Abstract
The generation of highly curved membranes is essential to cell growth, division and movement. Recent research in the field is focused to answer questions related to the consequences of changes in the topology of the membrane once it is created, broadly termed as membrane curvature sensing. Most probes that are used to study curvature sensing are intact membrane active proteins like DP1/Yop1p, ArfGAP1, BAR domains, and Synaptotagmin-I (Syt1). Taking a cue from nature, we created the cyclic peptide C2BL3C based on the membrane penetration C2B loop 3 of Syt1 via ‘Click’ chemistry. Using a combination of spectroscopic techniques, we investigated the peptide-lipid interactions of this peptide with synthetic phospholipid vesicles and exosomes from rat blood plasma. We found that the macrocycle peptide probe was selective for lipid vesicles with highly curved surfaces (d <100 nm). These results suggested that C2BL3C functions as a selective detector of highly curved phospholipid bilayers.
Membrane remodeling and vesiculation play pivotal roles in cell growth, division, and movement (1). Previous works have demonstrated that dynamic modulation of membrane curvature can be brought about by changes in lipid composition, scaffolding proteins, and insertion of protein regions into membranes (2–6). Proteins that were shown to bind, bend, and stabilize the plasma membrane include the endoplasmic reticulum associated DP1/Yop1p protein (2), Golgi-associated protein ArfGAP1 lipid packing sensor (ALPS) (5), Bin–Amphiphysin–Rvs (BAR) domain of amphiphysin (4), and C2B domain of Synaptotagmin-I (7). Nonetheless, there are few examples of using small peptides to selectively recognize highly curved membrane surfaces. Our goal is to develop peptides that bind to lipid vesicles with d <100 nm. Such peptides will have advantages over protein-based curvature sensors including stability, availability, and modifiability. We anticipate that these peptides might find applications as probes for detecting membranes with highly curved surfaces.
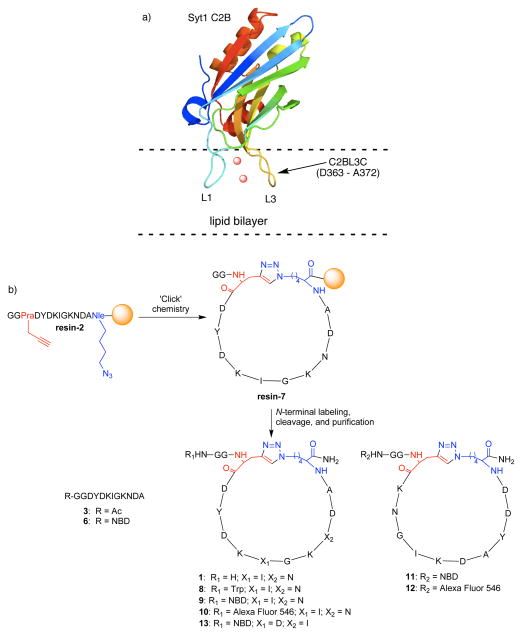
We chose the 10-residue membrane-penetrating loop 3 of the cytoplasmic C2B domain of the transmembrane protein Synaptotagmin-I (Syt1; pdb: 1uow) as the starting point (8). As a component of the Soluble N-ethylmaleimide-sensitive factor Attachment protein Receptor (SNARE) complex, Syt1 is believed to mediate calcium-dependent regulation of membrane trafficking and fusion (7). Chapman and co-workers previously found that loops 1 and 3 of Syt1 C2B domain insert into membrane bilayers at a depth of 2.14 ± 0.66 nm, which is suggested to be key in its physiological function (7). The ability of membrane binding and insertion of Syt1 C2 domains is facilitated by a strong attraction with the negatively charged phospholipid head groups upon the formation of a Ca2+-complex that includes the highly conserved basic residue of loop 3 (Lys366 in pdb: 1uow) (9). Furthermore, Syt1 prefers highly curved lipid vesicles (d = 105 nm) to larger vesicles (d = 252 nm) by sensing membrane curvature (7). Fluorescence quenching studies on C2B I367C mutant labeled at the C367 position showed that C2B loop 3 penetrates the membrane bilayer, demonstrating its phospholipid binding ability (10). Based on the observation that C2B loop 3 significantly contributes to membrane binding (9) while the rest of the protein provides a rigid scaffold (Fig. 1a), we designed a 12-residue cyclic peptide C2BL3C 1 by side chain-mediated head-to-tail cyclization of the loop 3 region (a.a. 363–372) of Syt1 C2B domain by ‘Click’ chemistry. A flexible -GG- dipeptide linker was appended to the N-terminus as a handle for functionalization.
