Abstract
MbtA is an adenylating enzyme from Mycobacterium tuberculosis that catalyzes the first step in the biosynthesis of the mycobactins. A potent bisubstrate inhibitor (Sal-AMS) of MbtA was previously described that displays potent antitubercular activity under iron-replete as well as iron-deficient growth conditions. This finding is surprising since mycobactin biosynthesis is not required under iron-replete conditions and suggests off-target inhibition of additional biochemical pathways. As a first step towards a complete understanding of the mechanism of action of Sal-AMS, we have designed and validated an activity-based probe (ABP) for studying Sal-AMS inhibition in M. tuberculosis. This probe labels pure MbtA as well as MbtA in mycobacterial lysate and labeling can be completely inhibited by preincubation with Sal-AMS. Furthermore, this probe provides a prototypical core scaffold for the creation of ABPs to profile any of the other 66 adenylating enzymes in Mtb or the multitude of adenylating enzymes in other pathogenic bacteria.
Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis (TB), is the leading cause of infectious disease mortality by a bacterial pathogen resulting in nearly two million deaths annually.(1) In the intervening four decades since the discovery of the first-line TB drugs, multidrug resistant TB (MDR-TB) strains have arisen that are resistant to both isoniazid and rifampin, the two most effective TB drugs.(2) Morevover, several recent case studies have identified totally drug resistant TB strains.(3) The development of new drugs, with novel mechanisms of action that target non-replicating bacilli and that are compatible with existing TB drugs, are desperately needed to treat drug-resistant TB as well as to shorten the treatment of drug-sensitive TB.
Inhibition of siderophore biosynthesis has emerged as a novel strategy for the development of new antibacterial agents for Mtb and other pathogenic bacteria.(4-6) Like virtually all pathogens, Mtb requires iron for its survival; however, the concentration of free iron is highly restricted in biological fluids due to the insolubility of iron under aerobic conditions and sequestration by iron-binding proteins such as transferrin.(7, 8) To survive under iron-limiting conditions, Mtb synthesizes a suite of structurally related small molecule iron chelating agents (i.e. siderophores) collectively known as the mycobactins that vary by the appended lipid residue on the central lysine moiety.(9) The mycobactins are exported across the mycobacterial cell envelope, where they scavenge non-heme iron and are then reinternalized to deliver the iron payload.(10) Several observations have provided evidence for the importance of mycobactins in Mtb survival. Targeted genetic inactivation of mbtB, a gene involved in the biosynthesis of mycobactins, resulted in a mutant able to replicate under iron-replete conditions, but unable to grow under iron-deficient conditions.(11) The ΔmbtB Mtb mutant is attenuated for growth in macrophages and incapable of establishing an infection in an immunocompromised mouse model.(11) In vivo gene expression profiles of Mtb show the iron-responsive gene mbtB is highly upregulated.(12) These findings collectively establish that the mycobactins are critical for pathogenesis of Mtb.
Biosynthesis of the mycobactins is initiated by the adenylating enzyme MbtA (Figure 1A), which activates salicylic acid (1), to form an acyl-adenylate intermediate (Sal-AMP, 2).(13) MbtA then catalyzes the transfer of the acyl-adenylate onto the N-terminal thiolation domain of MbtB. Elaboration of the MbtB-tethered salicylic acid to the mycobacins (3) is accomplished by a mixed non-ribosomal peptide synthetasepolyketide synthase (NRPS-PKS) assembly line.(13, 14) Our lab and others have developed a potent nanomolar bisubstrate inhibitor of MbtA, 5′-O-[N-(salicyl)sulfamoyl]adenosine (Sal-AMS, 4, Figure 1B), which mimics the acyl-adenylate intermediate and has impressive antitubercular activity under iron-deficient conditions with a minimum inhibitory concentration (MIC) of 0.39 μM.(4, 15, 16) Sal-AMS displays absolutely no mammalian toxicity providing therapeutic indices greater than 1000.(5) However, Sal-AMS does not phenocopy the ΔmbtB Mtb mutant, since it is also active in iron-replete conditions (MIC = 1.56 μM), suggesting Sal-AMS potentially possesses a secondary mechanism of action due to off-target binding.(5) Adenylation (activation of a carboxylic acid as the AMP ester) in Mtb is a ubiquitous process in both primary and secondary metabolic pathways including protein synthesis, glycolysis, lipid metabolism, and cofactor biosynthesis. Indeed, Mtb putatively encodes for at least 67 enzymes that catalyze adenylation, which may represent potential off-targets of Sal-AMS.(17) Furthermore, Sal-AMS could also potentially bind any one of the numerous adenosine-binding proteins in Mtb. A comprehenisve understanding of the mechanism of action of Sal-AMS will faciliate the design of improved chemical probes to unequivocally chemically validate siderophore biosynthesis as a virulence target.
