SUMMARY
Gene-distal enhancers are critical for tissue-specific gene expression, but their genomic determinants within a specific lineage at different stages of development are unknown. Here we profile chromatin state maps, transcription factor occupancy, and gene expression profiles during human erythroid development at fetal and adult stages. Comparative analyses of human erythropoiesis identify developmental stage-specific enhancers as primary determinants of stage-specific gene expression programs. We find that erythroid master regulators GATA1 and TAL1 act cooperatively within active enhancers but confer little predictive value for stage specificity. Instead, a set of stage-specific co-regulators collaborates with master regulators and contributes to differential gene expression. We further identify and validate IRF2, IRF6, and MYB as effectors of an adult-stage expression program. Thus, the combinatorial assembly of lineage-specific master regulators and transcriptional co-regulators within developmental stage-specific enhancers determines gene expression programs and temporal regulation of transcriptional networks in a mammalian genome.
INTRODUCTION
Erythropoiesis in mammals occurs in three waves consisting of primitive progenitors in the yolk sac, definitive precursors in the fetal liver and later in the postnatal bone marrow (McGrath and Palis, 2008; Orkin and Zon, 2008). Several transcription factors, such as GATA1 and TAL1 (or SCL), are essential for erythroid development and are recognized as the erythroid ‘master’ regulators (Cantor and Orkin, 2002). These lineage-specifying master regulators, together with other transcription factors and cofactors, act within complexes on chromatin, establish transcriptional networks, and orchestrate differentiation (Kim and Bresnick, 2007). Master regulators of different lineages often cross-antagonize each other’s activity during lineage specification (Graf and Enver, 2009). However, it is less clear how master regulators control programs at different stages of development within the same cell lineage.
A gene regulatory network consists of trans-acting regulators and cis-acting elements within core promoters and gene-distal enhancers whose interaction with each other control tissue- and developmental stage-specific programs (Bulger and Groudine, 2011). Genome-wide studies suggest that enhancers and promoters exhibit distinct chromatin ‘signatures’. The characteristic signature for enhancers consists of monomethylation of histone H3 lysine 4 (H3K4me1), acetylation of histones (H3K9ac and H3K27ac), and binding of the acetyltransferase p300 (Heintzman et al., 2007; Koch et al., 2007; Visel et al., 2009). Studies comparing various lineages indicate that enhancers are associated with highly cell-type-specific histone modifications and strongly correlate to global cell-type-specific programs (Blow et al., 2010; Creyghton et al., 2010; Ernst et al., 2011; Ghisletti et al., 2010; Heintzman et al., 2009; Mikkelsen et al., 2010; Rada-Iglesias et al., 2011). Hence, it has been suggested that enhancers are the primary determinants of cell type specificity in gene expression. However, the majority of studies have employed cells from very different lineages and/or immortalized cell lines. It remains unknown the degree of overlap between cells within a specific lineage at different stages of differentiation or similar cells at different stages of development.
We reasoned that comparative profiling of closely related cell types corresponding to distinct developmental stages should delineate regulatory networks directly related to the associated gene expression programs. Classification of the trans- and cis-regulatory elements that are either shared or stage-specific should clarify their relative importance and prioritize functional candidates. We have focused on an ex vivo maturation system for human fetal and adult erythropoiesis. Primary hematopoietic stem/progenitor cells (HSPCs) can be propagated and induced for erythroid differentiation with a set of defined cytokines ex vivo. This system has been widely used in molecular analysis of erythropoiesis (Migliaccio et al., 2009).
Here, we report the comparative investigation of genome-wide chromatin state maps, TF occupancy, and gene expression profiles from developing red cell precursors at two developmental stages. Distal enhancers, not promoters, are marked with highly stage-specific histone modifications and DNase I hypersensitivity, strongly correlate to stage-specific gene expression changes, and are functionally active in a stage-specific manner. Master regulators GATA1 and TAL1 act cooperatively within active enhancers but have little predictive value for stage-specific enhancer activity. In contrast, a set of stage-specific cofactors and signaling pathways collaborate with these regulators and account for the stage specificity. Two such cofactors, the interferon regulatory factors IRF2 and IRF6, are essential for activation of adult erythroid programs through cooperation with master regulators and cohesin-mediator complexes at distal enhancers.
RESULTS
Ex Vivo Maturation of Primary Human Fetal and Adult Erythroid Progenitors
We employed a serum-free two-phase liquid culture system to expand and differentiate primary fetal or adult stage human CD34+ hematopoietic stem/progenitor cells (HSPCs) ex vivo (Sankaran et al., 2008). In this experimental context, highly enriched populations of stage-matched, differentiating, primary proerythroblasts (ProEs) were generated (Figure 1A). We selected four time points (day 0, CD34+ HSPCs; day 3, 5, and 7, differentiating ProEs) that represented similar stages of differentiation (Figure 1A; Figure S1A,B). Adult CD34+ HSPC-derived ProEs expressed predominantly adult hemoglobin (β-globin). Conversely, fetal ProEs expressed largely fetal hemoglobin (γ-globin) (Figure 1B), indicating that the ex vivo system faithfully recapitulates stage-specificity.
Figure 1. Comparative Genomic Analyses of Human Erythropoiesis.
(A) Fetal and adult CD34+ HSCPs were differentiated into ProEs ex vivo. Cells at matched stages of differentiation were collected for gene expression profiling and ChIP-seq analyses.
(B) Expression of human embryonic (ε-), fetal (γ-), and adult (β-) globin mRNAs. Results are means ± SD of at least three independent experiments.
(C) Scatter plots of gene expression profiling between fetal and adult ProEs (F5 and A5). Dashed blue lines indicate the 1.5-fold differential expression cutoff to define the F5-high (increased expression in F5 relative to A5 ProEs) or A5-high (increased expression in A5 relative to F5 ProEs) genes. The numbers of differentially expressed genes are indicated.
(D) Numbers of differentially expressed genes conditional on changes in expression levels between A5 and F5 ProEs.
(E) The genome-wide distribution of the profiled histone marks and TFs. Total numbers of enriched regions in distal promoters (blue), proximal promoters (red), exons (green), introns (purple), and intergenic regions (light blue) were identified (Experimental Procedures). The graph shows the fraction of enriched regions for each histone mark and TF in fetal and adult ProEs, respectively.
See also Figures S1 and S2, and Tables S1 and S2.
Comparative Profiling of Gene Expression, Chromatin Signatures, and Transcription Factor Occupancy
We determined the mRNA expression profiles in fetal and adult HSPCs and differentiating ProEs by Affymetrix microarray (Figure 1A, Table S1). Upon differentiation of CD34+ HSPCs to day 5 ProEs, 858 and 1137 genes were significantly upregulated (fold change > 1.5, FDR < 0.05) at fetal and adult stages, respectively (Figure S1C). 333 genes were upregulated at both stages (Figure S1D). These genes are involved in heme biosynthesis, iron homeostasis, and erythrocyte differentiation (Figure S1E), suggesting that a common set of gene signatures required for erythroid functions is activated at both stages. More importantly, comparative transcriptome profiling revealed distinct gene expression programs at different stages of erythroid maturation. 1039 and 1291 genes linked to distinct functional annotations were differentially expressed in fetal and adult day 5 ProEs, respectively (Figure 1C; Figure S1F,G). 93.7% of the differentially expressed genes had expression changes less than 4-fold, suggesting that the majority of these genes differed to a modest extent between fetal and adult stages (Figure 1C,D).