Figure 1.
Design and synthesis of C2BL3C as a structural and functional mimic of Syt1 C2B loop 3. a) Pictorial rendition of Syt1 (pdb: 1uow) depicting the C2B loops 1 and 3 inserting in the lipid bilayer. Binding of Ca2+ (red spheres) to loops 1 and 3 facilitates Syt1 to adopt an active conformation to bind to membranes and sense curvature. b) Solid phase synthesis of designed peptides and their conjugates using Rink amide resin. Ac = acetyl, NBD = 7-nitrobenzo-2-oxa-1,3-diazole, Trp = tryptophan.
Peptide cyclization has been shown to increase binding to intended targets (11), which is mainly attributed to constrained conformation and backbone rigidification that lowers the entropic cost of binding. Cyclization of a peptide can be achieved through lactam formation (12), lactonization (13), ring closing metathesis (14), and disulfide bond formation (15). We chose ‘Click’ chemistry developed by Sharpless and co-workers (16) to achieve orthogonal, site-specific cyclization between azide- and alkyne-functionalized residues. Several groups have previously reported on adopting ‘Click’ chemistry to achieve peptide macrocyclization on solid support, which resulted in the preparation of i to i+5 (17), i to i+6 (18), i to i+8 (19), and i to i+10 (20) cyclic peptides.
We synthesized 1 from the resin-bound linear precursor GGPraDYDKIGKNDANle(ε-N3)(C2BL3L, resin-2), where Pra is L-propargylglycine and Nle(ε-N3) is ε-azido norleucine. Cyclization of resin-2 by ‘Click’ chemistry was done at positions i and i+11 (Fig. 1b) to give the cyclic peptide triazole resin-7. Progress of the reaction was monitored by taking small aliquots of the resin, performing a peptide cleavage, and analyzing by FT-IR spectroscopy to monitor the disappearance of the azide band at 2300 cm−1, which indicated the completion of the reaction. We found the following solid phase ‘Click’ chemistry conditions to be optimum: (1) low loading capacity resin of <0.2 mmol g−1 that prevented potential cross-linking and cyclodimerization (20); (2) use of [(CH3CN)4Cu]PF6 as the source of cuprous ion; and (3) 93:7 NMP/H2O that kept the sodium ascorbate in solution and ensured that the polystyrene solid support remained swollen. Analyses and comparisons of reversed phase HPLC chromatograms and 1H NMR and FT-IR spectra confirmed the successful synthesis of 1 (Supplementary Fig. S19). Portions of resin-bound cyclic peptide 7 were taken and N-terminally labeled with Trp or 7-nitrobenzo-2-oxa-1,3-diazole (NBD) to produce C2BL3C-Trp 8 and C2BL3C-NBD 9. For comparison, we prepared NBD-labeled linear peptide corresponding to the native C2B loop 3 region and an NBD-labeled scrambled analogue of 9, called C2BL3C-S-NBD (6 and 11, respectively). We also prepared a mutant of peptide 9 as an additional control, C2BL3C-I8D N11I-NBD 13, where the point mutations correspond to the residues found in canonical EF-hand calcium-binding loops (21).
An established fluorescence enhancement assay (5) was followed with slight modifications to investigate the lipid vesicle binding property of the fluorophore conjugates of 1, peptides 8 and 9, and their potential to differentiate lipid vesicles of different sizes. An increase in the observed fluorescence intensity and a blue shifted λem maximum are directly correlated to a change in the environment surrounding the peptide, i.e. from the polar aqueous solvent to the hydrophobic lipid vesicle, indicating a peptide-lipid interaction. Lipid vesicles composed of 8:2 palmitoyl oleoyl phosphatidylcholine (POPC) and palmitoyl oleoyl phosphatidylserine (POPS) were used to represent the approximate phospholipid composition of cellular membranes (22). These vesicles were prepared by extruding a suspension of multilamellar lipid vesicles in PBS buffer (pH = 7.4) through polycarbonate membranes. The diameters of these lipid vesicles were measured using dynamic light scattering (DLS) that was calibrated with 50 nm polystyrene beads. We used Syt1 as a positive control based on previous studies that it preferentially binds to highly curved lipid vesicles (7). We used peptides 6, 11, and 13 as negative controls. An optimal concentration of 0.5 μM was selected for all tested peptides (Supplementary Fig. S21).