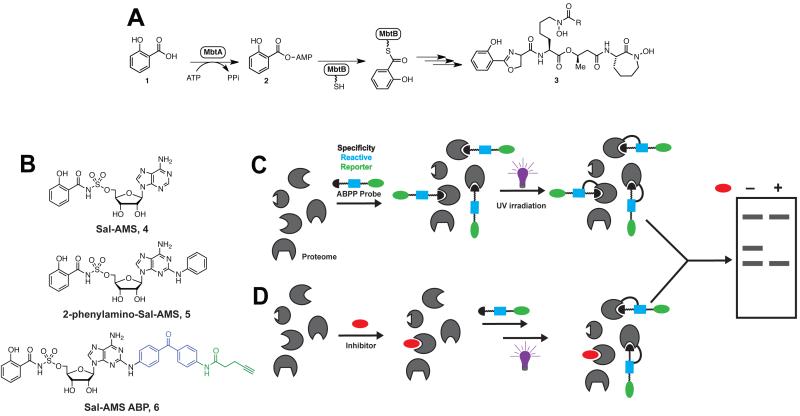
Figure 1.
A. Biosynthesis of the mycobactins. B. Chemical structures of Sal-AMS (4) inhibitor, 2-phenylamino-Sal-AMS (5) and Sal-AMS ABP (6). C. General strategy for photoaffinity labeling of an enzyme class using ABPs and gel electrophoresis as the detection method. D. Preincubation with excess inhibitor prior to photolysis with ABP.
Of the several strategies available for identifying drug targets, activity-based protein profiling (ABPP) has emerged as a promising new technique for target discovery.(18-21) Successful ABPP experiments begin with the design of a chemical probe that can bind and crosslink to the target enzyme (Figure 1C). In ABPP experiments designed to elucidate the protein targets of inhibitors, the activity-based probe (ABP) is first incubated with the proteome (either whole-cell or lysate) (Figure 1C). After UV photolysis, the labeled proteins are then analyzed by gel electrophoresis or liquid chromatography coupled with mass spectrometry (LC-MS). In a separate experiment, the proteome is pre-incubated with excess inhibitor prior to ABP incubation and photolysis (Figure 1D). Protein targets are identified by the disappearance of fluorescent labeling upon incubation with inhibitor. In order to successfully apply ABPP to target discovery, careful attention must be given to developing and validating the chemical ABP. The work described here details the design, synthesis, and evaluation of an ABP for MbtA. Not only will this probe be instrumental in identifying the protein targets of Sal-AMS, but it also represents the first ABP for an adenylating enzyme. Since Mtb and other pathogenic bacteria contain numerous adenylating enzymes (AEs) involved in a multitude of essential cellular functions, we expect the strategy outlined here for ABP development to be broadly useful for proteomic profiling of AEs.
