Abstract
Ascorbate peroxidases are important enzymes that detoxify hydrogen peroxide within the cytosol and chloroplasts of plant cells. To better understand their role in oxidative stress tolerance, the transcriptional regulation of the apx1 gene from Arabidopsis was studied. The apx1 gene was expressed in all tested organs of Arabidopsis; mRNA levels were low in roots, leaves, and stems and high in flowers. Steady-state mRNA levels in leaves or cell suspensions increased after treatment with methyl viologen, ethephon, high temperature, and illumination of etiolated seedlings. A putative heat-shock cis element found in the apx1 promoter was shown to be recognized by the tomato (Lycopersicon esculentum) heat-shock factor in vitro and to be responsible for the in vivo heat-shock induction of the gene. The heat-shock cis element also contributed partially to the induction of the gene by oxidative stress. By using in vivo dimethyl sulfate footprinting, we showed that proteins interacted with a G/C-rich element found in the apx1 promoter.
AOS such as superoxide radicals, hydrogen peroxide, and hydroxyl radicals are continuously formed in aerobic organisms. AOS cause oxidative damage of cell constituents (Halliwell and Gutteridge, 1989), and their involvement in different kinds of biotic or abiotic stresses such as chilling, drought, environmental pollution (ozone, sulfur dioxide), and pathogen attack is well documented (Bowler et al., 1992).
Plants have both nonenzymatic and enzymatic AOS-detoxification systems. Several small antioxidant molecules, such as ascorbic acid, glutathione, α-tocopherol, carotenoids, and flavonoids, can quench all kinds of AOS (Halliwell and Gutteridge, 1989). Because of the simple chemical nature of the quenching reactions, some of these molecules have to accumulate to very high concentrations within the cells to be effective (Loewus and Loewus, 1987). Several enzymes can efficiently detoxify AOS; whereas superoxide radicals are disproportionately detoxified by superoxide dismutases, hydrogen peroxide is destroyed by catalases and different kinds of peroxidases (Bowler et al., 1992).
A major hydrogen peroxide-detoxifying system in plant chloroplasts and cytosol is the so-called ascorbate-glutathione cycle, in which APXs are the key enzymes (Asada, 1994). So far, APX activity has been found in plants, algae, and some cyanobacteria, and it has also recently been identified in insects (Mathews et al., 1997). APX has been purified and characterized from several plant species. Functionally and structurally distinct from the typical peroxidase superfamily, APX is unique in having a high specificity toward ascorbic acid as an electron donor (for review, see Asada, 1994). The APXs are a fast-growing family of proteins. Several different protein isoforms are known: two soluble cytosolic forms, several cytosolic forms bound to membranes of glyoxisomes and peroxisomes, and two chloroplastic forms, one of which is stromal and the other is thylakoid bound (for review, see Jespersen et al., 1997). Analysis of the expressed sequence tag databases has allowed the distinction of as many as seven different types of APXs in Arabidopsis (Jespersen et al., 1997). Recently, APX activity in pea mitochondria has been reported (Jiménez et al., 1997).
APX is believed to be involved in the detoxification of photoproduced hydrogen peroxide. The activities of the cytosolic and chloroplastic APX increase in carotenoid-less mustard seedlings exposed to light because of the increased rate of AOS photoproduction (Thomsen et al., 1992). Furthermore, APX activity has been shown to increase in response to a number of stress conditions, such as drought (Smirnoff and Colombé, 1988; Tanaka et al., 1990; Mittler and Zilinskas, 1994), air pollution (Tanaka et al., 1985; Mehlhorn et al., 1987; Conklin and Last, 1995; Kubo et al., 1995; Rao et al., 1996), high light intensity combined with chilling (Schöner and Krause, 1990) or deficiency in microelements (Cakmak and Marschner, 1992), iron stress (Vansuyt et al., 1997), excessive light (Karpinski et al., 1997), UV-B light (Rao et al., 1996), and salt stress (Lopez et al., 1996). In some cases posttranslational components are involved in the regulation of the APX activity (Mittler and Zilinskas, 1994; Lopez et al., 1996). However, increases in APX activity are usually accompanied by transcriptional activation of the gene. In spite of this fact, surprisingly little is known about the mechanisms underlying this regulation or about the promoter organization of the APX genes.