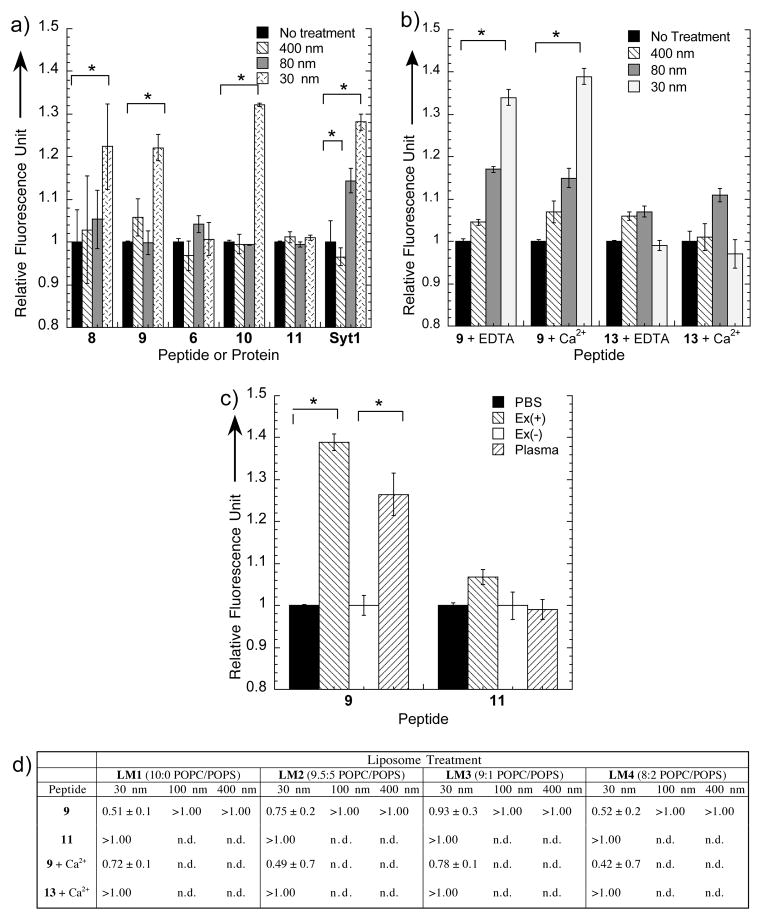
Lipid vesicles were prepared by extrusion through membranes (23) with pore sizes of 30, 80, and 400 nm to cover a wide range of curvatures. This process yielded vesicles with average diameters of 55 ± 6, 115 ± 6, and 511 ± 54 nm, respectively, the smallest representing vesicles with the highest curvature (Supplementary Fig. S20). As shown in Figure 2a, peptides 8 and 9 bound to the lipid surface with greatest curvature (d = 55 ± 6 nm) and with binding behaviors that are independent from the nature of the fluorescent tag. The linear peptide 6 and scrambled cyclic peptide 11 do not interact with lipid vesicles of any size (Fig. 2a), demonstrating that both cyclization and peptide sequence are essential for vesicle binding. The binding profiles to highly curved vesicles of 8 and 9 are comparable, indicating that the fluorescence enhancement is not dependent on different fluorophores. This fluorescence enhancement is comparable to established curvature-sensing proteins: Syt1 (1.28 ± 0.02 RFU with 30 nm vesicles, Fig. 2a) and the ALPS motif-bearing Golgi-microtubule-associated protein-2101–38 and nuclear pore complex protein-133245–267 (~1.25 RFU with 29 nm liposomes) (5). Last, unlike Syt1 (7, 24), the lipid vesicle binding by C2BL3C does not need Ca2+ as shown in Fig. 2b, suggesting that rigidification by the covalent constraints rendered the cyclic peptide C2BL3C in an active conformation for curvature detection of phospholipid bilayers. Peptide 13, the I8D N11I mutant of 9, does not bind to 8:2 POPC/POPS of any size in the presence of Ca2+.
Figure 2.
