Abstract
The microtubule associated protein tau (MAPT/tau) aberrantly accumulates in fifteen neurodegenerative diseases, termed tauopathies. One way to treat tauopathies may be to accelerate tau clearance, but the molecular mechanisms governing tau stability are not yet clear. We recently identified chemical probes that markedly accelerate the clearance of tau in cellular and animal models. In the current study, we used one of these probes in combination with immunoprecipitation and mass spectrometry to identify 48 proteins whose association with tau changes during the first 10 minutes after treatment. These proteins included known modifiers of tau proteotoxicity, such as ILF-2 (NFAT), ILF-3, and ataxin-2. A striking observation from the dataset was that tau binding to heat shock protein 70 (Hsp70) decreased while binding to Hsp90 significantly increased. Both chaperones have been linked to tau homeostasis, but their mechanisms have not been established. Using peptide arrays and binding assays, we found that Hsp70 and Hsp90 appeared to compete for binding to shared sites on tau. Further, the Hsp90-bound complex proved to be important in initiating tau clearance in cells. These results suggest that the relative levels of Hsp70 and Hsp90 may help determine whether tau is retained or degraded. Consistent with this model, analysis of reported microarray expression data from Alzheimer’s disease patients and age-matched controls showed that the levels of Hsp90 are reduced in the diseased hippocampus. These studies suggest that Hsp70 and Hsp90 work together to coordinate tau homeostasis.
INTRODUCTION
Tau is primarily expressed in neurons, where it plays a central role in stabilizing microtubules within axons (1–3). Tau homeostasis is regulated by its expression, phosphorylation, and turnover (4). In a series of tauopathies, including Alzheimer’s disease (AD), frontotemporal dementia (FTLD), progressive supranuclear palsy (PSP), and corticobasal degeneration (CBD), tau homeostasis is disrupted, leading to hyperphosphorylation and accumulation of intracellular aggregates (5–7). Genetic depletion of tau restores cognitive defects in several mouse models (8–12), suggesting that reducing tau levels may be an effective way to re-balance its homeostasis. Indeed, recent studies using compounds that reduce tau levels by increasing proteosomal and/or autophagic clearance (13) have shown that this strategy is able to partially recover cognitive defects in cellular and mouse models (14, 15). Together, these findings have focused attention on understanding which cellular pathways protect tau and which proteins are important for its degradation.
Important regulators of tau homeostasis include the molecular chaperones, heat shock protein 70 (Hsp70) and Hsp90. Hsp70 and its constitutively-expressed isoform, Hsc70, bind directly to tau in a region near the microtubule-binding domains upon release from the microtubule (16). Through these interactions, Hsp70s facilitate the rebinding of tau to microtubules and they have also been implicated in blocking tau aggregation and promoting its degradation (17–19). Similarly, Hsp90 and a number of other co-chaperones have been implicated in regulating tau phosphorylation, aggregation, and degradation (16, 17, 20–22). Thus, Hsp70 and Hsp90 appear to play roles during multiple processes involved in establishing tau homeostasis. However, these studies have not yet revealed the molecular mechanisms involved and it is unclear how these chaperones ultimately control tau stability.
Here, we have used a small molecule to acutely disrupt tau equilibrium and thereby promote degradation. A key feature of this approach is that this small molecule reduces tau levels rapidly (within ~ 15 minutes in HeLa cells), allowing identification of proteins that change in their association with tau during the first few minutes of triage. Using mass spectrometry and quantitative spectral analysis, we found that only 48 tau-associated proteins are released or enriched on tau during the switch to a degradation fate. Interestingly, Hsp70 is released during the early stages of tau degradation and is replaced by Hsp90. Knockdown analysis revealed that Hsp90 is required for degradation, suggesting that the switch to Hsp90 is an important step on the path to degradation. Further, we find that Hsp70 competes with Hsp90 for binding to tau, suggesting that the levels of Hsp70 and Hsp90, in some cases, may dictate tau stability, a result supported by analysis of expression data from AD patients and age matched controls. These studies suggest possible new strategies for the development of therapeutics that target tau for clearance.
RESULTS AND DISCUSSION
Identification of proteins involved in tau homeostasis
Tau is an intrinsically disordered protein (23) that is thought to engage in protein-protein interactions that govern its localization, activity, and stability. Thus, we reasoned that there are likely tau-associated proteins that stabilize tau within the cell, while other complexes may be critical for its clearance. We also reasoned that a better understanding of these tau-binding factors might reveal potential new drug targets and provide insights into the mechanisms of chaperone-mediated tau triage. Towards these goals, an important advance is the recent discovery of molecules that acutely disrupt tau equilibrium and favor a rapid change in tau stability. One such molecule, methylene blue (MB), has been shown to reduce tau levels in a variety of cellular and animal models of tauopathies (13–15) and reduce polyQ levels in polyglutamine-expansion disorders (24). To test whether this compound could be used to explore changes in the tau-associated proteome, we first confirmed that MB reduces total tau levels by ~80 % in HeLa (C3) cells stably expressing V5-4RON tau (Supplemental Figure 1A). Loss of tau in this model is rapid and a new equilibrium is reached within ~10 to 20 minutes (Supplemental Figure 1B). The speed of this switch is important because it allows insights into the acute changes that occur in the tau-associated protein complexes, while avoiding complications originating from global cellular responses to MB.
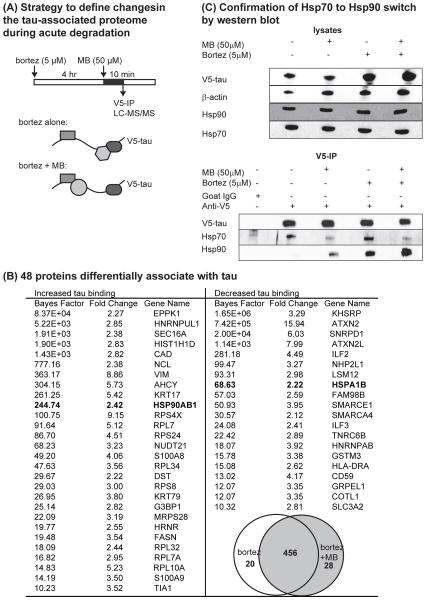
To identify proteins that change in their association with tau during the compound-initiated switch to a degradation fate, we pre-treated with the proteasome inhibitor, bortezomib, for 4 hours then applied MB (50 μM) or a vehicle control. Bortezomib was used to trap complexes destined for the proteasome, facilitating their subsequent identification by mass spectrometry (Supplemental Figure 1C). Immunoprecipitations with a V5 antibody were performed 10 minutes after MB or vehicle treatment and the precipitated material was subject to analysis by LC-MS/MS to identify the tau-associated proteome (Figure 1A). These experiments were performed in two independent, biological replicates and each replicate was analyzed by mass spectrometry in triplicate. Similar levels of tau (MAPT) were immunoprecipitated in each sample (Supplemental Figure 2) and a total of 504 proteins were identified to co-immunoprecipitate, including tubulin, TDP-43, Hsp70, and Hsp90 (Supplemental Table 1).
