfus New Faculty Awardee, a Camille & Henry Dreyfus Teacher-Scholar, and a Research Corporation Cottrell Scholar.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Atkins JF, Gesteland RF, Cech TR, editors. RNA Worlds: From Life’s Origins to Diversity in Gene Regulation. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- 2.Cooper TA, Wan LL, Dreyfuss G. RNA and Disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 4.Wilton SD, Fletcher S. RNA splicing manipulation: strategies to modify gene expression for a variety of therapeutic outcomes. Curr. Gene Ther. 2005;5:467–483. doi: 10.2174/156652305774329249. [DOI] [PubMed] [Google Scholar]
- 5.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 6.Mykowska A, Sobczak K, Wojciechowska M, Kozlowski P, Krzyzosiak WJ. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 2011;39:8938–8951. doi: 10.1093/nar/gkr608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat. Cell. Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 8.Bates G. Huntingtin aggregation and toxicity in Huntington's disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- 9.Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Hum. Mol. Genet. 2000;9:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 10.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 12.Kumar A, Parkesh R, Sznajder LJ, Childs-Disney JL, Sobczak K, Disney MD. Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chem. Biol. 2012;7:496–505. doi: 10.1021/cb200413a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman DE, Thornton CA, Disney MD. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database & chemical similarity searching. J. Am. Chem. Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Childs-Disney JL, Hoskins J, Rzuczek S, Thornton C, Disney MD. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem. Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho J, Rando RR. Specific binding of Hoechst 33258 to site 1 thymidylate synthase mRNA. Nucleic Acids Res. 2000;28:2158–2163. doi: 10.1093/nar/28.10.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bostrom J, Greenwood JR, Gottfries J. Assessing the performance of OMEGA with respect to retrieving bioactive conformations. J. Mol. Graph. Model. 2003;21:449–462. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 19.Grant JA, Gallardo MA, Pickup BT. A fast method of molecular shape comparison. A simple application of a Gaussian description of molecular shape. J. Comput. Chem. 1996;17:1653–1666. [Google Scholar]
- 20.Haigh JA, Pickup BT, Grant JA, Nicholls A. Small molecule shape-fingerprints. J. Chem. Inf. Model. 2005;45:673–684. doi: 10.1021/ci049651v. [DOI] [PubMed] [Google Scholar]
- 21.Mills JE, Dean PM. Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J. Comput. Aided Mol. Des. 1996;10:607–622. doi: 10.1007/BF00134183. [DOI] [PubMed] [Google Scholar]
- 22.Cho J, Hamasaki K, Rando RR. The binding site of a specific aminoglycoside binding RNA molecule. Biochemistry. 1998;37:4985–4992. doi: 10.1021/bi972757h. [DOI] [PubMed] [Google Scholar]
- 23.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tassone F, Hagerman RJ, Loesch DZ, Lachiewicz A, Taylor AK, Hagerman PJ. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am. J. Med. Genet. 2000;94:232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 25.Tassone F, Hagerman RJ, Taylor AK, Hagerman PJ. A majority of fragile X males with methylated, full mutation alleles have significant levels of FMR1 messenger RNA. J. Med. Genet. 2001;38:453–456. doi: 10.1136/jmg.38.7.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, van Unen L, Tassone F, Hoogeveen AT, Hagerman PJ, Mientjes EJ, Oostra BA. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum. Mol. Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- 27.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila . Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 28.Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tassone F, Hagerman RJ, Garcia-Arocena D, Khandjian EW, Greco CM, Hagerman PJ. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J Med Genet. 2004;41:e43. doi: 10.1136/jmg.2003.012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, Hessl D, Becker EJ, Papazian J, Leehey MA, Hagerman RJ, Hagerman PJ. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 31.Sellier C, Hagerman P, Willemsen R, Charlet-Berguerand N. DROSHA/DGCR8 sequestration by expanded CGG repeats leads to global micro-RNA processing alteration in FXTAS patients [abstract]. 12th International Fragile X Conference; Detroit, MI. [Google Scholar]
- 32.Chawla G, Lin CH, Han A, Shiue L, Ares M, Jr, Black DL. Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol. Cell. Biol. 2009;29:201–213. doi: 10.1128/MCB.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaires JB, Ragazzon PA, Garbett NC. A competition dialysis assay for the study of structure-selective ligand binding to nucleic acids. Curr. Protoc. Nucleic Acid Chem. Chapter 8. 2003 doi: 10.1002/0471142700.nc0803s11. Unit 8 3. [DOI] [PubMed] [Google Scholar]
- 34.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 35.McLennan Y, Polussa J, Tassone F, Hagerman R. Fragile x syndrome. Curr. Genomics. 2011;12:216–224. doi: 10.2174/138920211795677886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 37.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 39.Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J. Am. Chem. Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 40.Tran T, Disney MD. Two-dimensional combinatorial screening of a bacterial rRNA A-site-like motif library: defining privileged asymmetric internal loops that bind aminoglycosides. Biochemistry. 2010;49:1833–1842. doi: 10.1021/bi901998m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aminova O, Paul DJ, Childs-Disney JL, Disney MD. Two-dimensional combinatorial screening identifies specific 6'-acylated kanamycin A- and 6'-acylated neamine-RNA hairpin interactions. Biochemistry. 2008;47:12670–12679. doi: 10.1021/bi8012615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran T, Disney MD. Molecular recognition of 6'-N-5-hexynoate kanamycin A and RNA 1x1 internal loops containing CA mismatches. Biochemistry. 2011;50:962–969. doi: 10.1021/bi101724h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem. Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 44.Childs-Disney JL, Disney MD. A simple ligation-based method to increase the information density in sequencing reactions used to deconvolute nucleic acid selections. RNA. 2008;14:390–394. doi: 10.1261/rna.576908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul DJ, Seedhouse SJ, Disney MD. Two-dimensional combinatorial screening and the RNA Privileged Space Predictor program efficiently identify aminoglycoside-RNA hairpin loop interactions. Nucleic Acids Res. 2009;37:5894–5907. doi: 10.1093/nar/gkp594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velagapudi SP, Seedhouse SJ, Disney MD. Structure-activity relationships through sequencing (StARTS) defines optimal and suboptimal RNA motif targets for small molecules. Angew. Chem. Int. Ed. Engl. 2010;49:3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velagapudi SP, Seedhouse SJ, French J, Disney MD. Defining the RNA internal loops preferred by benzimidazole derivatives via 2D combinatorial screening and computational analysis. J. Am. Chem. Soc. 2011;133:10111–10118. doi: 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Rando RR. Specific binding of aminoglycoside antibiotics to RNA. Chem. Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 49.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.