Fluorescence assays probing the interaction of labeled-peptides on synthetic and biologically relevant lipid vesicles. a) Graph for peptide-liposome interactions of cyclic peptide-fluorophore conjugates and controls using 8:2 POPC/POPS vesicles extruded through 30, 80, and 400 nm pore membranes: C2BL3C-Trp 8, C2BL3C-NBD 9, linear C2B-NBD 6, C2BL3C-AF 10, C2BL3C-S-NBD 11, Synaptotagmin-I Syt1. b) Graph for peptide-liposome interactions of C2BL3C-NBD 9 and C2BL3C-I8D N11I-NBD with and without Ca2+ 13 using 8:2 POPC/POPS vesicles extruded through 30, 80, and 400 nm pore membranes. EDTA: 0.2 mM ethylenediaminetetraacetate, disodium salt; Ca2+: 1 mM CaCl2. c) Binding properties of C2BL3C-NBD 9 and C2BL3C-S-NBD 11 with exosomes in Ex(+) and complete blood plasma. Ex(+) = isolated exosomes. Ex(−) = exosome-depleted plasma supernatant and PBS = phosphate buffered saline, which both refer to solvents for peptides with no treatment. Error bars represent SEM (n = 3). *p <0.05 versus negative control. d) Dissociation constant (Kd) values, in mM, of C2BL3C-NBD 9, C2BL3C-S-NBD 11, and C2BL3C I8D N11I-NBD 13 with liposomes of varying POPC/POPS compositions (mol/mol) extruded through 30, 100 and 400 nm pore membranes are indicated in the table. Lower values indicate stronger binding. LM = liposome model; Ca2+: 1 mM CaCl2; n.d. = not determined.
Fluorescence anisotropy was performed to measure the binding affinity of the peptides to liposomes. Peptides were titrated with liposomes of varying compositions of 10:0, 9.5:5, 9:1, 8:2 POPC/POPS, hereinafter called liposome model (LM) 1, 2, 3, and 4. The peptide partitions between the hydrophobic lipid bilayer and the aqueous solvent (25). Since it is unlikely that the peptide forms a 1:1 complex with the lipid vesicle, the molar partition coefficient is often used. By definition, the Kd, apparent dissociation constant, is the reciprocal of the molar partition coefficient (26) described as the lipid concentration where 50% of the peptide is bound. As shown in Fig. 2d, the affinity of peptide 9 to lipid vesicles extruded through 30 nm pores is not affected by lipid composition, with Kd values of 0.51 ± 0.1, 0.75 ± 0.2, 0.93 ± 0.3, and 0.55 ± 0.2 mM for LM1, LM2, LM3, and LM4, respectively. Treatment with Ca2+ does not significantly affect the affinity of 9 for 30 nm lipid vesicles (Fig. 2d), consistent with our findings in the fluorescence enhancement assay. By contrast, peptides 11 and 13 did not show significant affinity to 30 nm lipid vesicles of any lipid composition tested. Peptide 9 showed fairly weak binding (> 1.00 mM) for 100 and 400 nm lipid vesicles. The observed binding of 9 for 30 nm vesicles is weaker relative to the property of Syt1 (Kd = 0.151 ± 0.6 and 0.263 ± 0.18 mM for 105 and 252 nm vesicles, respectively) (7). This is not surprising because peptide 9 was designed from only one of the components of the curvature sensing protein Syt1. Nonetheless, these findings show that 9 has the ability to distinguish lipid vesicles of different curvatures regardless of the liposome composition. Our hypothesis in the lack of effective electrostatic interaction as driving force in the peptide 9-liposome interaction, as shown in Fig. 2d, suggests that high curvature is a prerequisite for the observed size differentiation. Peptide 9 could distinguish lipid vesicle sizes perhaps due to its ability to recognize lipid packing defects in highly curved lipid bilayers, which is a consequence of the mismatch between the phospholipid geometry and the curvature of the bilayer (5).
To verify that the origin of the differential peptide interaction towards lipid vesicles is due to the innate property of the peptide and is not due to the fluorophores, we carried out 1H NMR spectrometry investigations to probe the interaction of peptide 1. We chose vesicles with d = 55 ± 6 nm for the NMR experiments due to the fact that these showed the highest increase in fluorescence intensity upon interacting with peptides 8 and 9. Visual inspection of the aromatic and amide region of the 1H NMR spectra showed that 1 treated with the lipid vesicles exhibited decreased peak intensity when compared to untreated 1 (Supplementary Fig. S24), indicating that the unlabeled peptide also interacts with liposomes (27, 28). By contrast, the spectra of liposome-treated, linear peptides 3 and 6 consistently showed significantly weaker binding in both fluorescence and NMR experiments (Supplementary Fig. S24). These findings further demonstrated that both peptide sequence and cyclization, but not the fluorophores, are crucial for the observed curvature-detecting property of C2BL3C.