Figure 1. Proteomic analysis of the proteins associated with tau during the acute switch to a degradation fate in response to methylene blue (MB) treatment.
(A) Schematic of the experimental strategy for identifying tau-associated proteins which alter tau stability. (B) Using spectral counting, 456 of co-immunoprecipitated proteins did not change in their association with tau in response to MB treatment. However, 20 proteins decreased their association with tau and 28 preferentially bound. (C) Western blots on freshly immunoprecipitated samples confirmed that MB causes Hsp70 to be released from tau, while Hsp90 binding increases.
Next, quantitative spectral counting was utilized to detect proteins that change in their association with tau in response to MB. To be considered differentially associated with tau, proteins with greater than a two-fold change in abundance and a Bayes factor greater than ten were identified. While the vast majority of interacting proteins, including tubulin, did not change their association with tau upon MB treatment, this criteria identified 48 differentially associated proteins, 20 of which decreased their binding to tau and 28 that increased their binding (Figure 1C, Supplemental Table 1). Interestingly, this list includes a number of factors previously identified as modifiers of tau toxicity, such as ataxin-2, IL-2, and IL-3 (25, 26), suggesting that some of these factors might, in part, alter tau proteotoxicity by influencing its turnover. This list also included a number of proteins involved in gene regulation, such as the SWI-SNF components SMARCE1 (BAF57) and SMARCA4 (BRG1), and ribosome-associated proteins, such as RPS4X.
Hsp90 is an important factor in tau degradation
One of the most striking observations from the tau interactome analysis was that the association with stress-inducible Hsp70 (HSPA1B) in response to MB was significantly reduced, while binding to Hsp90 (HSP90AB1) was increased (Figure 1C). To confirm this finding, the V5-immunoprecipitations were repeated on freshly treated HeLa(C3) cells and western blots for Hsp70 and Hsp90 were performed. Consistent with the mass spectrometry findings, Hsp70 decreased while Hsp90 increased its association with tau after MB treatment (Figure 1D). This switch also occurred in the absence of a proteasome inhibitor (Figure 1D). These results suggest that interplay between Hsp70 and Hsp90 may be involved in targeting tau for clearance. Consistent with this idea, it had been previously shown that over-expression of Hsp70 enhances MB-mediated clearance of tau (13).
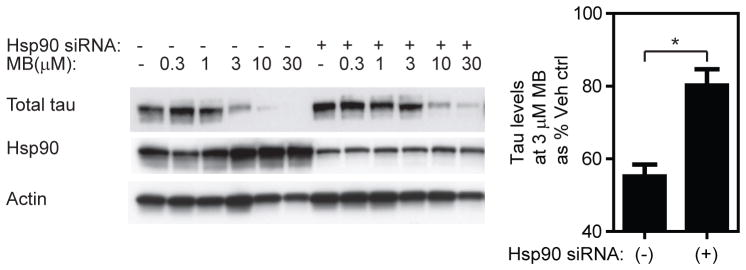
To better understand the specific role of Hsp90 in this system, we examined how changes in Hsp90 levels influence MB-initiated tau degradation. Hsp90 is best known for its ability to stabilize many substrates, such as nuclear hormone receptors and kinases, protecting them from degradation (27, 28). However, Hsp90 also promotes the degradation of other substrates, such as von Hippal Lindau factor and high-density lipoprotein (29, 30). To explore the role of Hsp90 within the tau system, we performed siRNA knock-down of Hsp90. Consistent with previous reports (17, 20, 31, 32), partial knockdown of Hsp90 (by ~80%) did not significantly change the levels of total tau. However, it did suppress the ability of MB to clear tau (Figure 2). This result supports a model in which Hsp90 is involved in targeting tau for degradation, at least in response to MB. This result is in agreement with a previous study using Hsp90 inhibitors, which suggested an active role for Hsp90 in targeting tau for degradation (20).
Figure 2. Methylene blue (MB)-initiated tau degradation is dependent on Hsp90.
Hsp90 siRNA does not cause a significant change in total tau levels in untreated cells, but it attenuates tau clearance in response to MB. Quantifications of band intensities from two independent experiments were performed using Image J. (* Student’s two-tailed unpaired t-test, p-value < 0.05).
Hsp90 competes with Hsp70 for binding to tau
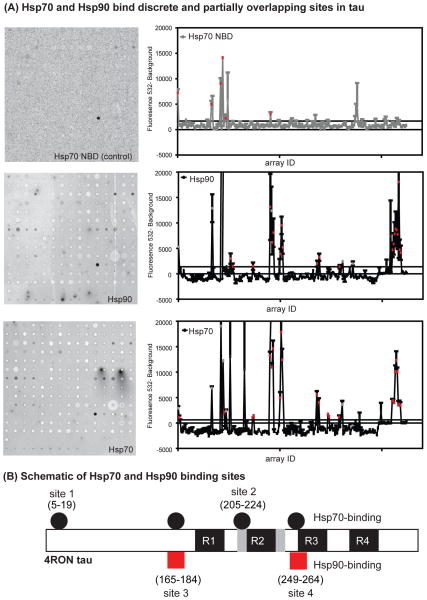
Based on these results, we hypothesized that exchange of Hsp70 for Hsp90 on tau might occur via competition for shared binding sites. At least two binding sites for Hsp70 have been identified on tau (33), but the binding sites for Hsp90 have not yet been described. To study this question, we developed peptide microarrays composed of 15-mer peptides covering the longest isoform of tau, which is expressed in the peripheral nervous system tau (4R2N). These tau peptides were assembled with 4 amino acid overlaps, for a total of 194 spots that were each present in triplicate on glass slides. Binding to Hsp70-His and Hsp90-His to these peptide arrays was measured using fluorescent anti-His antibodies and the negative controls were antibody alone and a nucleotide-binding domain (NBD) of Hsp70, which does not bind substrates. As expected, the negative controls bound non-specifically to only a few peptides, which were excluded from subsequent analyses (Figure 3A, Supplemental Table 2). In contrast, Hsp70 and Hsp90 were bound to a number of tau-derived peptides (Figure 3A). Restricting our analysis of potential binding sites to those present in the longest tau isoform of the central nervous system (4R0N), we identified four Hsp70 binding sites and two for Hsp90 (Figure 3B). Strikingly, both of the Hsp90 binding sites were shared by Hsp70. Previous work has determined that deletion of residues Ile219, Ile220 and Ile250, Val251 reduces Hsp70 binding to tau (33). Consistent with this result, these residues were present in the third and fourth Hsp70 binding sites, as measured by peptide microarray. These Hsp70 binding sites were further validated using a software program (34) developed to predict binding sites for the prokaryotic Hsp70, DnaK. Using this approach, sites 2, 3, and 4 were positively identified.