To investigate the basis of the distinct programs, we generated genome-wide maps for chromatin state and transcription factor (TF) occupancy by the ChIP-seq method. We profiled 9 histone modifications (H3K4me1/me2/me3, H3K9me3, H3K37me3, H3K36me2/me3, H3K9ac, and H3K27ac) and 6 TFs (GATA1, TAL1, NFE2, CTCF, RAD21, and RNA polymerase II) in fetal and adult day 5 ProEs. GATA1, TAL1, and NFE2 are critical hematopoietic regulators (Orkin and Zon, 2008). CTCF is required for transcriptional insulation (Phillips and Corces, 2009). RAD21 is a component of the cohesin complex and regulates chromatin architecture between enhancers and promoters (Kagey et al., 2010). Most TFs were expressed comparably between fetal and adult ProEs, except that TAL1 and NFE2 were slightly upregulated in adult ProEs (Figure S2A). We identified between 17106 and 71785 binding peaks for histone marks, and between 5664 and 22835 peaks for TFs (Table S2; Experimental Procedures). 95% (55 out of 58) and 97% (56 out of 58) of ChIP-seq peaks were validated by ChIP-qPCR in fetal and adult ProEs, respectively (Figure S2B–H).
The genomic distributions of the profiled histone marks were consistent with their known functions. H3K4me3, which is associated with transcriptional initiation, was distributed primarily near proximal promoters (Figure 1E). H3K4me1/me2, H3K9ac and H3K27ac, which are associated with open chromatin and distal elements, showed diverse distributions across promoters, intragenic, and intergenic regions. H3K36me3, which is associated with transcriptional elongation, was found primarily across gene bodies. Of note, the global distributions of histone marks were highly similar at fetal and adult stages. The distributions of profiled TFs were also comparable (Figure 1E; Table S2).
Comparative Analysis of the β-Globin Gene Cluster
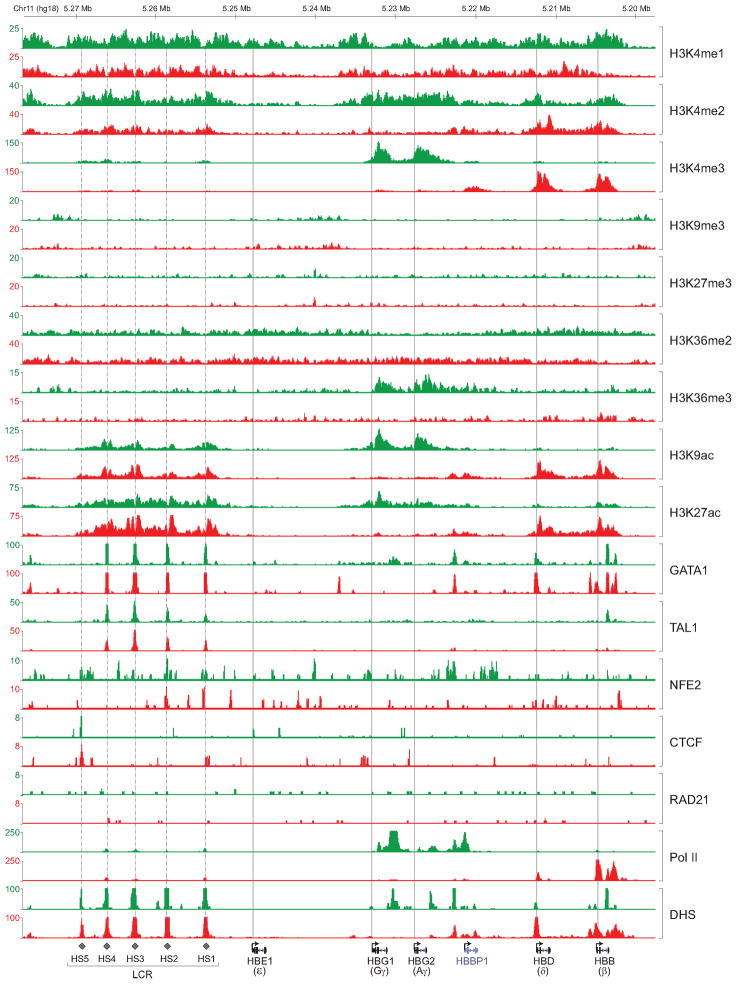
To explore the relationships between cis-regulatory elements, chromatin signatures, and TF occupancy, we focused on the β-globin gene cluster (Figure 2). The β-globin locus contains the embryonic (ε), fetal (Gγ, Aγ) and adult (δ, β) globin genes that are expressed sequentially during development. The switch from fetal (γ) to adult (β) expression provides a paradigm for tissue- and developmental stage-specific transcription (Sankaran et al., 2010). A locus control region (LCR) containing erythroid-specific cis-regulatory elements is located upstream and controls expression of the linked globin genes (Li et al., 2002a).
Figure 2. Chromatin State Maps and TF Occupancy within the Human β-Globin Gene Cluster.
ChIP-seq density plots were generated from raw read data and loaded into the UCSC genome browser as custom tracks. Profiles for histone marks, TFs, and DHS in fetal (green) and adult (red) ProEs are shown at the human β-globin gene cluster. The human β-globin locus is depicted at the bottom containing five β-like globin genes (ε, Gγ, Aγ, δ, and β). Dashed vertical line indicates the location of the upstream DHS (1–5) within the LCR. Solid vertical line indicates the TSS of each globin gene.
The profiled histone marks and TFs showed spatial and temporal density distributions that are qualitatively consistent with their known functions within the β-globin gene cluster (Figure 2). In fetal ProEs, the active fetal globin genes (Gγ and Aγ) were associated strongly with activating histone marks (H3K4me2/me3, H3K9ac, and H3K27ac). Conversely, these marks were enriched only at the adult globin genes (δ and β) in adult ProEs, indicating that the fetal-to-adult globin switch is reflected in a switch of chromatin landscape within the cluster (Figure 2). The upstream LCR was enriched with active chromatin marks in both fetal and adult ProEs. The peaks overlapped with known DNase I hypersensitive sites (DHS) (Figure 2), consistent with function of the LCR at both stages. In contrast with a dynamic chromatin profile within the β-globin locus, the occupancy of several TFs was largely comparable. GATA1 and TAL1 predominantly occupied HS1-4 in both fetal and adult ProEs. The profiles of several TFs were consistent with their known functions within the locus. NFE2 predominantly occupied the HS2 within the LCR and was associated with transcriptional activation (Andrews et al., 1993). CTCF occupied the HS5 at both stages, consistent with its insulator function within the LCR (Bell et al., 2001; Li et al., 2002b).