At present the sequences of three APX genes are available: two from Arabidopsis (apx1 and apx2; Kubo et al., 1993; Santos et al., 1996) and one from pea (apx1; Mittler and Zilinskas, 1992). All of them code for cytosol-soluble isoforms of APX that are highly homologous to each other (approximately 80% amino acid identity). However, apx2 from Arabidopsis seems to differ from the Arabidopsis apx1 gene as well as from the pea apx1 gene in many respects, particularly in the induction pattern, and represents another isoform of cytosol-soluble APX (Santos et al., 1996; Karpinski et al., 1997).
In pea apx1 gene expression is induced by oxidative stress, heat, and drought stress and by ethylene and ABA (Mittler and Zilinskas, 1992, 1994). In Arabidopsis the expression of the apx1 gene was shown to be induced by ozone, sulfur dioxide, and excessive light (Kubo et al., 1995; Karpinski et al., 1997). Sequence comparison of the promoter of the pea and Arabidopsis apx1 genes has revealed the presence of only one region of high homology that is located around the TATA box and contains several sequence motifs characteristic of the HSE identified in promoters of all heat-shock-inducible genes.
The heat-shock response is a general reaction of all organisms after exposure to elevated temperatures and is characterized by a rapid synthesis of a set of specific heat-shock proteins (for review, see Nover, 1991). However, the heat-shock response can be induced by other stresses, particularly by oxidative stress (Morgan et al., 1986; Courgeon et al., 1988; Liu and Thiele, 1996; McDuffee et al., 1997). Conversely, heat shock can result in an oxidative stress, which induces genes involved in the oxidative stress defense (Morgan et al., 1986).
We present an analysis of the Arabidopsis apx1 gene promoter. Data from in vitro interactions of a tomato (Lycopersicon esculentum) HSF with the apx1 promoter and mutational analysis confirmed that the HSE is responsible for the heat-shock induction of the gene and partially contributes to the induction by oxidative stress. Other putative regulatory cis elements were characterized by in vivo footprinting.
MATERIALS AND METHODS
Plant Material
Plants of Arabidopsis ecotype Columbia were grown in soil at 20°C under a 16-h light/8-h dark regime. The Arabidopsis suspension culture was grown on a rotary shaker in B5 Gamborg medium (GIBCO-BRL) supplemented with 1 mg L−1 2,4-D. For the expression analysis, plants were sprayed with 10−6 m methyl viologen or with 15 mm ethephon. For the light-induction experiments, 5- to 6-d-old etiolated seedlings were exposed to light. Protoplasts were prepared from soil-grown plants essentially as described previously (Altmann et al., 1992).
Screening of Arabidopsis cDNA and Genomic Libraries
Phages (5 × 105) of an Arabidopsis cDNA library in λgt10 were screened with a probe prepared from a spinach apx cDNA (S. Kushnir, unpublished results). Twelve positive plaques were purified to homogeneity and the two longest cDNAs were sequenced on both strands.
To clone the apx1 gene, 3 × 104 plaques of an Arabidopsis ecotype Landsberg erecta genomic library in λ Charon 35 were screened with the Arabidopsis apx1 cDNA probe. After the two overlapping clones, APAG7 and APAG3, were isolated and mapped, the apx1 gene sequence was determined on both strands, including 1.5 kb of the promoter sequence.
RNA Analysis
RNA was extracted from Arabidopsis organs according to the method of Shirzadegan et al. (1991). RNA gel-blot analysis of glyoxylated RNA was performed according to standard procedures (Ausubel et al., 1993). To ensure equal loading of RNA, blots were stained with methylene blue (Ausubel et al., 1993). Hybridizations of RNA blots were according to the method of Church and Gilbert (1984). Poly(A+) mRNA was prepared using oligo(dT)-beads (Dynabeads, Dynal, Oslo, Norway) as recommended by the manufacturer. The transcription start was mapped by T4 DNA polymerase primer extension mapping (Hu and Davidson, 1986) and by the PCR amplification of 5′ ends of the apx1 mRNA (Troutt et al., 1992).