Figure 3. Hsp70 and Hsp90 bind discrete and partially overlapping sites on tau, as measured by peptide microarrays.
(A) Representative array results using Hsp70 nucleotide binding domain (NBD), Hsp90, and Hsp70 (all 10 μM) are shown. Further, the fluorescence intensities at a wavelength of 532 nm, after background subtraction, for each array spot tested in triplicate are shown. Peptides with intensities 3 standard errors of mean (SEM) above the mean for the total dataset are colored in red. Peptides that bound either Hsp70 NBD or antibody alone are shown in gray. One false-positive spot (array ID: 161) was removed for clarity. (B) Schematic of the Hsp70 and Hsp90 binding sites mapped onto 4R0N tau, highlighting the four microtubule-binding repeats (R1–4).
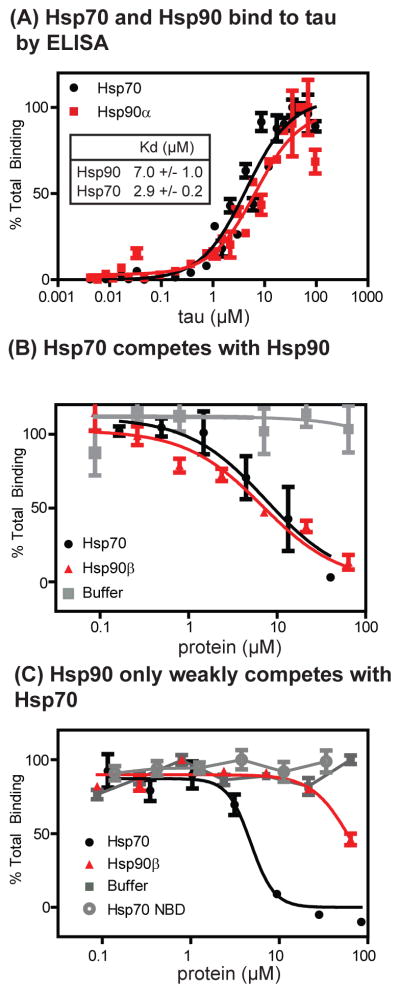
The peptide microarray findings indicate that Hsp90 binding sites are shared by Hsp70 and suggest that these chaperones might compete for binding to tau. To test this model, we first synthesized peptides (~7 to 15 amino acids) corresponding to the binding sites predicted by the microarray. However, these peptides showed weak (>100 μM) binding, which was difficult to accurately measure. Therefore, instead we aimed to validate this model by studying the binding of Hsp70 and Hsp90 in the context of 4RON tau in an ELISA-like platform. Immobilized Hsp70 bound tau with an affinity of 2.9 ± 0.2 μM and Hsp90 bound with a slightly weaker affinity of 7.0 ± 1.0 μM (Figure 4A). As predicted by the array results, Hsp70 competed for binding with Hsp90 (IC50 ~ 4 μM) (Figure 4B). Conversely, Hsp90 was less effective at competing with Hsp70 (IC50 > 50 μM), perhaps because of the two unique Hsp70 binding sites (Figure 4C). These same relationships were observed with the major stress-inducible (Hsp70, Hsp90α) and the constitutively expressed (Hsc70, Hsp90β) isoforms (Table 1). Based on these results, we concluded that the MB-initiated increase in Hsp90 binding observed by mass spectrometry may be the result of exchange of Hsp70 for Hsp90 on shared binding sites within tau.
Figure 4. Hsp70 competes with Hsp90 for binding to tau.
(A) Immobilized Hsp70 and Hsp90 bind to 4RON tau. (B) Soluble Hsp70 can inhibit binding of tau to immobilized Hsp90 with an IC50 value of 4 μM. As a control, free Hsp90 also competes with this interaction. (C) Soluble Hsp90 only weakly competes with immobilized Hsp70 for binding to tau with an IC50 value greater than 50 μM. As a control, free Hsp70 inhibits binding with an IC50 of 8 μM. Similar values were seen using different Hsp70 and Hsp90 isoforms (see Table 2). All of the experiments were performed in triplicate. Results are shown as the average and standard error of the mean.
Table 1.