Promoter Activities during Fetal and Adult Erythropoiesis
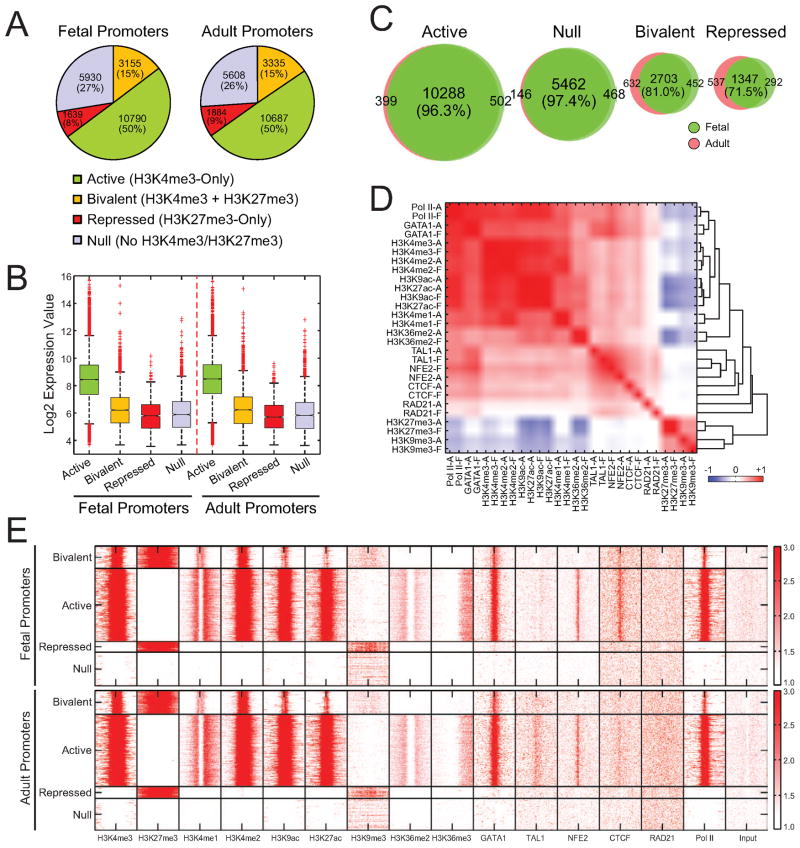
To investigate the regulatory mechanisms for stage-specificity, we first characterized the chromatin features around RefSeq-annotated promoters. We used H3K4me3 and H3K27me3 to categorize promoters into four groups: active (marked by H3K4me3-only), bivalent (marked by both H3K4me3 and H3K27me3) (Bernstein et al., 2006; Boyer et al., 2006), repressed (H3K27me3-only), and null (no H3K4me3/H3K27me3) (Figure 3A, Table S3). The promoter activities highly correlated with mRNA levels and were similar between fetal and adult ProEs (Figure 3A,B). Comparative analyses of the individual promoter categories revealed significant overlaps between fetal and adult cells (Figure 3C), indicating that the promoter activities are largely invariant. Clustering analysis of all ChIP-seq densities at proximal promoters indicated that the activating histone marks (H3K4me1/me2/me3, H3K9ac, and H3K27ac) highly correlated with each other and negatively correlated with repressive marks (H3K9me3 and H3K27me3). Most profiled histone marks and TFs displayed strong colocalization between fetal and adult cells (Figure 3D), indicating that their binding to the proximal promoters is largely invariant. Heatmap analysis revealed that several profiled TFs, such as GATA1, NFE2, and CTCF, were associated predominantly with active promoters at both stages (Figure 3E).
Figure 3. Promoter Activities in Fetal and Adult ProEs.
(A) Promoters were categorized into active, bivalent, repressed, and null promoters. The fraction of each promoter category is shown for both fetal and adult ProEs.
(B) mRNA expression values are shown for each promoter category in fetal and adult ProEs. Boxes show median line and quartiles. Whiskers show the boundary (1.5 times of the inter-quartile range from the first or third quartile) to define outliers (red dots).
(C) Venn diagrams show genome-wide overlaps between fetal and adult ProEs for each promoter category.
(D) Unsupervised hierarchical clustering of all ChIP-seq datasets within the proximal promoter regions (−2kb to +1kb of TSS) between fetal and adult ProEs. Heatmap depicting the Pearson correlation coefficient of ChIP-seq read densities of indicated TFs and histone marks is shown.
(E) ChIP-seq density heatmaps are shown for the profiled histone marks and TFs within each promoter category.
To directly examine the correlation between chromatin signatures, TF occupancy and mRNA expression, we focused on genes differentially expressed between fetal and adult day 5 ProEs. Surprisingly, no obvious changes in chromatin signatures and TF occupancy were found within the promoters of differentially expressed genes (Figure S3A), suggesting that the modest expression changes (Figure 1C,D) are not accompanied by changes in promoter activities. It is important to note that the small subset of genes showing the strongest differences in expression appear to be more likely than the rest to show changes in H3K4me3 in their promoters (data not shown). Similarly, no significant changes in TF occupancy were found in promoters that remained unchanged (‘invariant’ promoters) or changed their activities (‘variant’ promoters) at fetal and adult stages (Figure S3B,C). Thus, chromatin state and TF occupancy at proximal promoters are largely invariant on a global scale during fetal and adult erythropoiesis.
Identification of Developmental Stage-Specific Enhancers
Studies of cells representing different lineages have indicated that gene-distal enhancers are marked with highly cell-type-specific histone modifications and strongly correlate to cell-type-specific gene expression. We next investigated whether enhancers also contribute to stage-specific gene expression programs during fetal and adult erythropoiesis. To identify distal enhancers, we used histone marks H3K4me1, H3K9ac, H3K27ac, and H3K27me3 to distinguish distal elements from proximal promoters (Experimental Procedures). Genome-wide analysis identified 8947 and 11709 active enhancers (that is, marked by H3K4me1, H3K9ac or H3K27ac, DHS, and absence of H3K27me3) in fetal and adult ProEs, respectively. Enhancer regions that overlapped the same chromatin marks in both cells were identified as “common” enhancers (total 4360) and the remainder as “fetal-only” (total 2594) or “adult-only” (total 5730) enhancers (Figure 4A, Table S4). Representative common and developmental stage-specific enhancers are shown (Figure 4B; Figure S4D).
Figure 4. Identification of Developmental Stage-Specific Enhancers.
(A) Active enhancers were identified as those genomic regions harboring H3K4me1, H3K9ac or H3K27ac, DHS, and absence of H3K27me3. Venn diagram shows the overlap between fetal and adult active enhancers.
(B) Representative fetal-only, adult-only, and common enhancers are shown. The putative active enhancers are depicted by shaded lines.
(C) ChIP-seq density heatmaps are shown for the profiled histone marks and TFs within active (fetal-only, adult-only, and common) and bivalent (marked by H3K4me1, H3K9ac or H3K27ac, and H3K27me3) enhancers in both fetal (upper panel) and adult (lower panel) ProEs.
(D) The distribution of fetal-only, adult-only, and common enhancers around F5-high or A5-high genes, compared to all genes.
(E) Predicted enhancers were active in reporter assays in primary erythroid cells. Data are means ± SD of three independent experiments.
(F) 3C analysis within the representative fetal-only (COL4A5), adult-only (IRF2), and common (ZFPM1) enhancers. The interaction frequency between the anchoring point (black bar) and distal fragments (shaded bar) is shown. Results are means ± SD. ChIP-seq density plots for H3K4me1, H3K4me3, H3K27ac, and DNase-seq (DHS) density plots are shown.