Nanoparticle tracking analysis (NTA) was used to observe the individual lipid vesicle particles (29). NTA records videos in scatter (fluorescence independent) and fluorescence modes, the latter providing speciation to confirm the binding of fluorophore-labeled molecules on target particles. The NTA software analyzes the video and measures the size of each particle from direct observations of diffusion in a liquid medium, independent of particle refractive index or density (29). Although NTA can resolve and simultaneously measure a wide range of particle sizes at the same time, there is an inherent limitation of measuring a stochastic process (in this case Brownian motion) in a finite sampling time (limited by time at which each particle is tracked), which may result to lesser peak resolution quality. Nonetheless, the ability to observe and track nano-sized vesicles using our designed peptide will demonstrate the proof-of-concept of size selectivity.
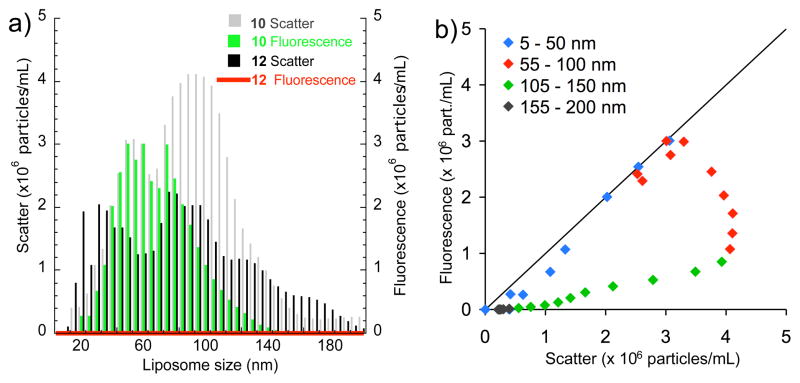
We labeled C2BL3C and the cyclic scrambled peptide C2BL3C-S with Alexa Fluor 546 (required for NTA detection) to yield C2BL3C-AF 10 and C2BL3C-S-AF 12, respectively. Heterogeneous lipid vesicles composed of 8:2 POPC/POPS were prepared and treated with peptides 10 and 12. Videos were recorded under light scatter mode using 532 nm laser and then under fluorescence mode using a 560 nm filter to investigate if the particles are tagged by the peptides (see Supplementary Videos for representative results). Figure 3a shows the NTA results in light scatter for heterogeneous lipid vesicles treated with 10. The total vesicle count was 3.50 × 108 mL−1, with majority size ranging from 35–140 nm. Under fluorescence mode, the total particle count was 2.00 × 108 mL−1 and the vesicle sizes that were clearly tagged by peptide 10 are in the range of 30–95 nm (Fig. 3a). The parametric plot (Fig. 3b) confirms that 10 preferentially tagged liposomes with a size range between 30 and 75 nm. In contrast, we did not observe peptide 10 to label large lipid vesicles, while peptide 12 did not tag any lipid vesicle size (Fig. 3a).
Figure 3.
Nanoparticle tracking analysis (NTA). a) Heterogeneous lipid vesicles composed of 8:2 POPC/POPS and treated with peptide 10 or 12 were studied by NTA. Green bars indicate the particle sizes that are preferred and labeled by peptide 10 and are visible under fluorescence mode. Compared to scatter mode, smaller particles (d < 75 nm) are selectively labeled by peptide 10. b) Parametric plot showing the liposome size range that was preferentially labeled by peptide 10. Liposomes that scatter and fluoresce are represented by points that are on or close to the diagonal.