Testing competition between Hsp70 and Hsp90 for binding to tau
| Hsp70-tau binding | Hsp90α-tau binding | |||
|---|---|---|---|---|
|
| ||||
| Protein | IC50 μM | SEM | IC50 μM | SEM |
| Hsp70 (HSPA1B) | 8 | 4 | 4 | 2 |
| Hsc70 (HSPA8) | 17 | 9 | 1.9 | 0.5 |
|
| ||||
| Hsp90α (HSP90AA1) | >100 | NA | 4 | 2 |
| Hsp90β (HSP90AB1) | 59 | 42 | 3 | 1 |
|
| ||||
| buffer | >100 | NA | >100 | NA |
| Hsp70 NBD | >100 | NA | NA | NA |
IC50 denotes the concentration which reaches 50% inhibition
SEM denotes the standard error of the mean
Hsp90 is decreased in human brains with Alzheimer’s disease
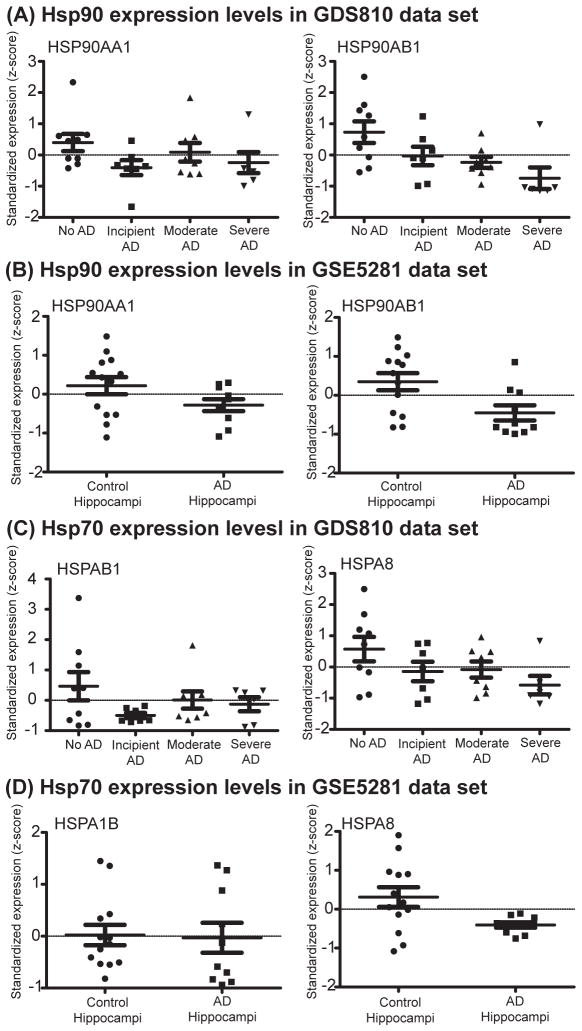
These results suggest that tau degradation may be sensitive to the relative levels of Hsp70 and Hsp90. More specifically, reduced Hsp90 levels might favor net retention of tau, perhaps unbalancing tau homeostasis. To explore this idea, we examined the GDS810 dataset, which collected microarray expression data from the hippocampal tissue of patients with incipient, moderate, and severe AD and age matched controls (35). Similarly, we queried the GSE5281 dataset that includes expression results from dissected, histopathologically normal, hippocampal neurons of AD patients and age matched controls (36, 37). Previous analyses of these datasets had already noted AD-related changes in protein folding genes, such as chaperones. However, we wanted to specifically ask how the relative levels of Hsp70 and Hsp90 might change as a function of disease. Accordingly, we analyzed both datasets (Supplemental Figure 3) and observed statistically significant changes and trends that indicated an AD-associated decrease in Hsp90 (Figure 5). Although the samples sizes are relatively low, we did not observe consistent changes in stress-inducible Hsp70 levels (HSPA1B, HSPA2, HSPA6, HSPA14), but Hsc70 (HSPA8) was decreased in both datasets (Table 2). Importantly for the current study, the ratio of stress-inducible Hsp70s and Hsp90s was significantly changed in AD, suggesting that imbalance in this chaperone ratio might contribute to the accumulation of tau. Interestingly, these finding are consistent with an observation made in an AD mouse model in which Hsp90 protein levels were found to be inversely correlated with tau levels (17). Taken together, a model emerges in which Hsp90 levels and the Hsp70:Hsp90 ratio help determine tau stability.
Figure 5. Hsp90 levels are decreased in hippocampal samples from Alzheimer’s disease (AD) patients.
Analysis of the (A) GDS810 and (B) GSE5281 datasets reveals a decrease in the mRNA levels of both isoforms of Hsp90 (HSPAA1and HSP90AB1) in AD samples. There were no statistically significant changes in the levels of most the stress-inducible Hsp70s, such as HSPAB1 (C and D), HSPA1B, HSPA2, HSPA4, or HSPA14 (Table 2 and Supplemental Fig 4). However, the levels of Hsc70 (HSPA8) were decreased in AD patient samples (C and D).
Table 2.
Gene expression level changes for Hsp70 and Hsp90 in Alzheimer’s disease (AD).
| GSE5281 | GDS810 | ||||
|---|---|---|---|---|---|
|
| |||||
| Gene | Array IDa | AD p-value |
Incipient AD p-value |
Moderate AD p-value |
Severe AD p-value |
| HSP90AA1 | 210211_s_at, 211968_s_at, 214328_s_at | d0.088 | d0.031 | d0.236 | d0.088 |
| 200064_at, 214359_s_at for both and 1557910_at for | |||||
| HSP90AB1 | GSE5281 | d0.017 | d0.114 | d0.046 | d0.008 |
|
| |||||
| HSPA1B | 200799_at, 200800_s_at, 202581_at | 0.438 | d0.252 | 0.743 | 0.272 |
| HSPA8 | 208687_x_at, 210338_s_at, 221891_x_at for both and 224187_x_at for GSE5281 | d0.028 | d0.211 | d0.236 | d0.066 |
| HSPA2 | 211538_s_at | 0.227 | d0.470 | 0.370 | u0.224 |
| HSPA4 | 208815_x_at, 211015_s_at, 211016_x_at | 0.515 | 1.000 | 0.321 | 0.864 |
| HSPA6 | 117_at, 213418_at | u0.032 | d0.252 | 0.321 | 0.181 |
| 219212_at for both and 226887_at, 227650_at for | |||||
| HSPA14 | GSE5281 | 0.687 | 0.711 | d0.167 | d0.114 |
|
| |||||
| NUP37 | 218622_at | 0.403 | 0.837 | 0.321 | 0.776 |
Unless otherwise noted the Array IDs given were present in both datasets.
Abbreviations are used to indicate the direction of change in gene expression; d denotes a decrease in gene expression in AD patients and u denotes an increase. Grey denotes gene expression changes with a Mann Whitney U test p-value of <0.05.
Discussion
Tau homeostasis is important in neurodegenerative tauopathies and enhancing tau degradation may be a promising therapeutic strategy (10, 13–16). Thus, it is important to understand the pathways that stabilize tau and those that favor its turnover. Using a chemical biology approach, we specifically studied the early changes in the tau-associated proteome that occur during the acute switch to a degradation fate.