HSF Expression
A tomato (Lycopersicon esculentum) HSF cDNA was amplified by PCR from tomato cDNA using Pfu polymerase (Stratagene) and the primers CCAACTTCACCTCAGTTACAAACC and GGATCCCATATGTCGCAAAGAACAGCGCCGGCG. Primers were designed according to the known cDNA sequence of the tomato HSF B2 (Scharf et al., 1990; Nover et al., 1996). The cloned PCR fragment was sequenced and the encoded protein was identical to HSF B2 except for a few differences in amino acid sequence (data not shown). The tomato HSF B2 cDNA was subcloned in the pQE8 (Qiagen, Chatsworth, CA) and pET11a (Novagen, Madison, WI) expression vectors to produce an in-frame amino-terminal fusion with a stretch of six His residues (the fusion protein referred to as hHSF) and with a T7 immunotag (the fusion protein referred to as tHSF), respectively. The expression of proteins was in Escherichia coli strains M15 (Qiagen) for pQE8 or BL21 for pET11a. The protein synthesis was induced in logarithmically grown E. coli cultures in Luria-Bertani medium (37°C) by the addition of 1 mm isopropyl-β-d-thiogalacto-pyranoside, and the cultures were grown for an additional 4 to 5 h at different temperatures. HSF active in DNA binding was obtained only when E. coli cells were grown at 25°C to 28°C but not at 37°C. Most of the hHSF fusion protein was found in inclusion bodies regardless of the temperature at which cells were grown, whereas the tHSF was detected in soluble form and with high activity in DNA binding.
Both HSF proteins were purified under native conditions. The hHSF was purified as recommended by the manufacturer on a nickel-chelating column (Qiagen), dialyzed against TM buffer (Kroeger et al., 1993), and stored at −70°C.
For the tHSF preparation 200 mL of induced E. coli culture was used. The tHSF was purified as follows: after induction, cells were harvested by centrifugation and the pellet was resuspended in TM buffer supplemented with 20 mg mL−1 leupeptin. Cells were disrupted by sonication and the homogenate was cleared by centrifugation in a SW27 rotor (Beckman) for 30 min at 4°C. The supernatant was mixed with 5 mL of CM Sephadex G-50 (Pharmacia, Uppsala, Sweden) equilibrated in the same buffer, and proteins were allowed to absorb for 30 min at 4°C. Sephadex was then packed in a small column and washed with TM buffer. Bound proteins were eluted by TM buffer containing 0.4 m KCl. The eluate was diluted 2-fold with TM buffer, applied to a heparin-Sepharose column (5 mL) equilibrated with TM buffer containing 0.2 m KCl, washed, and subsequently eluted with 0.4 m KCl in TM buffer. At this step, the tHSF was approximately 80% pure and the major contaminating proteins were truncated forms of the recombinant protein that were efficiently removed by gel filtration in TM buffer on an Ultrahydrogel 500 column (7.8 mm × 30 cm [Waters]). The tHSF peak fractions were separated into aliquots and stored at −70°C.
Gel-Shift Analysis and in Vitro Footprinting
Two apx1 promoter fragments were amplified by PCR and subcloned in pBluescript KS+ (Stratagene). Fragment A contained sequences from −274 to +42 (primers CCCTCCACACGAAGCATGTATCC and CTGGAGAAATGCGAGTGG), and fragment B contained sequences from −246 to −499 (primers ATTGGAGGATACATGCTTCGTGTGG and GGTGAGAAACCTAATAACACTG). The 3′ and 5′ end-labeled fragments were prepared according to standard procedures (Ausubel et al., 1993).
DNase I footprinting was done essentially as described previously (Kroeger et al., 1993). Labeled probe (20,000-50,000 cpm), 1 μg of poly(dIdC)·poly(dIdC), and tHSF were incubated in binding buffer (50 mm Tris-HCl, pH 7.3, 50 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 5% Suc, and 5% glycerol) in a total volume of 20 μL. After the sample was incubated for 20 to 30 min at room temperature, 2 μL of a Ca/Mg mixture (10 mm CaCl2 and 10 mm MgCl2) and 2 μL of DNase I (Pharmacia) dilution were added to the binding buffer. After 1 min of DNase I digestion, samples were processed as described previously (Kroeger et al., 1993). The same labeled fragment was chemically cleaved at the G and A bases following standard procedures (Ausubel et al., 1993) to provide molecular mass markers.