The Sal-AMS ABP (6, Figure 1B) mimics the structure of Sal-AMS. The black portion of the probe (Sal-AMS) imparts binding selectivity for MbtA (and potential off target proteins). Several structure activity relationships (SAR) studies undertaken in our lab have indicated that MbtA tolerates modification of Sal-AMS at the C-2 position of the adenosine ring.(22) In fact, 2-phenylamino-Sal-AMS 5 (Figure 1B) is the most potent Sal-AMS derivative yet identified.(22) Therefore, the reactive and reporter groups were tethered to the 2-position. For a reactive group, we chose to use the photoreactive benzophenone moiety that has seen previous success in cross-linking ABPs to proteins.(23) Prior to UV-activation, this photogroup is stable and allows the probe to bind its protein target(s); subsequent exposure of the benzophenone group to UV light (365 nm) generates a reactive species that covalently crosslinks to amino acid residues in close proximity.(24, 25) Furthermore, a benzophenone moiety was selected as the photoreactive group, as it is generally accepted that its photocrosslinking efficiency is higher than that of aryl azides.(24) As for the reporter group, we chose a small alkyne handle onto which a fluorescent- or biotin-azide could be installed via the click reaction post photolysis to aid in visualization and enrichment, respectively.(26) ABP 6 was synthesized in 6 steps as described in the Supporting Information (Figure S1).
In order for an ABP to be a useful tool compound in studying the properties of the parent inhibitor, it must recapitulate both the in vitro and in vivo activities. Therefore, the ability of probe 6 to inhibit MbtA was measured using a [32P]PPi-ATP exchange assay.(15) The apparent inhibition constant () of probe 6 with MbtA using this assay is 0.94 nM (Figure S2), equivalent to 2-phenylamino-Sal-AMS 5 and roughly 7-fold lower than Sal-AMS (4).(15) This result is not surprising, since it was found that C-2 modifications of Sal-AMS led to enhanced binding affinity.(22) To ensure that the ABP displays a similar phenotype to Sal-AMS (4) and 2-phenylamino-Sal-AMS (5), the antitubercular activity of probe 6 was evaluated against Mtb H37Rv. Under iron-deficient conditions, the minimum inhibitory concentration that results in complete inhibition of observable cell growth (MIC) is 3–6 μM, as compared to the MIC of Sal-AMS (4) of 0.39 μM.(22) Under iron-rich conditions, the MIC is 50 μM for probe 6, as compared to 1.56 μM for Sal-AMS (4). From these data, one can extract the selectivity factor (S), which is the ratio of activity under iron-rich over iron-deplete conditions. If Sal-AMS had no off target-inhibition effects (and only targeted MbtA), this selectivity factor would be high. However, the selectivity is relatively low (S = 1.56/0.39 = 4), indicating Sal-AMS (4) may inhibit other targets. The selectivity factor for probe 6 is 8–17, which is comparable to 2-phenylamino-Sal-AMS (5) (S = 8). These data confirm probe 6 displays a similar phenotype to the parent inhibitor, suggesting 6 is an acceptable probe for proteomic profiling.
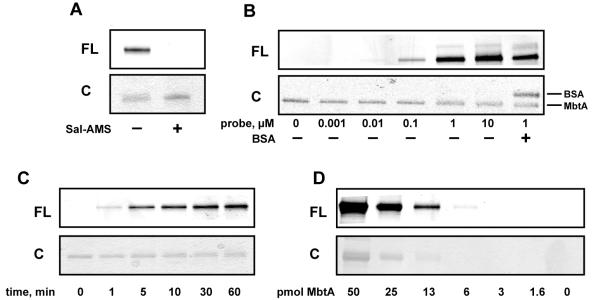
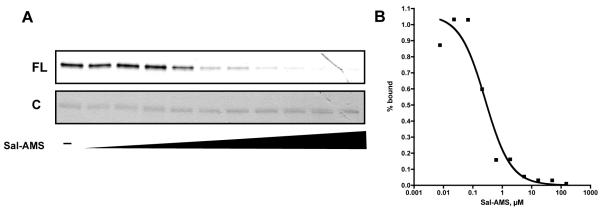