To find out if our findings from synthetic liposomes would translate to biological specimens, we also tested plasma samples from rats that underwent inescapable tailshock stress as a model of exosome release (30). Exosomes are nano-sized particles with diameters of ≤100 nm shed by various cell types, therefore providing a good biological model for testing our curvature-detecting peptides (31). The blood plasma was fractionated to separate the exosomes, hereinafter called Ex(+), from the plasma supernatant, hereinafter called Ex(−). The size range of exosomes is known (32) and exosomes are characterized by the presence of a membrane-bound tetraspanin called CD63 (33). The transmission electron microscopy images of Ex(+) showed vesicles with d<150 nm with the majority of the particles having d<100 nm while the blood plasma showed a more heterogeneous mixture of a wide range of sizes from different kinds of particles (Supplementary Fig. S29). The exosomes in Ex(+) were confirmed by enzyme-linked immunosorbent assay (Supplementary Fig. S28). We tested the exosome-detecting ability of C2BL3C by fluorescence enhancement using either PBS buffer or the exosome depleted plasma supernatant, Ex(−), as controls for untreated peptide. Figure 2b showed that the Ex(+) treated peptide renders a higher fluorescence intensity at 1.39 ± 0.02 RFU than the untreated peptide, which demonstrated that the synthetic phospholipid binding property of 9 is translated to the detection of exosomes. We also tested if we could use peptide 9 to detect exosomes in blood plasma, and found that plasma-treated 9 showed a fluorescence intensity that is remarkably higher than the untreated 9 (1.27 ± 0.05 RFU). In contrast, scrambled peptide 11 showed negligible binding. This demonstrates that the complex blood plasma matrix does not compromise the exosome-detecting ability of peptide 9.
In this report we demonstrated the synthesis and characterization of a cyclic peptide probe (1) designed from Syt1 C2B loop 3 that selectively binds highly curved phospholipid bilayers. Our efforts led to the successful preparation and functionalization of this 12-residue macrocycle that, to the best of our knowledge, is the largest monomeric cyclic peptide to date prepared on solid support by ‘Click’ chemistry. C2BL3C favorably bound to nano-sized synthetic lipid vesicles and blood-borne exosomes with highly curved surfaces. This peptide selects curvature perhaps due to its ability to insert into lipid packing defects in highly curved lipid bilayers. The analogues of 9, peptides 11 and 13, does not bind to lipid vesicles, which shows the sequence specificity of lipid vesicle recognition. Furthermore, NTA experiments showed that the Alexa Fluor 546 fluorophore conjugate 10, but not the scrambled control 12, favorably bound to highly curved surfaces of synthetic phospholipid bilayers with d≤100 nm, which correlates with our observations in fluorescence and NMR assays. In summary, our results showed that C2BL3C possesses curvature-detecting properties, thereby providing a potential tool to study membrane morphology and peptide-lipid interactions.
METHODS
Solid Phase Peptide Syntheses
Peptide resin-2 was synthesized using Rink amide resin and microwave-assisted solid phase synthesizer following the standard Fmoc chemistry. To make peptide resin-7, linear peptide precursor resin-2 (0.05 mmol) was swelled in 2.8 mL NMP for 5 min. A solution of Cu(I) tetrakis(acetonitrile) hexafluorophosphate (18.6 mg, 0.05 mmol) and Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, 26.5 mg, 0.05 mmol) in 0.5 mL NMP was added to the resin, followed by sodium ascorbate (20 mg, 0.1 mmol in 250 μL H2O) and 2,6-lutidine (10 μL, 0.1 mmol). Portions of the resin-7 were taken and labeled at the N-terminus with Trp and 4-Chloro-7-nitrobenzo-2-oxa-1,3-diazole (NBD) to yield 8 and 9, respectively. Peptide 6 was prepared following the same solid phase synthesis and labeling as described above. Peptides 12 and 13 were prepared following the same solid phase synthesis, cyclization, and labeling as described above to yield C2BL3C-S-NBD (12) and C2BL3c-I8D N11I-NBD (13). Resin-bound peptide 7 and its scrambled analogue were labeled with Alexa Fluor 546™ to yield C2BL3C-AF (10) and C2BL3C-S-AF (12), respectively.
Lipid Vesicle Preparation
Lipid vesicles composed of POPC and 9.5:5, 9:1, and 8:2 mixture (mol/mol) of POPC and POPS were prepared as described previously (23).