For the electrophoretic mobility-shift assays, labeled fragments were incubated with tHSF or hHSF under the same conditions as for the DNase I footprinting, and DNA-protein complexes were resolved by native 5% PAGE in 0.5× Tris-borate buffer at room temperature. Variables were the amount of nonspecific competitor DNA, the type of competitor DNA (salmon-sperm), and the presence or absence of specific competitors (fragments A and B).
apx1 Promoter-gus Fusion Construction, Site-Directed Mutagenesis of HSE, and Plant Transformation
Fusion at the ATG start codon of the gus reporter gene was obtained after site-directed mutagenesis by PCR. The apx1 gene-specific primer CCATGGTAGCTAAGCTCTGGAACAA was used to introduce a NcoI site at the ATG start codon. The amplified 1.2-kb fragment was digested with NcoI and HindIII and subcloned into NcoI plus HindIII-cleaved pGUS1 plasmid (Peleman et al., 1989). apx1 promoter-gus fusions were subcloned as EcoRI-HindIII fragments into the binary vector pGSV6 (Plant Genetic Systems N.V., Gent, Belgium). Binary vectors were transformed into Agrobacterium tumefaciens C58C1RifR (pGV2260) according to the method of De Block et al. (1987). Arabidopsis C24 was transformed according to the method of Valvekens et al. (1988).
To introduce point mutations into the HSE, site-directed mutagenesis was done by PCR. The HSE-specific primer CAGATCTACCATAACATTATCATTAATGAC with two mutated bases was used as a mutagenic PCR primer, whereas the above-mentioned apx1 gene-specific primer generating the NcoI site at the ATG codon was utilized as the second PCR primer. The wild-type sequence in the apx1 promoter-gus fusion was replaced with an amplified genomic fragment with mutated HSE using BglII and NcoI sites.
To study the induction of the gene with mutated HSE, 2-week-old, in vitro-grown Arabidopsis seedlings of four independent transgenic lines transformed with the mutant or wild-type apx1 promoter-gus fusion were pooled and infiltrated with 10−5 m methyl viologen. To induce heat shock, plants were grown in a chamber (Weiss Klimatechnik GmbH, Reiskirchen, Germany) at 22°C and then subjected to single-step increases in temperature to 37°C.
In Vivo Footprinting
The ligation-mediated PCR version of in vivo footprinting was used (Mueller and Wold, 1989; Pfeifer et al., 1989). Arabidopsis protoplast suspensions (10 mL) were treated with 0.2% DMS in W5 salt solution (Altmann et al., 1992) at room temperature for 5 and 10 min. Protoplasts were then pelleted and washed with 10 mL of ice-cold W5 solution. The protoplast pellet was lysed with a lysis buffer and DNA was purified as described above. The DNA samples were processed as described previously (Mueller and Wold, 1989; Pfeifer et al., 1989). Three specific overlapping oligonucleotides (CCTTAGTCCAATTGGGATCTTCGCC at position −415, TTGGGATCTTCGCCTGCGTGAGACG, and CGCCTGCGTGAGACGCGTCACACCTGCG) were used in the ligation-mediated PCR footprinting of the apx1 promoter. The adaptor for the ligation was prepared by annealing two oligonucleotides (GCGGTGACCCGGGAGATCTGAATTC and GAATTCAGATC) (Mueller and Wold, 1989).
RESULTS
Previously, a cDNA and a gene encoding the cytosolic APX from Arabidopsis were cloned (Kubo et al., 1992, 1993). The independent isolation, and sequencing of the gene, and mapping of the transcription start confirmed the published data (data not shown).
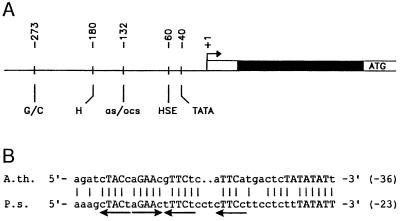
A comparison of the promoter sequences showed that the Arabidopsis apx1 gene shared only one region (approximately 44 bp) with strong homology to the promoter of the pea apx1 gene (80% identity over 44 bp; Fig. 1B). This region, located around the TATA box, contains a putative HSE. The promoter sequence of another APX gene from Arabidopsis, apx2, had no extended sequence homology with the promoter of apx1. However, two HSE-like sequences were also present in the promoter of the apx2. A first HSE was identified by Santos et al. (1996) at position −293, but careful sequence inspection exposed a second HSE at position −205.