We next evaluated the ability of the ABP to photolabel pure recombinant MbtA. MbtA was first incubated with probe 6 for 10 minutes, followed by UV-photolysis at 365 nm for 30 minutes. A rhodamine-azide conjugate (TAMRA-N3) was then coupled to the terminal alkyne using standard click chemistry conditions(26), and the proteins were separated by denaturing gel electrophoresis and visualized by in-gel fluorescence scanning. As shown in Figure 2A, a clear fluorescent protein band is observed. When excess Sal-AMS (4) is preincubated with MbtA prior to addition of 6, no fluorescent band is observed (Figure 2A) indicating that probe 6 binds to the active site of MbtA. In order to determine the optimal probe concentration, varying concentrations of 6 were incubated with a constant concentration of MbtA. Figure 2B shows that a probe concentration of 1.0 μM is sufficient to provide a robust fluorescent signal. Additionally, to ensure that probe 6 is specific for labeling MbtA, BSA was also included in the photolabeling reaction. As shown in the last lane of Figure 2B, no photolabeling of BSA is observed. A UV-photolysis time course showed that maximum signal is achieved in 30 minutes (Figure 2C). When MbtA was incubated with a stochiometric amount of probe 6, irradiated for 30 minutes and subjected to TAMRA conjugation using the click reaction, it was found that the labeling efficiency was 20% (Figure S3). Assuming that the click reaction proceeds to completion in the 1 hour reaction time, this percent labeling value can be attributed to the photo-crosslinking efficiency of the benzophenone moiety of probe 6 to residues within the active site of MbtA. To assess the sensitivity of probe 6, we used in-gel fluorescence scanning to determine the lower limit of MbtA that could be detected. We found that probe 6 could detect as low as 6 picomoles of MbtA (Figure 2D). Lastly, a dose-response curve was created by incubating varying concentrations of Sal-AMS (4) with a fixed concentration of MbtA and probe 6 prior to photolysis and TAMRA conjugation (Figure 3A). The fluorescent band intensities were plotted against Sal-AMS (4) concentration (Figure 3B) to provide an IC50 value of 0.26 μM for Sal-AMS (4).
Figure 2.
In vitro labeling of pure MbtA with probe 6. A. Labeling of MbtA andcompetition with excess Sal-AMS. B. Concentration dependence of probe 6. C. UV photolysis time studies. D. Limit of detection of MbtA labeling.
Figure 3.
A. Dose response curve of Sal-AMS (4) competition with probe 6. B. Doseresponse of fractional amount of probe 6 bound (calculated from band intensities in A.) versus Sal-AMS concentration.
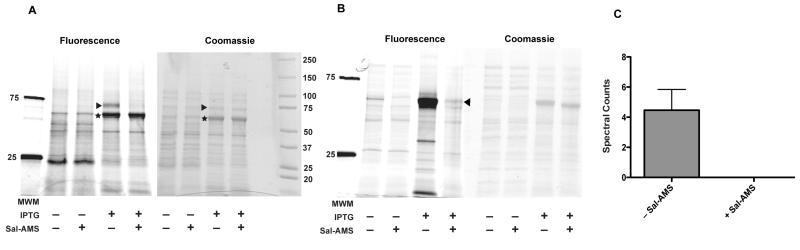
Having optimized probe concentration and UV-irradiation time using pure recombinant MbtA, we next assessed the ability of probe 6 to label overexpressed MbtA in E. coli cell lysates. E. coli was transformed with an IPTG-inducible pET-SUMO-MbtA plasmid and the pGRO7 plasmid. The pGRO7 plasmid contains the chaperone proteins GroEL and GroES and inclusion of these chaperones increases MbtA expression levels 10–20-fold.(15) E. coli cells were then grown either in the absence or presence of IPTG, photolabeled with probe 6 and subjected to the click reaction with TAMRA-N3. In the absence of IPTG, MbtA is not expressed, as evidenced by the lack of fluorescent MbtA in lane 1 of Figure 4A. However, when IPTG is added to the growth media, a strong fluorescent band corresponding to the molecular weight of MbtA-SUMO (~73 kDa) is observed (lane 3, Figure 4A, triangle). Remarkably, probe labeling can be completely inhibited by pre-incubation with excess Sal-AMS (4) (lane 4, Figure 4A). Interestingly, probe 6 also labels the GroEL as indicated by the star in lane 3 of Figure 4A. However, this labeling is non-specific, as Sal-AMS (4) does not inhibit photolabeling. These results highlight the remarkable specificity of ABP 6 towards MbtA in crude mixtures.