From the list of tau-associated proteins, a striking observation was the loss of Hsp70 (HSPA1B) binding, coupled with an increase in Hsp90 (HSP90AB1) binding. What role is Hsp70 playing in this process? Previous findings suggest that Hsp70 is important in stabilizing tau. For example, Hsc70 is required to recycle tau on microtubules, perhaps restricting its availability to the degradation pathways (19). However, it is also clear that Hsp70s are not exclusively devoted to stabilizing tau. For example, if this were the case, then Hsp70 knockdown should reduce tau levels, phenocopying MB treatment. Instead, only mild changes in tau levels are typically observed upon knockdown or over-expression of Hsp70 (17). One possible way to account for these observations is that Hsp70 could play an active role in “hand-off” of tau to the degradation pathway. This process might be initiated by MB in these studies, because, while MB is certainly not a selective Hsp70 inhibitor (38, 39), over-expression of Hsp70 enhances compound-induced degradation of tau (13). Thus, MB might cause a structural transition in Hsp70, linking the chaperone to the degradation pathway. Such a mechanism could involve co-chaperones, such as Hop, which bridge the Hsp70 and Hsp90 systems or, alternatively, acute release of tau from Hsp70 may generate a type of tau structure that binds particularly well to Hsp90. In this model, the activity of Hsp70 that is favored by chemical inhibitors might not produce the same phenotype as knockdown or over-expression, which removes the entire protein. Rather, the transition might require a chemical trigger, such as MB or other Hsp70 inhibitors, which alter Hsp70’s nucleotide state, substrate affinity and/or conformation. (16, 40)
What role is Hsp90 playing in this process? Our observations point to a role of Hsp90 in the degradation of tau (see Figure 2). Hsp90 is best known for its ability to stabilize ~200 “client” proteins, such as the glucocorticoid receptor (GR) (28). In those systems, Hsp90 is normally found in the final, high affinity complex that protects the active protein fold (27, 41–43). Accordingly, inhibitors of Hsp90 relieve the protective effect and favor degradation of the clients, usually through a process involving Hsp70. Thus, Hsp90 is typically considered a protective chaperone, while Hsp70 is considered a triage chaperone. However, there are clear examples of clients in the literature that do not follow this paradigm. For example, Hsp90 promotes the degradation of the von Hippel-Lindau tumor-suppressor protein and high-density lipoprotein (29, 30) and, in those systems, Hsp70 appears to play an upstream role in folding. Our results suggest that tau might fall into this latter category, for which Hsp90 assumes the task of promoting turnover. However, tau seems to be differ in some important ways. For example, Hsp90 inhibitors block the degradation of Von Hippel-Lindau tumor-suppressor protein and high-density lipoprotein, whereas these same compounds reduce mutant and phosphorylated tau in a pathway dependent on CHIP, but not HSF-1 or Hsp70 (20). These findings point to a model in which Hsp90 stabilizes some forms of tau, protecting them from clearance (20, 31, 44), while also participating in the clearance of tau when Hsp90 or Hsp70 is inhibited. Clearly, the roles played by Hsp90 in tau homeostasis are complex.
One compelling way to rationalize these observations is to invoke different “pools” of Hsp90. Hsp90 engages in protein-protein interactions with many different co-chaperones and recent work has elegantly shown that distinct Hsp90 complexes exist in the cytosol (45). We can speculate that some of these Hsp90 complexes may be involved in stabilizing tau while others, such as the ones apparently favored by MB treatment, might target it for degradation. Indeed, some Hsp90 co-chaperones are known to promote tau stabilization, such as cdc37 and FKBP51, while others accelerate degradation, such as FKBP52 and CHIP (16). Further, the acetylation state of Hsp90 may also influence whether it targets tau for degradation (46, 47). Unfortunately, there was no obvious change in co-chaperone levels in the tau mass spectrometry studies (Supplemental Table 1), but these factors may not have been abundant enough or they might bind weakly. Further work, perhaps using small molecules that target individual Hsp90 complexes, will likely be required to better understand this mechanism.
Some of the biggest unresolved issues in understanding the tau degradation are what happens during the handoff of tau from Hsp70 to Hsp90 and what happens after formation of the Hsp90-tau complex? One key observation could be that Hsp70 efficiently competes with Hsp90 for binding tau, but not the inverse (see Figure 4 and Table 1). Thus, Hsp90-mediated degradation might depend on a prior signal to release Hsp70 and, moreover, this signal might induce a conformation of Hsp70 that actively recruits Hsp90. Despite these speculations and open questions, the work here provides an unexpected insight into how Hsp90 is involved in degrading tau in response to MB. Moreover, these findings suggest that exchange of Hsp70 for Hsp90 is one key aspect of the mechanism. This model is supported by patient expression data and results from mouse models (17), in which decreased Hsp90 transcript level is linked to tau accumulation and AD. Additional insights into this process will likely accelerate drug discovery in tauopathies, perhaps by suggesting ways of using Hsp70 and Hsp90 inhibitors to maximize the restoration of tau homeostasis. We suggest that rapidly altering the fate of tau using chemical probes will be a particularly valuable strategy towards that goal.
Finally, the list of tau-associated proteins sensitive to compound treatment (see Figure 1B) included a number of factors, including ILF-2 (NFAT), ILF-3 and ataxin 2, that were previously linked to tau proteotoxicity. While our work focused on the contributions of Hsp70 and Hsp90 in regulating tau protein homeostasis, this list of tau-associated factors may be enriched in other proteins that contribute to the regulation of tau homeostasis. For example, although not the focus of this work, the results with ataxin-2 (ATXN2) and ataxin-2-like protein (ATXN2L) were particularly striking, with ~16- and 8-fold reductions in binding to tau during degradation, respectively. These observations are interesting because ataxin-2 is linked to spinocerebellar ataxia type 2 (SCA2), a polyglutamine expansion neurodegenerative disorder(48), so direct contacts between these disease-associated proteins might be speculated to be important in their pathophysiology.
METHODS
Reagents, cell lines, and general methods
Tetramethylthionine (methylene blue) was purchased from Sigma.. siRNA (Qiagen) was transfected at 20 nM (20, 21). Antibodies utilized are as follows: V5 (Sigma, V8137), β-actin (Anaspec, 54591), tau-WB/ELISA (Santa Cruz Biotech, sc5587), pTau (pS396/S404, provided by Dr. Peter Davies Albert Einstein College of Medicine), Hsp70 (Assay Designs), Hsp90 (Santa Cruz Biotech, sc7947), goat anti-rabbit HRP (Anaspec, 28177), and goat anti-mouse HRP (Anaspec 28173). All cells were maintained according to ATCC guidelines. Stably transfected HeLa (C3) cells overexpressing wild-type 4RON human tau were previously generated by clonal selection with G418 (Invitrogen) (13). Absorbance measurements were performed using a SpectraMax M5 multimode plate reader (Molecular Devices).
Western blot analysis
Samples were separated under reducing and denaturing conditions using 10–20 % Tris-Tricine gels (Invitrogen). After transfer to nitrocellulose membrane (Whatman), the membranes were blocked with 5 % w/v milk in TBS-T (25 mM Tris–HCl, pH 7.4, 140 mM NaCl, 0.1 % v/v Tween 20) for at least 3 hours. Protein levels were detected by incubating the membrane overnight at 4 °C with primary antibodies diluted to 1:1,000, unless otherwise noted, in TBS-T containing 2 % w/v bovine serum albumin (BSA, Sigma). Finally, the membrane was incubated for 1 hr with the appropriate HRP-conjugated secondary antibody diluted 1:10,000 in TBS-T with 5 % w/v BSA. Membranes were washed three times for 5 minutes with TBS-T at each step. Membranes were developed using Supersignal West Pico chemiluminescence kit (Thermo Scientific). For the western blot analysis of immunoprecipitated samples titers of 1:250 Hsp70 antibody and 1:500 Hsp90 antibody were utilized.