Heatmap analysis revealed that most of the profiled TFs associated strongly with active enhancers at both stages (Figure 4C). The binding intensities of the master regulators, such as GATA1 and TAL1, were higher at common enhancers as compared to fetal- or adult-only enhancers (Figure 4C). We also detected preferential association of activating histone marks (H3K4me1/2, H3K9ac, and H3K27ac) and RNA Pol II with common enhancers at both stages. To determine whether the chromatin signatures for distal enhancers correlate with transcriptional activity, we profiled enhancer peak densities around differentially expressed genes (Figure 4D) (Experimental Procedures). Remarkably, the peak densities of fetal-only enhancers were significantly higher at genes upregulated in fetal day 5 ProEs (F5-high genes) compared to adult day 5 (A5)-high genes, and vice versa. Common enhancers associated comparably with both F5- and A5-high genes (Figure 4D). Both common and stage-specific enhancers were associated positively with mRNA levels in F5 and A5 ProEs (Figure S4A), and were highly enriched among genes activated during erythropoiesis (Figure S4B). This analysis also indicated that enhancer peaks were highly enriched in both intragenic regions (TSS to TES) and gene-distal regions, indicating that a significant subset of enhancers is located within gene bodies.
To functionally test whether the distal open chromatin marks identified erythroid cell- and developmental stage-specific enhancers, we performed transient enhancer reporter assays in erythroleukemia K562 cells and stable reporter assays in primary fetal or adult erythroid progenitor cells. Genomic fragments containing putative enhancers markedly enhanced reporter expression in K562 cells (Figure S4C). More importantly, fetal- or adult-only enhancers displayed differential activities consistent with their stage-specificity (Figure 4E). Thus, distal regions that show changes in open chromatin marks during fetal and adult erythropoiesis are likely to be enriched for stage-specific enhancers.
Enhancers often function through direct enhancer-promoter contact by DNA loop formation. We next determined whether the identified enhancers interact with proximal promoters by chromosome conformation capture (Dekker et al., 2002). The frequency of interaction between the IRF2 promoter and a putative downstream enhancer was significantly higher than other tested regions (Figure 4F). Importantly, the interaction frequency increased 2-fold in adult relative to fetal ProEs, consistent with an adult stage-specificity. Conversely, a representative fetal-only enhancer within the COL4A5 gene displayed increased interaction frequency in fetal ProEs. In contrast, a representative common enhancer within the ZFPM1 gene had comparable interaction frequency in both cells (Figure 4F; Figure S4D).
Enhancers Correlate with Differential Gene Expression
To directly assess whether stage-specific enhancers contribute to differential gene expression patterns, we mapped enhancers to target genes. We reasoned that selecting enhancers with the most ‘biased’ (for stage-specific enhancers) or ‘unbiased’ (for common enhancers) activities would illustrate how distal enhancers and the associated trans-acting regulators contribute to the developmental stage-specific gene regulatory networks. To this end, we first normalized the ChIP-seq datasets by ‘MAnorm’ (Shao et al., 2012). In this approach, ChIP-seq datasets from different samples were quantitatively compared after rescaling of ChIP-seq signals by a set of common peaks (Experimental Procedures). We then selected biased stage-specific enhancers by a more stringent threshold requiring a fold change ≥ 2, which represents the ratio of enhancer binding intensities between fetal and adult samples or vice versa. These analyses identified of 967 and 2024 high-confidence (HC) fetal-only and adult-only enhancers, respectively (Figure S5A,B, Table S5). Similarly, we identified 2970 high-confidence common enhancers (fold change < 2). Compared to enhancers from previous overlap analysis (Figure 4A), the high-confidence stage-specific enhancers displayed more distinct stage-specific patterns for histone marks (Figure S5C,D), consistent with their stage specificity. Finally, we mapped enhancers to target genes by two approaches including ‘nearest neighbor gene’ and ‘proximal neighbor genes’ (Figure S6A,B). By comparing the gene expression patterns of enhancer targets, we observed no significant difference between two approaches and choose the ‘nearest neighbor gene’ approach for the remaining analyses (Figure S6A; Table S7).
We then assessed the relationship between enhancer binding and mRNA expression. Strikingly, genes targeted by HC-fetal-only enhancers were significantly enriched for F5-high genes and depleted for A5-high genes (P = 4.8 × 10−22). Genes targeted by HC-adult-only enhancers were significantly enriched for A5-high genes (P = 1.9 × 10−48). In contrast, genes targeted by HC-common enhancers were enriched for both F5- and A5-high genes and had little predictive value (Figure 5A). These analyses demonstrate that the stage-specific enhancers highly correlate with changes in expression of the linked genes. Notably, we found that the greater the expression level of a gene increased in fetal or adult ProEs, the more likely it was to be associated with fetal- or adult-stage-specific enhancers (Figure 5B).
Figure 5. Enhancers Control Developmental Stage-Specific Gene Expression Programs.
(A) Target genes of HC-fetal-only, HC-adult-only, and HC-common enhancers were compared with genes differentially expressed between F5 and A5 ProEs. The enrichment scores represent the fold change of the number of overlapped genes between enhancer target genes and differentially expressed genes compared to the number expected at random using all genes as background. P-values are calculated by Fisher’s exact test to quantify the significance of the relative bias towards F5-high or A5-high genes using all differentially expressed genes as background.
(B) Fractions of genes associated with HC-fetal-only, HC-adult-only, or HC-common enhancers, conditional on changes in expression levels between F5 and A5 ProEs.
(C) Heatmap of DNase-seq intensities within the HC-fetal-only, HC-adult-only, or HC-common enhancers in fetal and adult erythroid progenitors, respectively.
(D) TF motifs associated with HC-fetal-only or HC-adult-only enhancers. Top 10 most enriched motifs from mammalian TRANSFAC and JASPAR databases are shown. The ratios represent the fold change of the frequency to observe motif targets in HC-adult-only enhancers compared to HC-fetal-only enhancers. P-values are calculated by hypergeometric distribution to compare the motif presence between HC-adult-only and HC-fetal-only enhancers.
(E) Expression of IRF2, IRF6, and MYB proteins in F5 and A5 ProEs.
(F,G) IRF2 and IRF6 proteins were depleted by lentiviral shRNAs in adult ProEs, respectively.
(H) Boxplots show mRNA expression changes of A5-high or F5-high genes upon depletion of IRF2, IRF6, or MYB in adult ProEs, respectively. Boxplots are constructed as described in Figure 3B.
(I) Gene set enrichment analysis (GSEA) of A5-high signature genes using the expression array data of shIRF2, shIRF6, or shMYB relative to controls, respectively.
(J) ChIP-seq density plot is shown for IRF2 occupancy within the HC-fetal-only and HC-adult-only enhancers. The average read density in unit of RPKM (number of reads per kilo base-pairs per million mapped reads) is shown for the 4kb region surrounding the enhancer summit.
(K) Venn diagrams show genome-wide overlaps between IRF2 peaks, fetal, and adult enhancers.
(L) Fractions of IRF2-occupied HC-fetal-only, HC-adult-only, and HC-common enhancers are shown, respectively.