Figure 4.
Labeling of adenylating enzymes in crude cell lysates. A. Labeling of overexpressed MbtA in E. coli cell lysate. B. Labeling of overexpressed EntE in E. coli cell lysate. C. Mass spec detection of labeled MbtA from M. smegmatis. C. LC-MS/MS analysis of labeling of M.smegmatis MbtA by probe 6 and competition with Sal-AMS.
Encouraged by the ability of probe 6 to label MbtA in crude cell lyaste, we next assessed the ability of the ABP 6 to label an adenylating enzyme from an organism other than Mtb. EntE is the E. coli aryl acid adenylating enzyme involved in the biosynthesis of enterobactin, which adenylates 2,3-dihydroxybenzoic acid. Sal-AMS (4) is a slow tight-binding inhibitor of EntE with a Ki value of 0.47 nM.(27) We therefore tested whether ABP 6 could label EntE in crude cell lysate and if this labeling could be inhibited by competition with excess Sal-AMS (4). E. coli was transformed with an IPTG-inducible pET15b-EntE plasmid and grown in the presence or absence of IPTG. After photolabeling with probe 6 and subsequent click reaction with TAMRA-N3, tagged proteins were separated by gel electrophoresis and imaged for fluorescence (Figure 4B). Similar to MbtA, minimal labeling of EntE occurs in the absence of IPTG. However, when EntE expression is induced using IPTG, a strong fluorescent signal is observed (lane 3, Figure 4B, triangle). When lysate containing overexpressed EntE is preincubated with excess Sal-AMS (4), the fluorescence signal of EntE is largely attenuated (lane 4, Figure 4B). These studies indicate the potential utility of our ABP in studying aryl acid adenylating enzymes from many different organisms.
We next tested the ability of probe 6 to label native (uninduced) MbtA in mycobacterial cell lysate. Mycobacterium smegmatis cultures were grown in iron deficient media, lysed, and incubated with either DMSO or Sal-AMS. After UV irradiation in the presence of ABP probe 6, alkyne labeled proteins were modified with a biotin-azide group via click chemistry. Biotin-tagged proteins were enriched using streptavidin beads, trypsin digested, and analyzed by LC–MS/MS.(28) In the absence of Sal-AMS, MbtA was identified by LC-MS/MS with an average spectral count of 4 from triplicate experiments (Figure 4C). In the presence of excess Sal-AMS, MbtA was not enriched and identified by LC-MS/MS as evidence by zero spectral counts in all three replicates tested. These results indicate that MbtA, present at low concentrations in its native lysate, can be labeled in Mycobacterial species.
Activity-based probe 6 was designed in order to study the mechanism of action of Sal-AMS (4), a bisubstrate inhibitor that shows potent antitubercular activity under both iron-rich and iron-deficient conditions. After successful identification of Sal-AMS (4) off targets in mycobacteria using ABPP, this probe will aid in guiding future medicinal chemistry efforts that focus on developing selective inhibitors of MbtA to unequivocally chemically validate siderophore biosynthesis as a virulence target. Knowledge of additional targets, may enable design of improved multitarget inhibitors with more potent activity that can potentially evade the development of resistance. Finally, ABP 6 represents a scaffold for the development of activity probes for other AEs including BirA, FadD22, FadD32, MenE, MshC, and PanC, which are being pursued as potential drug targets due to their confirmed essentiality or requirement for virulence.(17)
Methods
General
MbtA purification and the ATP/PPi exchange assay were performed as described.(15)
In vitro labeling of pure recombinant MbtA