Fluorescence Assays
Fluorescence enhancement measurements were performed by measuring the fluorescence emission spectra of 300 μL solutions of fluorophore-peptide conjugates (0.5 μM) in PBS, pH 7.4, at room temperature with λex of 280 and 470 nm and λem scan of 300–500 and 500–650 nm for Trp and NBD, respectively. Separate peptide-conjugate solutions were also prepared in the presence of 0.5 mM liposomes that were incubated at 4 °C overnight, and their fluorescence emission spectra recorded under identical conditions as above. For Ca2+-dependence assays, the samples were prepared in HEPES buffer, pH 7.4, and treated with CaCl2 before fluorescence measurements. As a positive control, 0.20 μM of rat Syt1 (G374, residues 96–421) was treated with Ca2+ and liposomes that were extruded through 30, 80, and 400 nm pores using λex of 280 nm to excite the tyrosine in the membrane insertion loop 3 (7). Similar experiments were also conducted using Ex(+) and blood plasma as sources of lipid vesicles, and PBS, pH 7.4, and Ex(−) as solvents for untreated peptides. For the anisotropy titrations, the NBD-labeled peptides (1 μM) were individually titrated with 2 mM liposomes. Anisotropy was monitored with the excitation wavelength set to 480 nm and the emission wavelength set to 545 nm. Titrations were made to produce the [Total Lipid] on the x-axis versus the anisotropy values on the y-axis. The plots were fitted as previously described (24).
Isolation of Exosomes
The care and treatment of the rats were in accordance with protocols approved by the University of Colorado Institutional Animal Care and Use Committee. Exosomes were isolated from rat blood plasma according to the procedure provided with the ExoQuick™ kit. This process yielded a supernatant that was aspirated off and labeled as the exosome depleted fraction, called Ex(−). The remaining pellet that contained the exosomes were resuspended PBS, pH 7.4, and was called Ex(+).
Nanoparticle Tracking Analyses
An aliquot of 150 μL heterogeneous lipid vesicles were taken and diluted with PBS, pH 7.4 to give a particle count between 1 – 8 × 108 mL−1, following a previous work on nano-sized vesicles (29). The vesicles were treated with 10 nM of peptides 10 and 12, incubated, and diluted to a final volume of 300 μL. Nanoparticle Tracking Analyses (NTA) were performed using NanoSight LM10-HS equipped with a 532 nm laser at scatter and fluorescence modes (filter = 560 nm). The instrument was calibrated using commercially available 50 nm polystyrene beads (Polysciences, Warrington, Pennsylvania).
Supplementary Material
Acknowledgments
H.Y. thanks the Howard Hughes Medical Institute (HHMI) Collaborative Innovation Award. E. R. C. thanks the National Institute of Health (MH061876) for support. M. F. thanks the National Science Foundation (NSF IOS 1022451) for financial support. E.R.C. is an Investigator of the Howard Hughes Medical Institute. L.A.M. has been supported by a Ruth L. Kirschstein National Research Service Award (F31 CA165349) and a NIH training grant (T32 GM008759). We thank M. Stowell (CU Boulder), D. Rees and R. Phillips (Caltech) for their invaluable comments; R. Shoemaker (CU Boulder NMR Facility) for his help in NMR experiments; and T. Maslanik (CU Boulder) for technical support.
Footnotes
Supporting Information Available. Further experimental details, 1H NMR, MALDI-TOF-MS, and infrared spectra, DLS data, NTA videos, and exosome isolation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 2.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerberg J, McLaughlin S. Membrane curvature: How BAR domains bend bilayers. Curr Biol. 2004;14:R250–R252. doi: 10.1016/j.cub.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 5.Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 6.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 7.Hui EF, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca2+-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Y, Sequeira SM, Malinina L, Tereshko V, Sollner TH, Patel DJ. Crystallographic identification of Ca2+ and Sr2+ coordination sites in synaptotagmin I C2B domain. Protein Sci. 2004;13:2665–2672. doi: 10.1110/ps.04832604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paddock BE, Striegel AR, Hui E, Chapman ER, Reist NE. Ca2+-dependent, phospholipid-binding residues of synaptotagmin are critical for excitation-secretion coupling in vivo. J Neurosci. 2008;28:7458–7466. doi: 10.1523/JNEUROSCI.0197-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai J, Wang P, Chapman ER. C2A activates a cryptic Ca2 -triggered membrane penetration activity within the C2B domain of Synaptotagmin I. Proc Natl Acad Sci U S A. 2002;99:1665–1670. doi: 10.1073/pnas.032541099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilon C, Halle D, Chorev M, Selincer Z, Byk G. Backbone cyclization: A new method for conferring conformational constraint on peptides. Biopolymers. 1991;31:745–750. doi: 10.1002/bip.360310619. [DOI] [PubMed] [Google Scholar]