Figure 1.
A, Schematic representation of the apx1 promoter organization. ATG indicates the initiating translation codon. An intron located in the 5′ untranslated region and the transcription start are marked by a black box and an arrowhead, respectively. The positions (in bp) of the putative and identified cis elements as related to the transcription start are indicated above the line. as/ocs, Sequences similar to the cis elements as-1 from the 35S cauliflower mosaic virus promoter (Benfey and Chua, 1990) and ocs from the octopine synthase promoter (Singh et al., 1990), which are recognized by a member of the b-Zip family of DNA-binding proteins; H, sequence similar to the H box, recognized by proteins from the myb family (Sablovski et al., 1994); G/C, G- and C-rich sequence for which in vivo footprints have been identified in this study. B, Comparison of the heat-shock elements from pea (P.s.) and Arabidopsis (A.th.) apx1. Sequences matching the nGAAn, the basic 5-bp HSE motif, are indicated in uppercase letters. The two central motifs are in reverse orientation and perfectly match requirements for the minimal HSF-binding motif nGAAnnTTCn. They are flanked by two other motifs, of which the upstream motif has one tolerated substitution and the downstream motif perfectly fits the nGAAn consensus except that it is shifted on 1 bp and has a direct instead of a reverse orientation to the adjacent 5-bp motif. Orientations of the nGAAn-like motifs are indicated by arrows. The sequence of the TATA box is shown in bold uppercase letters.
Computer analysis of the Arabidopsis apx1 gene also revealed the presence of two other elements, the H box and the as-1/ocs box, which are possibly recognized by Myb and b-Zip proteins, respectively (Fig. 1A). The as-1/ocs-like motifs were also found in the pea apx1 promoter and were similar to the xenobiotic responsive element that is recognized by AP-1-like b-ZIP transcription factors in mammalian cells (Mittler and Zilinskas, 1992).
Analysis of apx1 mRNA Levels in Response to Stress
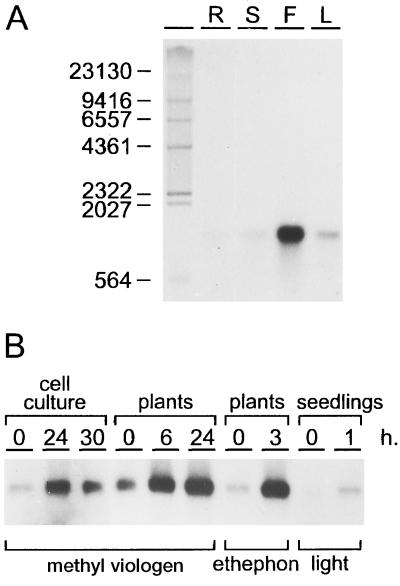
Expression analysis was undertaken to characterize the apx1 gene. A single transcript of approximately 1080 nucleotides hybridizing to the apx1 probe could be detected by RNA gel-blot analysis of total Arabidopsis RNA. Figure 2A shows that the apx1 mRNA of Arabidopsis was present in all tested organs, with the highest level in flowers and lower levels in leaves, stems, and roots.
Figure 2.
RNA gel-blot analysis of APX mRNA. A, Size and levels of apx1 mRNA in roots (R), stems (S), flowers (F), and leaves (L) of soil-grown Arabidopsis. Labeled and glyoxylated λHindIII fragments were loaded to evaluate the size of the mRNA, which is indicated in bases. B, The effect of methyl viologen (10−6 m), ethephon (15 mm), and light on the apx1 mRNA levels of in vitro-grown cells, mature plants, and etiolated seedlings, respectively. Hours after treatment are given at the top of the lanes.