Immunoprecipitation of V5-tau
HeLa cells stably transfected with V5-4RON tau (13) were grown to 90 % confluence and subsequently treated with 5 μM Bortezimib for four hours, followed by a ten minute treatment with either 50 μM MB or vehicle (DMSO) control (1 % v/v). Cells were lysed with M-Per lysis buffer with protease and phosphatase inhibitor cocktails (19) and 5 mg of lysate was immunoprecipitated by incubating with 75 μl of goat anti-V5 conjugated to agarose beads (Bethyl Laboratories, S190-119) at 4 °C overnight in the dark. Beads were subsequently washed with 100 μL of PBS (10mM Na2HPO4, 2 mM KH2PO4, pH 7.4, 137 mM NaCl, and 2.7 mM KCl) + 0.1 % v/v Tween 20, followed by washes with 100 μL and then 200 μL of PBS. Finally proteins associated were eluted using 45 μL of 0.1 M glycine (pH 2.7) and neutralized by adding 5 μL 1M Tris-HCl (pH 8.0). 25 μL of these samples were separated on 10–20 % Tris-Tricine gel (Invitrogen) for western blot or mass spectrometry analysis. Previous work utilizing this HeLa model has demonstrated that the V5-4R0N tau is present in several phosphorylated forms including pS212/p231, pS262/S456, and pS396/S404 (13, 21). Further, MB reduces both total tau and tau phosphorylated at S396/S404 (13). Thus, the V5 immunoprecipitated samples likely include a number of tau and phospho-tau variants, providing an overview of the changes in the tau-associated proteome.
Mass spectrometry
Immunoprecipitations were performed on two independent biological samples and separated, as described, using 10–20 % Tris-Tricine gels (Invitrogen). Each sample was subjected to in-gel digestion with trypsin and proteins were identified by liquid chromatography-high resolution tandem MS (LTQ-Orbitrap XL, ThermoFisher) at the University of Michigan’s Proteomics Resource facility, using standard protocol as previously described (49). Mass spec analysis was performed in technical triplicates on each sample, providing a total of six datasets per condition. ThermoFisher RAW files were converted to mzXML format using msconvert (50) and were searched against the human UniProt database (release version: 2010_09). This database fasta file contained 20286 protein entries concatenated with reverse (decoy) sequences and common contaminants. Searches were performed using X!Tandem with the k-score plug-in (51, 52). Search results were post-processed using PeptideProphet and ProteinProphet from the Trans-Proteomic Pipeline (TPP) software suite (Supplemental Table 1).
Quantitative spectral analysis
Total spectral counts were obtained from each of the six datasets using Abacus (55). The abacus parameters used were: maxIniProbTH = 0.97, minCombinedFilePw=0.5, all other parameters were kept at their default values. Using these options, a protein-level false-discovery rate (FDR) of 0.05 was achieved. At this threshold 504 proteins were identified to co-immunoprecipitate with V5-tau across all six data sets. Differential protein expression was assessed by label-free quantification using QSpec (56). From the QSpec results, a protein was considered to be differentially associated with tau if it had a Bayes Factor greater than ten and exhibited a fold change greater than two. Using this definition, 48 of the 504 co-immunoprecipitating proteins were found to be differentially associated upon MB treatment (Figure 1C, Supplemental Table 1).
Protein purification
Human Hsp70 (HSPA1A), Hsc70 (HSPA8), and Hsp70 NBD (1–383) were purified as previously described for DnaK using a His column, including cleavage of the His tag via TEV protease as indicated, and a final purification on an ATP-agarose column (57, 58). Human Hsp90-beta (HSP90AB1) and Hsp90-alpha (HSP90AA1) were purified as previously described (59). N-terminal His-tagged Human 4RON tau was purified according to the previously developed protocol (60).
Tau peptide microarray
The tau peptide microarray was designed using 15-mer peptides with 4 amino acid overhangs, spanning the full sequence of PNS-tau (P10636–9). In addition full-length proteins expected to bind Hsp70 and Hsp90 were printed as positive controls, including human and mouse IgG and tau. Empty spots were used as negative controls. The arrays were printed on single microscope slides in triplicate (JPT peptide technologies). Binding was tested per manufactor’s protocol using 10 μM Hsp70-His, Hsp90-His, or Hsp70 nucleotide binding domain (NBD) in binding buffer (25 mM HEPES pH 7.2, 150 mM NaCl, 20 mM KCl, 5 mM MgCl2, and 0.5 mM DTT). Binding was detected using 1:1,000 titer of Hilyte555 anti-His antibody (Anaspec) in TBS-T with 1 % w/v BSA (Sigma) and scanning the arrays at a fluorescence emission of 532 nM using a GenePix 4100A Microarray Scanner (Molecular Devices) and standard local background subtraction. Binding was defined as peptides with fluorescence signals greater than 3 standard errors above the mean of the total data set. Peptides that appear as positive in the Hsp70 NBD or antibody alone tests were excluded as false positives.
ELISA-based tau binding
The procedure for chaperone binding to tau was adapted from a previous report (61). Briefly, 0.1 mg mL−1 of either human Hsp70 or Hsp90 (30 μL) in immobilization buffer (50 mM MES, pH 5.5, and 0.5 mM DTT) was added to each well of 96-well plates (Thermo Fisher, clear, nonsterile, flat bottom) and these plates incubated for 6 hrs at 37 °C. The wells were then washed with 100 μL of TBS-T (3×3 min, rocking). To these wells, 50 μL of 4RON tau solution (at indicated concentrations) in binding buffer (25 mM HEPES, pH 7.2, 150 mM NaCl, 20 mM KCl, 5 mM MgCl2, and 0.5 mM DTT) with 1 mM ATP was added and the plates were incubated at room temperature with gentle rocking overnight. Competition experiments were performed in the presence of 7.5 μM tau. Plates were developed using the rabbit anti-tau primary antibody (Santa Cruz, sc5587) (1:2,000 dilution, in TBS-T, 50 μL/well) and the goat anti-rabbit HRP conjugated secondary antibody (Anaspec, 28177) (1:2,000 dilution in TBS-T, 50 μL/well). The TMB substrate kit (Cell Signaling Technology) was used to detect binding. In each experiment, the signal from non-specific binding of 4RON to empty control wells was used as a negative control, but this signal was minimal. Binding curves were fit using hyperbolic fits with a non-zero intercept in GraphPad Prism version 5.0 for Windows (GraphPad Software).