- 12.Cudic M, Wade JD, Otvos L. Convenient synthesis of a head-to-tail cyclic peptide containing an expanded ring. Tetrahedron Lett. 2000;41:4527–4531. [Google Scholar]
- 13.Meutermans WDF, Golding SW, Bourne GT, Miranda LP, Dooley MJ, Alewood PF, Smythe ML. Synthesis of difficult cyclic peptides by inclusion of a novel photolabile auxiliary in a ring contraction strategy. J Am Chem Soc. 1999;121:9790–9796. [Google Scholar]
- 14.Miller SJ, Grubbs RH. Synthesis of conformationally restricted amino acids and peptides employing olefin metathesis. J Am Chem Soc. 1995;117:5855–5856. [Google Scholar]
- 15.Ranganathan D, Haridas V, Nagaraj R, Karle IL. Double-helical cyclic peptides: Design, synthesis, and crystal structure of figure-eight mirror-image conformers of adamantane-constrained cystine-containing cyclic peptide cyclo (Adm-Cyst)3. J Org Chem. 2000;65:4415–4422. doi: 10.1021/jo0003807. [DOI] [PubMed] [Google Scholar]
- 16.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Roice M, Johannsen I, Meldal M. High capacity poly(ethylene glycol) based amino polymers for peptide and organic synthesis. QSAR Comb Sci. 2004;23:662–673. [Google Scholar]
- 18.Turner RA, Oliver AG, Lokey RS. Click chemistry as a macrocyclization tool in the solid-phase synthesis of small cyclic peptides. Org Lett. 2007;9:5011–5014. doi: 10.1021/ol702228u. [DOI] [PubMed] [Google Scholar]
- 19.Goncalves V, Gautier B, Regazzetti A, Coric P, Bouaziz S, Garbay C, Vidal M, Inguimbert N. On-resin cyclization of peptide ligands of the Vascular Endothelial Growth Factor Receptor 1 by copper(I)-catalyzed 1,3-dipolar azide-alkyne cycloaddition. Bioorg Med Chem Lett. 2007;17:5590–5594. doi: 10.1016/j.bmcl.2007.07.087. [DOI] [PubMed] [Google Scholar]
- 20.Punna S, Kuzelka J, Wang Q, Finn MG. Head-to-tail peptide cyclodimerization by copper-catalyzed azide-alkyne cycloaddition. Angew Chem-Int Edit. 2005;44:2215–2220. doi: 10.1002/anie.200461656. [DOI] [PubMed] [Google Scholar]
- 21.Kirberger M, Wang X, Deng H, Yang W, Chen G, Yang JJ. Statistical analysis of structural characteristics of protein Ca2+-binding sites. Biol Inorg Chem. 2008;13:1169–1181. doi: 10.1007/s00775-008-0402-7. [DOI] [PubMed] [Google Scholar]
- 22.Rothman JE, Lenard J. Membrane asymmetry. Science. 1977;195:743–753. doi: 10.1126/science.402030. [DOI] [PubMed] [Google Scholar]
- 23.Morton LA, Saludes JP, Yin H. Constant Pressure-Controlled Extrusion Method for the Preparation of Nano-sized Liposomes. J Vis Exp. 2012;64:e4151. doi: 10.3791/4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007:316. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 25.Kim JY, Blackshear PJ, Johnson JD, McLaughlin S. Phosphorylation reverses the membrane association of peptides that correspond to the basic domains of MARCKS and neuromodulin. Biophys J. 1994;67:227–237. doi: 10.1016/S0006-3495(94)80473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rusu L, Gambhir A, McLaughlin S, Radler J. Fluorescence correlation spectroscopy studies of peptide and protein binding to phospholipid vesicles. Biophys J. 2004;87:1044–1053. doi: 10.1529/biophysj.104.039958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overduin M, Kutateladze T. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 28.Bonechi C, Ristori S, Martini G, Rossi C. Study of bradykinin conformation in the presence of model membrane by nuclear magnetic resonance and molecular modeling. Biochim Biophys Acta. 2009;1788:708–716. doi: 10.1016/j.bbamem.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 29.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJP, Hole P, Carr B, Redman CWG, Harris AL, Dobson PJ, Harrison P, Sargent IL. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine: NBM. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson JD, Fleshner M. Releasing signals, secretory pathways, and immune function of endogenous extracellular heat shock protein 72. J Leukoc Biol. 2006;79:425–434. doi: 10.1189/jlb.0905523. [DOI] [PubMed] [Google Scholar]
- 31.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 33.Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. 2009;315:1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.