Photo-labeling experiments were performed in 96-well clear plates at a final volume of 25 μL in phosphate buffered saline (PBS, BioRad). MbtA (final concentration of 1.0 μM) was preincubated with either DMSO (0.25 μL) or 500 μM Sal-AMS (4) (0.25 μL of a 50 mM DMSO stock) at room temperature for 10 min. ABP 6 (0.25 μL of a 100 μM DMSO stock) was added to provide a final probe concentration of 1.0 μM. After 10 min at room temperature, the 96-well plate was placed on ice and irradiated at 365 nm for 30 min using a Spectroline UV handheld lamp (Model ENF-280C). To initiate the click reaction, TAMRA-N3 (Invitrogen), TCEP, TBTA ligand, and CuSO4 (Sigma Aldrich) were added to provide final concentrations of 100 μM, 1.0 mM, 100 μM, and 1.0 mM, respectively. After 1 hour at room temperature, 25 μL of 2× SDS gel loading buffer was added and the samples were heated at 95°C for 5 min. Samples (20 μL) were separated by 1D SDS-PAGE (4–20% Tris-HCl Rgels, BioRad) and fluorescent proteins were visualized by in-gel fluorescence scanning using a FMBIO III flatbed scanner (excitation laser = 532 nm, emission filter = 605 nm) (Hitachi MiraiBio Group, South San Francisco, CA). For optimal probe concentration studies, MbtA or BSA (final concentration of 1.0 μM) were incubated with either DMSO (0.25 μL) or Sal-AMS (4) (0.25 μL of 100× stock in DMSO) in a final volume of 25 μL for 10 min at room temperature before photolysis on ice and reaction with TAMRA-N3. For UV photolysis time studies, MbtA (1.0 μM) was incubated with 6 (1.0 μM) in a final volume of 25 μL for 10 min at room temperature before photolysis. After UV photolysis on ice for the respective time, the 25 μL reaction was removed from the 96-well plate and reacted with TAMRA-N3 in a separate microcentrifuge tube. The samples were then quenched and separated by SDS-PAGE gel as described above. For limit of detection studies, MbtA (1.0 μL of 100× stock in PBS) was incubated with either DMSO (0.25 μL) or 6 (0.25 μL 100× stock in DMSO) in a final volume of 25 μL for 10 min at room temperature before photolysis on ice and reaction with TAMRA-N3.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grants AI070219 (to C.C.A.), F32 AI084326 (to B.P.D.) and the Intrmural Research Program of the NIAID, NIH (to C.E.B.). Funding for the 850 MHz NMR instrumentation was provided by the University of Minnesota Office of the Vice President for Research, the Medical School, the College of Biological Science, NIH, NSF, and the Minnesota Medical Foundation.
Footnotes
Supporting Information Synthesis of probe 6, determination for probe 6 with MbtA, M. tuberculosis H37Rv MIC assays, MbtA labeling efficiency, Sal-AMS dose response curve, E. coli and M. smegmatis cell lysate labeling studies. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.World Health Organization . Global tuberculosis control: WHO report 2011. Geneva (Switzerland): 2011. [Google Scholar]
- 2.Chan ED, Iseman MD. Multidrug-resistant and extensively drug-resistant tuberculosis: a review. Curr. Opin. Infect. Dis. 2008;21:587–595. doi: 10.1097/QCO.0b013e328319bce6. [DOI] [PubMed] [Google Scholar]
- 3.Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. Totally Drug-Resistant Tuberculosis in India. Clin. Infect. Dis. 2012;54:579–581. doi: 10.1093/cid/cir889. [DOI] [PubMed] [Google Scholar]
- 4.Ferreras JA, Ryu JS, Di Lello F, Tan DS, Quadri LE. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 2005;1:29–32. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- 5.Somu RV, Boshoff H, Qiao C, Bennett EM, Barry CE, 3rd, Aldrich CC. Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. J. Med. Chem. 2006;49:31–34. doi: 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- 6.Quadri LE. Strategic paradigm shifts in the antimicrobial drug discovery process of the 21st century. Infect. Disord. Drug Targets. 2007;7:230–237. doi: 10.2174/187152607782110040. [DOI] [PubMed] [Google Scholar]
- 7.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergne AF, Walz AJ, Miller MJ. Iron chelators from mycobacteria (1954-1999) and potential therapeutic applications. Nat. Prod. Rep. 2000;17:99–116. doi: 10.1039/a809397k. [DOI] [PubMed] [Google Scholar]