Analysis of Hsp70 and Hsp90 expression levels in AD samples
The GDS810 (35) and GSE5281 (36) gene expression microarray datasets were downloaded from the NCBI Gene Expression Omnibus. Normalized data for relevant microarray probes in all samples were extracted for the genes listed in Table 2. The sample GSM21206 was excluded from the GDS810 dataset due to artifact rendering which made this sample incomparable to the rest. Probe expression values were standardized as a z-score (z = (value − mean)/(standard deviation)). Individual probe z-scores belonging to a given gene were averaged within a single patient’s sample. Further, the average and variance of expression for a given gene was compared across sample groups (control patients versus patients with disease). Statistical significance (p-value <0.05) for gene expression changes between two sample groups was determined with a Mann Whitney U test (see Supplemental Figure 3).
Supplementary Material
Acknowledgments
The authors would to thank W. Pratt, A. Lieberman, and Y. Osawa for helpful discussions. We would also like to acknowledge D. Southworth for help in purifying Hsp90. The University of Michigan’s Department of Pathology Proteomics Core performed the mass spectrometry studies. A. Thompson was supported by a pre-doctoral fellowship from the National Institute of Health (F30AG035464). This work was additionally supported by NIH R01NS059690 and an anonymous gift from the Michigan Alzheimer’s Research Center (MADC). Additional support was from NIH R01NS073899, CurePSP and the Alzheimer’s Association to C. Dickey.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Drechsel DN, Hyman AA, Cobb MH, Kirschner MW. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Biol Cell. 1992;3:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drubin DG, Feinstein SC, Shooter EM, Kirschner MW. Nerve growth factor-induced neurite outgrowth in PC12 cells involves the coordinate induction of microtubule assembly and assembly-promoting factors. J Cell Biol. 1985;101:1799–1807. doi: 10.1083/jcb.101.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kempf M, Clement A, Faissner A, Lee G, Brandt R. Tau binds to the distal axon early in development of polarity in a microtubule- and microfilament-dependent manner. J Neurosci. 1996;16:5583–5592. doi: 10.1523/JNEUROSCI.16-18-05583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 5.Spillantini MG, Goedert M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 1998;21:428–433. doi: 10.1016/s0166-2236(98)01337-x. [DOI] [PubMed] [Google Scholar]
- 6.Williams DR. Tauopathies: classification and clinical update on neurodegenerative diseases associated with microtubule-associated protein tau. Intern Med J. 2006;36:652–660. doi: 10.1111/j.1445-5994.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 7.Kidd M. Paired helical filaments in electron microscopy of Alzheimer’s disease. Nature. 1963;197:192–193. doi: 10.1038/197192b0. [DOI] [PubMed] [Google Scholar]
- 8.Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, Forster C, Yue M, Orne J, Janus C, Mariash A, Kuskowski M, Hyman B, Hutton M, Ashe KH. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sydow A, Van der Jeugd A, Zheng F, Ahmed T, Balschun D, Petrova O, Drexler D, Zhou L, Rune G, Mandelkow E, D’Hooge R, Alzheimer C, Mandelkow EM. Tau-induced defects in synaptic plasticity, learning, and memory are reversible in transgenic mice after switching off the toxic Tau mutant. J Neurosci. 2011;31:2511–2525. doi: 10.1523/JNEUROSCI.5245-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberson ED, Halabisky B, Yoo JW, Yao J, Chin J, Yan F, Wu T, Hamto P, Devidze N, Yu GQ, Palop JJ, Noebels JL, Mucke L. Amyloid-beta/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer’s disease. J Neurosci. 2011;31:700–711. doi: 10.1523/JNEUROSCI.4152-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu GQ, Mucke L. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 12.Vossel KA, Zhang K, Brodbeck J, Daub AC, Sharma P, Finkbeiner S, Cui B, Mucke L. Tau reduction prevents Abeta-induced defects in axonal transport. Science. 2010;330:198. doi: 10.1126/science.1194653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, O’Leary J, Morgan D, Lee DC, Shults CL, Rousaki A, Weeber EJ, Zuiderweg ER, Gestwicki JE, Dickey CA. Chemical manipulation of hsp70 ATPase activity regulates tau stability. J Neurosci. 2009;29:12079–12088. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Congdon EE, Wu JW, Myeku N, Figueroa YH, Herman M, Marinec PS, Gestwicki JE, Dickey CA, Yu WH, Duff K. Methylthioninium chloride (methylene blue) induces autophagy and attenuates tauopathy in vitro and in vivo. Autophagy. 2012:8. doi: 10.4161/auto.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Leary JC, 3rd, Li Q, Marinec P, Blair LJ, Congdon EE, Johnson AG, Jinwal UK, Koren J, 3rd, Jones JR, Kraft C, Peters M, Abisambra JF, Duff KE, Weeber EJ, Gestwicki JE, Dickey CA. Phenothiazine-mediated rescue of cognition in tau transgenic mice requires neuroprotection and reduced soluble tau burden. Mol Neurodegener. 2010;5:45. doi: 10.1186/1750-1326-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyata Y, Koren J, Kiray J, Dickey CA, Gestwicki JE. Molecular chaperones and regulation of tau quality control: strategies for drug discovery in tauopathies. Future Med Chem. 2011;3:1523–1537. doi: 10.4155/fmc.11.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou F, Netzer WJ, Tanemura K, Li F, Hartl FU, Takashima A, Gouras GK, Greengard P, Xu H. Chaperones increase association of tau protein with microtubules. Proc Natl Acad Sci U S A. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, Kim J, Dillmann WH, Browne SE, Hall A, Voellmy R, Tsuboi Y, Dawson TM, Wolozin B, Hardy J, Hutton M. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 19.Jinwal UK, O’Leary JC, 3rd, Borysov SI, Jones JR, Li Q, Koren J, 3rd, Abisambra JF, Vestal GD, Lawson LY, Johnson AG, Blair LJ, Jin Y, Miyata Y, Gestwicki JE, Dickey CA. Hsc70 rapidly engages tau after microtubule destabilization. J Biol Chem. 2010;285:16798–16805. doi: 10.1074/jbc.M110.113753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, Patterson C, Dickson DW, Nahman NS, Jr, Hutton M, Burrows F, Petrucelli L. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinwal UK, Koren J, 3rd, Borysov SI, Schmid AB, Abisambra JF, Blair LJ, Johnson AG, Jones JR, Shults CL, O’Leary JC, 3rd, Jin Y, Buchner J, Cox MB, Dickey CA. The Hsp90 cochaperone, FKBP51, increases Tau stability and polymerizes microtubules. J Neurosci. 2010;30:591–599. doi: 10.1523/JNEUROSCI.4815-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimura H, Miura-Shimura Y, Kosik KS. Binding of tau to heat shock protein 27 leads to decreased concentration of hyperphosphorylated tau and enhanced cell survival. J Biol Chem. 2004;279:17957–17962. doi: 10.1074/jbc.M400351200. [DOI] [PubMed] [Google Scholar]