(M) Representative IRF2-occupied adult-only (AIG1 and ANKFY1) and common (TCTN3) enhancers. ChIP-seq density plots for H3K4me1, H3K4me3, H3K27ac, GATA1, TAL1, IRF2, and DHS density plots are shown.
See also Figures S5, S6, and S7, and Tables S5 and S7.
Active regulatory elements often contain low nucleosome density and DNase I hypersensitive sites (DHS). We examined whether stage-specific enhancers correlate with DHS at global scale using DNase-seq (Hesselberth et al., 2009). Strikingly, the enhancer activities highly correlate to DNase-seq intensities in a stage-dependent manner (Figure 5C). DNase-seq intensities were markedly increased in the HC-fetal-only enhancers as compared to the HC-adult-only enhancers in fetal erythroid cells, and vice versa. HC-common enhancers displayed comparable levels, suggesting that they are largely invariant (Figure 5C).
We next explored whether enhancer maps might reveal novel trans-regulatory factors involved in the stage-specific gene expression. We first observed that the most enriched TF motifs are recognized by erythroid master regulators (TAL::GATA1 and GATA1) in all three enhancer classes (Figure S6C), suggesting that binding of these regulators does not distinguish and/or predict stage selectivity. To identify TF motifs differentially enriched between the stage-specific enhancers, we enumerated instances of all known TF motifs and ranked them according to their relative enrichment in fetal- or adult-only enhancers. Among the motifs most enriched within the HC-fetal-only enhancers are recognition sites for known regulators of hematopoiesis, including EVI1, GATA2, and GATA1 (Figure 5D). The list of motifs contained other known regulators of cell cycle regulation, proliferation and apoptosis, such as the SRF, MAF, and AP1 families. The most enriched motifs within the HC-adult-only enhancers are recognized by the interferon regulatory factors (IRF2, IRF1, and IRF8), CBF, EGR, and MYB proteins (Figure 5D).
Candidate Regulators of Adult Enhancers
The motif enrichment analyses of developmental stage-specific enhancers identified candidate regulators whose roles in human erythropoiesis remain unexplored. We examined whether these candidate regulators contribute to erythroid enhancer functions. Expression of two of the nine IRF family genes, IRF2 and IRF6, was specifically and progressively activated during adult erythropoiesis (Figure S7A). We confirmed mRNA and protein expression in CD34+ HSPCs and differentiating ProEs (Figure 5E; Figure S7B). Interestingly, studies in knockout mice have implicated IRF2 in aspects of hematopoiesis (Matsuyama et al., 1993). Besides defective lymphoid development, adult Irf2-deficient mice are anemic due to ineffective erythropoiesis (Mizutani et al., 2008). IRF2 can activate or repress gene transcription depending on the cellular context (Harada et al., 1990; Lohoff et al., 2000). IRF6 has been linked to Van der Woude and popliteral pterygium syndromes but its role in hematopoiesis has not been explored (Kondo et al., 2002). The involvement of the proto-oncogene MYB in regulation of erythropoiesis has been established in mice containing null or hypomorphic Myb alleles (Emambokus et al., 2003; Mucenski et al., 1991). Although the mechanism remains elusive, MYB controls erythropoiesis by coordinating cell cycle regulation, differentiation, and globin gene expression (Emambokus et al., 2003; Sankaran et al., 2011). MYB expression was significantly higher in adult relative to fetal ProEs (Figure 5E; Figure S7A,B).
To establish whether these factors also regulate human erythropoiesis, we used loss-of-function assays in primary erythroid progenitors. Upon shRNA-mediated depletion of IRF2 or IRF6 expression, differentiating adult ProEs failed to activate a large portion of A5-high signature genes, whereas F5-high signature genes were slightly derepressed (Figure 5H; Figure S7C,D). Similarly, depletion of MYB expression downregulated A5-high genes. Moreover, loss of IRF2, IRF6 or MYB expression led to downregulation of the signature genes induced during adult erythropoiesis (A0-to-A5-high genes; Figure S7E), whereas the F0-to-F5-high signature genes remained largely unaffected (Figure S7F). Gene set enrichment analysis (GSEA) was used with A5-high signature genes. A marked decrease in expression of A5-high signature genes was notable in the shRNA-treated cells compared with controls, suggesting that these candidate regulators are indispensable for activation of adult-high signature genes (Figure 5I).
To directly assess the function of IRF2 in adult cells, we performed ChIP-seq analysis of IRF2 in adult ProEs. IRF2 is preferentially associated with adult-only enhancers, consistent with its role in adult enhancer functions (Figure 5J,K). IRF2 occupied 11.4% and 13.8% of HC-Adult-Only and HC-Common enhancers, respectively, whereas only 2.8% of HC-Fetal-Only enhancers were occupied (Figure 5L,M). Taken together, these data demonstrate crucial roles of several candidate regulators for the adult stage-specific program, and indicate that trans-regulatory factors can be identified by an integrative approach combining genomic profiling and motif enrichment analysis.
Combinatorial Regulation of Stage-Specific Erythroid Enhancers
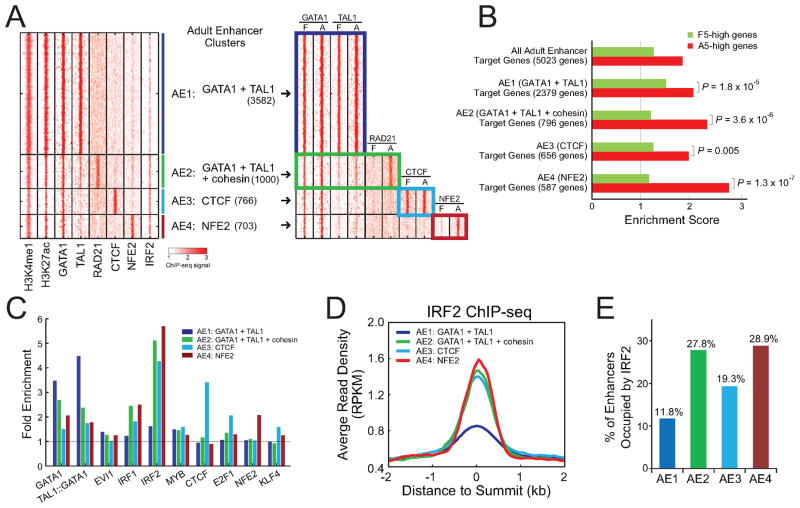
Our comparative investigation of epigenetic state maps and TF occupancy provided a unique system to explore the combinatorial regulation of erythroid enhancers. We performed k-means clustering to classify each enhancer on the basis of TF occupancy (Experimental Procedures). We observed four distinct clusters (AE1 to AE4) within adult enhancers characterized by distinct patterns of TF occupancy (Figure 6A, Table S6). Notably, AE1 was the largest cluster and occupied by the master regulators GATA1 and TAL1. AE2 was co-occupied by GATA1 and TAL1, together with the cohesin subunit RAD21. AE3 and AE4 were predominantly associated with CTCF and NFE2, respectively. To assess the temporal regulation of adult enhancers, we next compared TF occupancy between fetal and adult ProEs. Profiles of TF occupancy within the AE1 and AE3 clusters were largely invariant at both stages. In contrast, RAD21 and NFE2 profiles within the AE2 and AE4 clusters were variable, increasing from little or no binding in fetal ProEs to strong binding in adult ProEs (Figure 6A; Figure S8A). These analyses indicate that the variant enhancer clusters are associated with temporal changes in combinatorial regulatory patterns and may contribute to stage-specific transcription.