Methyl viologen is commonly used in experiments to induce oxidative stress. Apx1 mRNA levels increased within a few hours after Arabidopsis plants were sprayed with this herbicide (Fig. 2B). No visible damage of the leaves was observed 24 h after 10−6 m methyl viologen was sprayed. Elevated apx1 mRNA levels could also be detected after treatment of an Arabidopsis cell-suspension culture with methyl viologen (Fig. 2B).
APX activity was shown to increase in plants after ethylene treatment (Mehlhorn et al., 1987). To determine whether ethylene had an effect on the apx1 mRNA levels, Arabidopsis plants were sprayed with a 15 mm solution of ethephon, which releases ethylene after decomposition. Figure 2B shows that such a treatment led to a severalfold increase of the apx1 mRNA within 3 h. Furthermore, light had positive effects on the amount of the apx1 mRNA in etiolated Arabidopsis seedlings (Fig. 2B).
Because the pea apx1 gene was induced by heat, and the putative HSE was present in the apx1 gene promoter of both pea and Arabidopsis genes, we analyzed the expression of the apx1 gene after heat shock and found that the steady-state Arabidopsis apx1 mRNA level was also induced by heat treatment (see below).
In Vitro Interactions of the HSF with the apx1 promoter
Heat-shock induction of the gene strongly suggested that the putative HSE might be responsible for the induction. A necessary condition for the heat-shock-activated transcription was a binding of the HSF trimer(s) to the sequence of HSE (for review, see Wu, 1995).
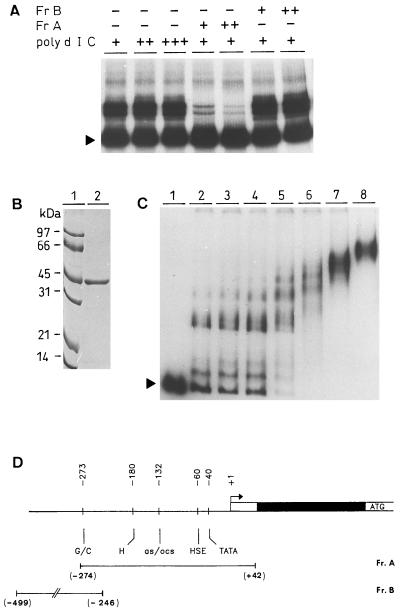
In vitro binding assays were performed to obtain evidence for the possible binding of an HSF to the putative HSE. The full-length cDNA encoding the tomato HSF B2 (Scharf et al., 1990; Nover et al., 1996) was cloned after PCR amplification, and the encoded HSF was expressed, albeit at low amounts, in the soluble fraction as hHSF in E. coli. The interaction of the hHSF with apx1 promoter fragments was analyzed by an electrophoresis mobility-shift assay. As shown in Figure 3A, incubation of the 316-bp fragment containing the HSE of the apx1 promoter (fragment A) with the hHSF resulted in the formation of DNA-protein complexes that were insensitive to increasing amounts of nonspecific competitor nucleic acids. This hHSF-binding activity could be competed out by only a 50- and 100-fold excess of the same unlabeled fragment but not with fragment B from the apx1 promoter.
Figure 3.
Expression and analysis of DNA-binding activities of the HSF. A, Electrophoresis mobility-shift assay of the DNA-protein complexes between hHSF and fragment A from the apx1 promoter. Labeled fragment A (40,000 cpm; see Methods) was incubated with hHSF in the presence of the nonspecific DNA competitor poly(dIdC)·poly(dIdC) in increasing amounts (0.5, 1.0, and 2.0 μg) with unlabeled, cold fragment A (Fr A) at 20 ng (+) and 100 ng (++) as a specific competitor, and with fragment B (Fr B) at 20 ng (+) and 100 ng (++) as an unspecific competitor. After 30 min of incubation, DNA-protein complexes were resolved by 5% PAGE and the gel was dried and exposed to radiographic film. B, Purification of tHSF. Approximately 2 μg of the tHSF (lane 2) after the gel-filtration step was mixed with loading buffer, boiled, and resolved by the denatured 12% SDS-PAGE aside a molecular mass marker (lane 1). Molecular masses are indicated on the left in kD. C, Saturation binding of the tHSF to the fragment A from apx1 promoter. Fragment A was incubated with increasing amounts of the tHSF. Lanes 2 to 8 contain 1, 5, 10, 50, 100, 200, and 300 ng of tHSF, respectively, in the binding buffer in the presence of poly(dIdC)·poly(dIdC) as a nonspecific competitor. The DNA-protein complexes were resolved by native PAGE. In lane 1, no protein was added. Arrowheads in A and C indicate migration of the free probe. D, Schematic representation of fragments A and B used in the experiment with respect to the apx1 sequence.