- 23.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 24.Wang AM, Morishima Y, Clapp KM, Peng HM, Pratt WB, Gestwicki JE, Osawa Y, Lieberman AP. Inhibition of hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J Biol Chem. 2010;285:15714–15723. doi: 10.1074/jbc.M109.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulman JM, Feany MB. Genetic modifiers of tauopathy in Drosophila. Genetics. 2003;165:1233–1242. doi: 10.1093/genetics/165.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006:111–138. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- 29.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 30.Gusarova V, Caplan AJ, Brodsky JL, Fisher EA. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J Biol Chem. 2001;276:24891–24900. doi: 10.1074/jbc.M100633200. [DOI] [PubMed] [Google Scholar]
- 31.Luo W, Dou F, Rodina A, Chip S, Kim J, Zhao Q, Moulick K, Aguirre J, Wu N, Greengard P, Chiosis G. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc Natl Acad Sci U S A. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahara N, Maeda S, Yoshiike Y, Mizoroki T, Yamashita S, Murayama M, Park JM, Saito Y, Murayama S, Takashima A. Molecular chaperone-mediated tau protein metabolism counteracts the formation of granular tau oligomers in human brain. J Neurosci Res. 2007;85:3098–3108. doi: 10.1002/jnr.21417. [DOI] [PubMed] [Google Scholar]
- 33.Sarkar M, Kuret J, Lee G. Two motifs within the tau microtubule-binding domain mediate its association with the hsc70 molecular chaperone. J Neurosci Res. 2008;86:2763–2773. doi: 10.1002/jnr.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang WS, Reiman EM, Valla J, Dunckley T, Beach TG, Grover A, Niedzielko TL, Schneider LE, Mastroeni D, Caselli R, Kukull W, Morris JC, Hulette CM, Schmechel D, Rogers J, Stephan DA. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105:4441–4446. doi: 10.1073/pnas.0709259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette C, Schmechel D, Alexander GE, Reiman EM, Rogers J, Stephan DA. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28:311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oz M, Lorke DE, Petroianu GA. Methylene blue and Alzheimer’s disease. Biochem Pharmacol. 2009;78:927–932. doi: 10.1016/j.bcp.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 39.Schirmer RH, Adler H, Pickhardt M, Mandelkow E. Lest we forget you--methylene blue…. Neurobiol Aging. 2011;32:2325 e2327–2316. doi: 10.1016/j.neurobiolaging.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Rousaki A, Miyata Y, Jinwal UK, Dickey CA, Gestwicki JE, Zuiderweg ER. Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J Mol Biol. 2011;411:614–632. doi: 10.1016/j.jmb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morishima Y, Murphy PJ, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor. hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J Biol Chem. 2000;275:18054–18060. doi: 10.1074/jbc.M000434200. [DOI] [PubMed] [Google Scholar]
- 42.Billecke SS, Draganov DI, Morishima Y, Murphy PJ, Dunbar AY, Pratt WB, Osawa Y. The role of hsp90 in heme-dependent activation of apo-neuronal nitric-oxide synthase. J Biol Chem. 2004;279:30252–30258. doi: 10.1074/jbc.M403864200. [DOI] [PubMed] [Google Scholar]
- 43.Schumacher RJ, Hurst R, Sullivan WP, McMahon NJ, Toft DO, Matts RL. ATP-dependent chaperoning activity of reticulocyte lysate. J Biol Chem. 1994;269:9493–9499. [PubMed] [Google Scholar]
- 44.Dickey CA, Dunmore J, Lu B, Wang JW, Lee WC, Kamal A, Burrows F, Eckman C, Hutton M, Petrucelli L. HSP induction mediates selective clearance of tau phosphorylated at proline-directed Ser/Thr sites but not KXGS (MARK) sites. FASEB J. 2006;20:753–755. doi: 10.1096/fj.05-5343fje. [DOI] [PubMed] [Google Scholar]
- 45.Moulick K, Ahn JH, Zong H, Rodina A, Cerchietti L, Gomes DaGama EM, Caldas-Lopes E, Beebe K, Perna F, Hatzi K, Vu LP, Zhao X, Zatorska D, Taldone T, Smith-Jones P, Alpaugh M, Gross SS, Pillarsetty N, Ku T, Lewis JS, Larson SM, Levine R, Erdjument-Bromage H, Guzman ML, Nimer SD, Melnick A, Neckers L, Chiosis G. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7:818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cook C, Gendron TF, Scheffel K, Carlomagno Y, Dunmore J, Deture M, Petrucelli L. Loss of HDAC6, a novel CHIP substrate, alleviates abnormal tau accumulation. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 48.Lastres-Becker I, Rub U, Auburger G. Spinocerebellar ataxia 2 (SCA2) Cerebellum. 2008;7:115–124. doi: 10.1007/s12311-008-0019-y. [DOI] [PubMed] [Google Scholar]
- 49.Brady GF, Galban S, Liu X, Basrur V, Gitlin JD, Elenitoba-Johnson KS, Wilson TE, Duckett CS. Regulation of the copper chaperone CCS by XIAP-mediated ubiquitination. Mol Cell Biol. 2010;30:1923–1936. doi: 10.1128/MCB.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 52.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22:2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 53.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 54.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 55.Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. Abacus: a computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics. 2011;11:1340–1345. doi: 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK) J Biol Chem. 2010;285:21282–21291. doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang L, Bertelsen EB, Wisen S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 59.Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barghorn S, Biernat J, Mandelkow E. Purification of recombinant tau protein and preparation of Alzheimer-paired helical filaments in vitro. Methods Mol Biol. 2005;299:35–51. doi: 10.1385/1-59259-874-9:035. [DOI] [PubMed] [Google Scholar]
- 61.Miyata Y, Chang L, Bainor A, McQuade TJ, Walczak CP, Zhang Y, Larsen MJ, Kirchhoff P, Gestwicki JE. High-throughput screen for Escherichia coli heat shock protein 70 (Hsp70/DnaK): ATPase assay in low volume by exploiting energy transfer. J Biomol Screen. 2010;15:1211–1219. doi: 10.1177/1087057110380571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.