Figure 6. Combinatorial Regulation of Enhancer Functions.
(A) K-means clustering of all TF-associated adult enhancers (see Experimental Procedures). The heatmap on the left shows ChIP-seq read density of all TFs used for clustering in adult ProEs. The heatmap on the right shows a side-by-side comparison of ChIP-seq read densities between fetal and adult ProEs within the same enhancer clusters (AE1 to AE4).
(B) Target genes of all adult enhancers and each enhancer cluster were compared with genes differentially expressed between F5 and A5 ProEs. The enrichment scores and P-values were calculated as in Figure 5A.
(C) Enrichment of selected JASPAR motifs in each enhancer cluster. The fold enrichment represents the frequency to observe motif targets in the selected enhancer cluster compared to randomly selected regions from human genome.
(D) ChIP-seq density plot is shown for IRF2 within each enhancer cluster in adult ProEs.
(E) Fraction of IRF2-occupied enhancers within each enhancer cluster is shown.
To test this hypothesis, we mapped each enhancer to its target gene and assessed the relationship between enhancer clustering and mRNA expression. Genes targeted by AE2 (GATA1 + TAL1 + cohesin) and AE4 (NFE2) were significantly enriched for A5-high genes relative to F5-high genes (P = 3.6 × 10−6 and 1.3 × 10−7, respectively) (Figure 6B, Table S7), suggesting that these variant enhancer clusters are strongly associated with adult-specific transcription. In contrast, the invariant clusters (AE1 and AE3) showed less enrichment and had little predictive value for stage selectivity (Figure 6B). To explore other candidate regulators that may contribute to the temporal changes in enhancer activity, we examined the enriched TF motifs within the enhancer clusters. Consistent with the TF occupancy patterns, motif analyses identified strong enrichment of motifs for the corresponding TFs in each enhancer cluster (Figure 6C). Notably, the IRF2 motif was more enriched in variant clusters (AE2 and AE4) than invariant clusters (Figure 6C), consistent with increased IRF2 occupancy within the variant clusters (Figure 6D,E), suggesting that it may contribute to the temporal changes in enhancer activity by differential enhancer association.
To gain further insight into the mechanistic role of IRF2 and IRF6 in regulating adult enhancers, we identified their interacting protein complexes by a proteomic screen in human erythroid cells (Figure 7A). In addition to known interacting proteins such as IRF2BP1, IRF2BP2, RELA, STAT1, and STAT2 (Childs and Goodbourn, 2003; Rouyez et al., 2005), we identified subunits of the mediator and cohesin complexes, implicating the association between IRF2/IRF6 and the mediator and cohesin complexes for function. Physical association between IRF2/IRF6 and the mediator subunits (MED1 and MED12) or the cohesin subunits (RAD21 and SMC1A) was confirmed by independent co-IP experiments in K562 cells and primary erythroid cells (Figure 7B,C). Of note, interaction between IRF2/IRF6 and the master regulators (GATA1 and TAL1), but not NFE2 or CTCF, was also observed, indicating that they may function in the same complexes. Importantly, IRF2 co-occupied a portion of GATA1 and TAL1 peaks at a global scale (Figure 7D). The overlap was significantly higher within adult enhancers (Figure 7E). In addition, both IRF2 and IRF6 genes were associated with adult-only enhancers occupied by GATA1 and TAL1 in a stage-dependent manner (Figure 7F). It remains unclear how IRF proteins contribute to adult-stage selectivity of CTCF- or NFE2-enriched enhancer cluster (AE3 and AE4; Figure 6A). We did not observe interaction between IRF proteins, CTCF, and NFE2 (Figure 7A,B). However, IRF proteins may act indirectly on NFE2 activity. For example, enforced expression of IRF2 in myeloid progenitor cells induced megakaryocytic maturation and enhanced NFE2 binding activity to its consensus DNA sequences (Stellacci et al., 2004).
Figure 7. IRF Proteins Contribute to Adult Enhancer Activity.
(A) IRF2 or IRF6 interacting proteins identified by proteomic screen in K562 stable cell lines. The number of peptides obtained from at least two independent experiments is shown.
(B) Validation of protein interaction by co-IP experiments in K562 stable cell lines.
(C) Validation of protein interaction by co-IP experiments in primary adult ProEs.
(D) Venn diagrams show genome-wide overlaps between IRF2, GATA1, and TAL1 ChIP-seq peaks.
(E) Venn diagrams show overlaps between IRF2, GATA1, and TAL1 ChIP-seq peaks within adult enhancers.
(F) ChIP-seq density plots for H3K4me1, H3K4me3, H3K27ac, GATA1, and TAL1, and DHS density plots are shown for both IRF2 and IRF6 loci in fetal (green) and adult (red) ProEs, respectively. Putative adult-specific enhancers are indicated by shaded vertical lines.
(G) ChIP-qPCR analysis of representative GATA1/IRF2 co-bound enhancers and GATA1 only bound enhancers in control and IRF2 lentiviral shRNA (sh1 and sh2) transduced adult ProEs. Results are means ± SD of three independent experiments.
(H) Model of the temporal regulation of adult-high genes through gene-distal enhancers.
To further explore how IRF2 cooperates with the erythroid regulators within adult enhancers, we examined the TF occupancy and chromatin state at a set of representative adult enhancers (GATA1/IRF2 co-bound or GATA1 only bound enhancers; Figure 7G) in adult ProEs in the presence or absence of IRF2. Upon depletion of IRF2 expression, GATA1 occupancy was diminished at 7 out of 8 GATA1/IRF2 co-bound enhancers whereas it remained associated with GATA1 only bound enhancers (Figure 7G). Binding of cohesin subunit (RAD21) and mediator subunit (SMC1A) was abolished at GATA1/IRF2 co-bound enhancers, suggesting that IRF2 is indispensable for maintaining the configuration of at least a subset of adult stage-specific enhancers and contributes directly or indirectly to the recruitment of master regulators. Taken together, these data support a model in which the erythroid regulators GATA1 and TAL1 trans-activate IRF2 and IRF6 by occupying their distal enhancers during adult erythropoiesis. IRF2 and IRF6 then cooperate with these regulators, as well as the mediator and cohesin complexes, on a set of adult stage-specific enhancers and contribute to the differential activation of an adult program (Figure 7H).