- 10.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. USA. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA. (3rd) 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timm J, Post FA, Bekker LG, Walther GB, Wainwright HC, Manganelli R, Chan WT, Tsenova L, Gold B, Smith I, Kaplan G, McKinney JD. Differential expression of iron-, carbon-, and oxygenresponsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. USA. 2003;100:14321–14326. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quadri LE, Sello J, Keating TA, Weinreb PH, Walsh CT. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 14.Chavadi SS, Stirrett KL, Edupuganti UR, Vergnolle O, Sadhanandan G, Marchiano E, Martin C, Qiu WG, Soll CE, Quadri LE. Mutational and phylogenetic analyses of the mycobacterial mbt gene cluster. J. Bacteriol. 2011;193:5905–5913. doi: 10.1128/JB.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somu RV, Wilson DJ, Bennett EM, Boshoff HI, Celia L, Beck BJ, Barry CE, 3rd, Aldrich CC. Antitubercular nucleosides that inhibit siderophore biosynthesis: SAR of the glycosyl domain. J. Med. Chem. 2006;49:7623–7635. doi: 10.1021/jm061068d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miethke M, Bisseret P, Beckering CL, Vignard D, Eustache J, Marahiel MA. Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. FEBS. J. 2006;273:409–419. doi: 10.1111/j.1742-4658.2005.05077.x. [DOI] [PubMed] [Google Scholar]
- 17.Duckworth BP, Nelson KM, Aldrich CC. Adenylating enzymes in Mycobacterium tuberculosis as drug targets. Curr. Top. Med. Chem. 2012;12:766–796. doi: 10.2174/156802612799984571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Shreder KR, Gai W, Corral S, Ferris DK, Rosenblum JS. Wortmannin, a widely used phosphoinositide 3-kinase inhibitor, also potently inhibits mammalian polo-like kinase. Chem. Biol. 2005;12:99–107. doi: 10.1016/j.chembiol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 19.MacKinnon AL, Garrison JL, Hegde RS, Taunton J. Photoleucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J. Am. Chem. Soc. 2007;129:14560–14561. doi: 10.1021/ja076250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puri AW, Bogyo M. Using small molecules to dissect mechanisms of microbial pathogenesis. ACS Chem. Biol. 2009;4:603–616. doi: 10.1021/cb9001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eirich J, Orth R, Sieber SA. Unraveling the protein targets of vancomycin in living S. aureus and E. faecalis cells. J. Am. Chem. Soc. 2011;133:12144–12153. doi: 10.1021/ja2039979. [DOI] [PubMed] [Google Scholar]
- 22.Neres J, Labello NP, Somu RV, Boshoff HI, Wilson DJ, Vannada J, Chen L, Barry C. E. r., Bennett EM, Aldrich CC. Inhibition of siderophore biosynthesis in Mycobacterium tuberculosis with nucleoside bisubstrate analogues: Structure activity relationships of the nucleobase domain of 5′-O-[N-(salicyl)sulfamoyl]adenosine. J. Med. Chem. 2008;51:5349–5370. doi: 10.1021/jm800567v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. USA. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie BJ, Hergenrother PJ. Identification of the cellular targets of bioactive small organic molecules using affinity reagents. Chem. Soc. Rev. 2008;37:1347–1360. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- 25.Geurink PP, Prely LM, van der Marel GA, Bischoff R, Overkleeft HS. Photoaffinity labeling in activity-based protein profiling (2012) Top. Curr. Chem. 2011;324:85–113. doi: 10.1007/128_2011_286. [DOI] [PubMed] [Google Scholar]
- 26.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Sikora AL, Wilson DJ, Aldrich CC, Blanchard JS. Kinetic and inhibition studies of dihydroxybenzoate-AMP ligase from Escherichia coli. Biochemistry. 2010;49:3648–3657. doi: 10.1021/bi100350c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)–a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.