Transcriptional Networks of Erythroid Differentiation
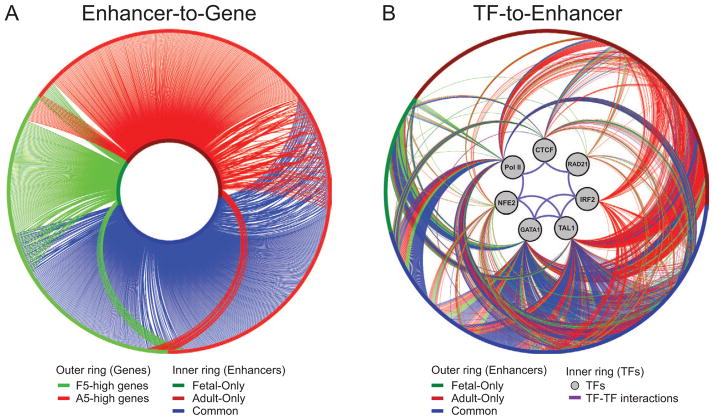
To illustrate the temporal and stage-specific regulation of distal enhancers, gene expression patterns, and TF occupancy during fetal and adult erythropoiesis, we assembled the gene regulatory networks by connecting enhancer activity and gene expression changes (Enhancer-to-Gene) or TF occupancy and enhancer activity (TF-to-Enhancer). In the Enhancer-to-Gene network, we connected an edge from each enhancer to its corresponding target gene and focused on those enhancers associated with genes differentially expressed between fetal and adult ProEs (Figure 8A, Table S8). The Enhancer-to-Gene connections were largely exclusive and reflected stage-specificity. Adult stage-specific enhancers were predominantly associated with genes upregulated in adult erythropoiesis. Conversely, fetal stage-specific enhancers were highly associated with genes upregulated in fetal erythropoiesis. Common enhancers were associated with both gene groups and had little predictive value for gene expression changes.
Figure 8. Gene Regulatory Networks underlying Human Erythropoiesis.
(A) Enhancer-to-Gene network. The edges represent the mapping of enhancers to their target genes. The inner ring of the network represents enhancers, colored according to whether they were identified in fetal (green), adult (red), or both (blue) samples. The outer ring represents differentially expressed genes that are upregulated in fetal (green) or adult (red) ProEs. Edges between enhancers and genes are colored according to their originating enhancers.
(B) TF-to-Enhancer network. The outer ring represents the same set of enhancers as shown in the inner ring of panel (A). The inner ring contains the seven profiled TFs. Edges extending from these TFs to an enhancer represent the presence of an identified TF binding site in this enhancer region in fetal (green), adult (red), or both (blue) samples. Known protein-protein interactions (purple edges) between the profiled TFs are also indicated.
See also Table S8.
To further delineate the relationship between TF occupancy and enhancer activity, we generated the TF-to-Enhancer network in which each edge was defined when at least one enrichment peak for the profiled TFs was present within the enhancer region (Figure 8B). Subsets of the common and stage-specific enhancers were associated with TFs in a highly stage-specific manner, suggesting that the temporal changes in TF occupancy control differential enhancer activity. Moreover, the high degree of co-occupancy by multiple TFs within each enhancer cluster suggests that they are coregulated by several TFs. Of note, because a large portion of enhancers were associated with yet to be determined TFs, the density and complexity of the combinatorial regulation is likely even greater than observed. Thus, our data suggest that gene-distal enhancers are major determinants of developmental stage specific expression. The combinatorial assembly of master regulators and transcriptional co-regulators at developmental stage-specific enhancers controls gene expression programs and temporal regulation of transcriptional networks during human erythropoiesis.
DISCUSSION
Role of Enhancers in Developmental Stage-Specific Transcription
Our comparative analyses of erythropoiesis at two developmental stages has identified a set of developmental stage-specific enhancers that are marked with highly stage-specific histone marks and functionally active in a stage-specific manner. Compared to common enhancers, the stage-specific enhancers are more weakly associated with master regulators (GATA1 and TAL1) as well as active histone marks (Figure 4C). These data suggest that stage-specific enhancers may function to modulate expression of target genes (“fine-tuning”) rather than to control genes “on” or “off”. Consistent with this hypothesis, the vast majority of the differentially expressed genes display only modest changes in expression between fetal and adult stages (Figure 1C,D). Alternatively, the stage-specific enhancers may be bound/activated in only a proportion of the cells, while the common enhancers are bound/activated in all cells. Single cell analysis is needed to determine the frequency of expression of stage-specific genes and enhancer activities. Nevertheless, our findings suggest that distal enhancers are the major determinants of developmental stage-specific gene expression programs. Further characterization of these stage-specific enhancers provides a path to discovery of novel candidate regulators that contribute to the differential enhancer activity required for erythroid development.
Role of Master Regulators in Stage-Specific Gene Expression
Lineage-specifying master regulators are required for programming of global gene expression towards specific lineages and often exhibit dual functions by promoting their own lineage decisions while antagonizing factors favoring other choices. Genome-wide studies in multiple cell types have revealed pivotal roles of master regulators in establishing gene expression programs by diverse mechanisms including cooperation with other cofactors and chromatin modifying complexes (Cheng et al., 2009; Fujiwara et al., 2009; Kassouf et al., 2010; Pilon et al., 2011; Tijssen et al., 2011; Yu et al., 2009). Moreover, studies utilizing embryonic stem (ES) cells and multiple hematopoietic cell types have shown that master regulators of various cell lineages direct signaling transcription factors to cell-type-specific gene programs during differentiation and regeneration (Chen et al., 2008; Mullen et al., 2011; Trompouki et al., 2011). Here we demonstrate that master regulators act cooperatively within active enhancers but have little predictive value for stage-specific transcription. Instead, a set of stage-specific cofactors and signaling pathways, such as the interferon regulatory factors IRF2 and IRF6, collaborate with master regulators and confer stage specificity. Thus, we provide a model in which master regulators are actively involved in cell-type and developmental stage-specific transcriptional programs, but their specific roles are instructed and refined by developmental stage-specific cofactors to establish stage-specificity.
Potential Implications in Hematologic Malignancies and ‘Reprogramming’ Hematopoietic Differentiation
Mutation or altered regulation of master TFs has been linked to hematologic malignancies. Our findings suggest that transcriptional co-regulators or signaling pathways may be essential for the proper functionality of master regulators, and thus may be potential targets for therapeutic interventions. Particularly, cell-type and stage-specific activity may offer a therapeutic option that is more selective and effective. The stage-specific activity of co-regulators may provide an opportunity to ‘reprogram’ hematopoietic differentiation by switching of gene expression programs. For example, LIN28B gene is specifically expressed in fetal cells and harbors a fetal stage-specific enhancer (Table S1; Figures 4E and S4C). Ectopic expression of LIN28B reprograms adult hematopoietic progenitors to fetal-like lymphocytes in a cell autonomous manner (Yuan et al., 2012), suggesting that LIN28B plays a critical role in regulating fetal identity. Our studies indicate that the developmental stage-specific enhancers may contribute to the differential expression of LIN28B gene, and interference with enhancer functionality may lead to altered gene expression. Manipulation of stage-selective cis- or trans-regulatory elements can highlight therapeutic interventions in hematologic disorders, such as sickle cell anemia and β-thalassemias, in which the fetal-to-adult hemoglobin switch is important for pathogenesis and its reversal can ameliorate disease severity (Xu et al., 2011). Thus, comparative analyses of enhancer signatures provide critical insights into global networks regulating transcription during the ontogeny and pathogenesis of human hematopoiesis.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
Primary human fetal CD34+ HSPCs were isolated from second trimester fetal livers as described (Van Handel et al., 2010). Primary human adult CD34+ HSPCs were obtained from Yale Center of Excellence in Molecular Hematology (YCEMH). Primary fetal or adult erythroblasts were generated ex vivo as described (Sankaran et al., 2008; Xu et al., 2010). The K562-BirA, K562-FLAG-Bio-IRF2 (FB-IRF2), and K562-FLAG-Bio-IRF6 (FB-IRF6) stable cell lines were generated as described (Kim et al., 2009).
ChIP
ChIP was performed as described (Xu et al., 2010). See Supplemental Experimental Procedures.
ChIP-seq Analysis
For ChIP-seq analysis using the HeliScope™ Single Molecule Sequencer, ChIP DNA was processed for 3′ polyA tailing, followed by 3′ ddATP-blocking as described (Hart et al., 2010). Processing samples by ligation, amplification and size selection are not required by Helicos sequencing. Peaks were called using Model-based Analysis for ChIP-Seq (MACS) (Zhang et al., 2008). See Supplemental Experimental Procedures.
Gene Expression Analysis
mRNA profiling of primary fetal and adult HSPCs (CD34+; F0 and A0) and maturing erythroid cells (ProE; F3, F5, F7, A3, A5, and A7) were performed using Affymetrix human genome U133 Plus 2.0 arrays. See Supplemental Experimental Procedures.
Chromatin Conformation Capture (3C)
3C assay was performed as described (Dekker et al., 2002; Xu et al., 2010). See Supplemental Experimental Procedures.
Lentiviral RNAi and Enhancer Reporter Assay
Lentiviral RNAi was performed as described (Xu et al., 2010). For enhancer reporter assay in primary human erythroid progenitors, genomic fragments were cloned into a lentiviral vector in front of a TK minimal promoter driving GFP expression. See Supplemental Experimental Procedures.
Multiprotein Complex Purification and Proteomic Analysis
IRF2- and IRF6-containing multiprotein complexes were purified as described (Kim et al., 2009). See Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Comparative genomic profiling of human fetal and adult erythropoiesis
Developmental stage-specific enhancers determine gene expression programs
Master regulators cooperate within enhancers but confer little stage specificity
Co-regulators collaborate with master regulators in establishing gene networks
Acknowledgments
We thank V.G. Sankaran, D. Higgs, D.A. Williams, L.I. Zon, E.H. Bresnick, and members of the Orkin laboratory for discussions. We thank Z. Herbert for assistance with Helicos sequencing, F. Abderazzaq and R. Rubio at CCCB sequencing facility at DFCI for assistance with Illumina HiSeq2000, E. Fox at Microarray Core at DFCI for microarray analyses, and R. Tomaino and S. Gygi at the Taplin Mass Spectrometry Facility for assistance with protein identification. This work was supported by funding from the National Institutes of Health (NIH) (S.H.O., G.C.Y., J.A.S., and H.K.A.M.), and NIH ARRA grant RCHL101553 (S.H.O.). S.H.O. is an Investigator of the Howard Hughes Medical Institute (HHMI). J.X. is an HHMI-Helen Hay Whitney Foundation fellow and supported by a NIDDK Career Development Award K01DK093543.
Footnotes
ACCESSION NUMBERS
The cDNA microarray and ChIP-seq data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE36994.
Supplemental Information includes 8 figures, 8 tables, and Supplemental Experimental Procedures and can be found with this article online.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KS, Goodbourn S. Identification of novel co-repressor molecules for Interferon Regulatory Factor-2. Nucleic Acids Res. 2003;31:3016–3026. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. Embo J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- Hart C, Lipson D, Ozsolak F, Raz T, Steinmann K, Thompson J, Milos PM. Single-molecule sequencing: sequence methods to enable accurate quantitation. Methods Enzymol. 2010;472:407–430. doi: 10.1016/S0076-6879(10)72002-4. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nature methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassouf MT, Hughes JR, Taylor S, McGowan SJ, Soneji S, Green AL, Vyas P, Porcher C. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20:1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26:6777–6794. doi: 10.1038/sj.onc.1210761. [DOI] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002a;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang M, Han H, Rohde A, Stamatoyannopoulos G. Evidence that DNase I hypersensitive site 5 of the human beta-globin locus control region functions as a chromosomal insulator in transgenic mice. Nucleic Acids Res. 2002b;30:2484–2491. doi: 10.1093/nar/30.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff M, Duncan GS, Ferrick D, Mittrucker HW, Bischof S, Prechtl S, Rollinghoff M, Schmitt E, Pahl A, Mak TW. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. The Journal of experimental medicine. 2000;192:325–336. doi: 10.1084/jem.192.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- McGrath K, Palis J. Ontogeny of erythropoiesis in the mammalian embryo. Curr Top Dev Biol. 2008;82:1–22. doi: 10.1016/S0070-2153(07)00001-4. [DOI] [PubMed] [Google Scholar]
- Migliaccio AR, Whitsett C, Migliaccio G. Erythroid cells in vitro: from developmental biology to blood transfusion products. Curr Opin Hematol. 2009;16:259–268. doi: 10.1097/MOH.0b013e32832bcaa2. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Tsuji K, Ebihara Y, Taki S, Ohba Y, Taniguchi T, Honda K. Homeostatic erythropoiesis by the transcription factor IRF2 through attenuation of type I interferon signaling. Exp Hematol. 2008;36:255–264. doi: 10.1016/j.exphem.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, Anderson SM, Mullikin JC, Gallagher PG, Hardison RC, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118:e139–148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyez MC, Lestingi M, Charon M, Fichelson S, Buzyn A, Dusanter-Fourt I. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. Journal of immunology (Baltimore, Md : 1950) 2005;174:3948–3958. doi: 10.4049/jimmunol.174.7.3948. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Scepanovic D, Vergilio JA, Ji P, Kim J, Thiru P, Orkin SH, Lander ES, Lodish HF. MicroRNA-15a and -16–1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc Natl Acad Sci U S A. 2011;108:1519–1524. doi: 10.1073/pnas.1018384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH. Human Fetal Hemoglobin Expression Is Regulated by the Developmental Stage-Specific Repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br J Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellacci E, Testa U, Petrucci E, Benedetti E, Orsatti R, Feccia T, Stafsnes M, Coccia EM, Marziali G, Battistini A. Interferon regulatory factor-2 drives megakaryocytic differentiation. The Biochemical journal. 2004;377:367–378. doi: 10.1042/BJ20031166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijssen MR, Cvejic A, Joshi A, Hannah RL, Ferreira R, Forrai A, Bellissimo DC, Oram SH, Smethurst PA, Wilson NK, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Developmental cell. 2011;20:597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Handel B, Prashad SL, Hassanzadeh-Kiabi N, Huang A, Magnusson M, Atanassova B, Chen A, Hamalainen EI, Mikkola HK. The first trimester human placenta is a site for terminal maturation of primitive erythroid cells. Blood. 2010;116:3321–3330. doi: 10.1182/blood-2010-04-279489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Peng C, Sankaran VG, Shao Z, Esrick EB, Chong BG, Ippolito GC, Fujiwara Y, Ebert BL, Tucker PW, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334:993–996. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sankaran VG, Ni M, Menne TF, Puram RV, Kim W, Orkin SH. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.