To optimize the expression of the HSF we expressed it as an amino-terminal fusion in the pET11a plasmid in E. coli BL21 (see Methods). The expressed protein (tHSF), which was mainly present in the soluble fraction, was purified to near homogeneity by three chromatography steps (Fig. 3B). In a dilution series of tHSF, complete saturation of the binding sites in the apx1 promoter could be reached (Fig. 3C). Although in vivo HSF binds DNA as a trimer (Scharf et al., 1990; Lis and Wu, 1993), trimerization increases only the affinity of HSF to DNA, and only the monomeric HSF and the DNA-binding domain of the HSF are able to specifically bind DNA (Flick et al., 1994). Retention time of the tHSF peak elution in the HPLC gel-filtration step indicated that the tHSF was in a monomeric form. Because the tHSF protein existed as a monomer in the solution, and several binding sites for the protein were present in a HSE, we observed the formation of several DNA-protein complexes.
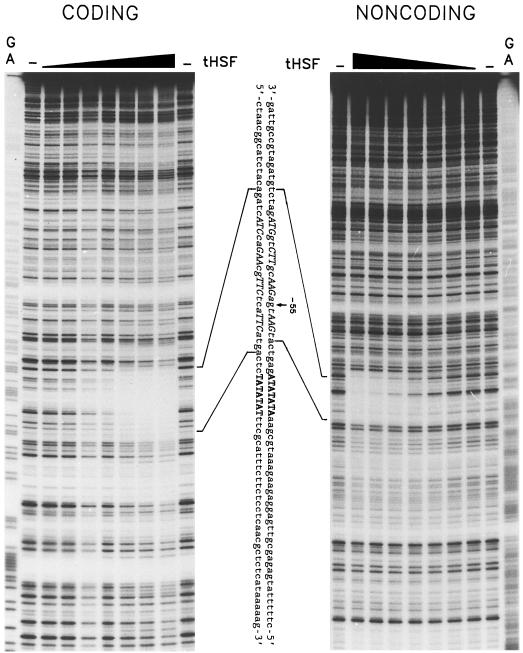
To prove that the interaction of tHSF is specific and to delineate the HSE in the apx1 gene, we performed in vitro DNase I-footprinting analysis. As shown in Figure 4, incubation of the apx1 promoter fragment with increasing amounts of tHSF resulted in the complete protection of a 30-bp sequence on both strands. This sequence contained two 5-bp core HSE motifs arranged in reverse orientation, aGAAcgTTCt, which perfectly fit a defined minimal unit nGAAnnTTCn necessary for HSF trimer binding (Perisic et al., 1989). Two other similar 5-bp motifs were found on both sides of the central inverted repeat, which served as additional binding sites for the HSF; it has been shown that at least three such 5-bp basic motifs are necessary for high-affinity binding of the HSF (Perisic et al., 1989; Kroeger et al., 1993).
Figure 4.
In vitro DNase I-footprinting analysis of the tHSF interactions with apx1 promoter. Fragment A was labeled at the 3′ (coding strand) or 5′ (noncoding strand) ends. Labeled fragments were incubated with increasing amounts of tHSF (1, 5, 10, 50, 100, 200, and 300 ng) or alone in the binding buffer, followed by DNase I digestion and separation on a 5% sequencing gel. The fragment was chemically cleaved at the A/G bases to provide a molecular mass marker. Triangles over the lanes indicate increasing concentrations of tHSF. The sequence of the apx1 promoter with the putative HSE is in between the two sequencing gels. Regions protected from DNase I digestion are indicated by bars. All sequences that fit the nGAAn 5-bp HSE motif are indicated by italic uppercase letters, and the expected TATA box is indicated by boldface uppercase letters.
Stress Responsiveness of the apx1 Promoter Mutated in HSE
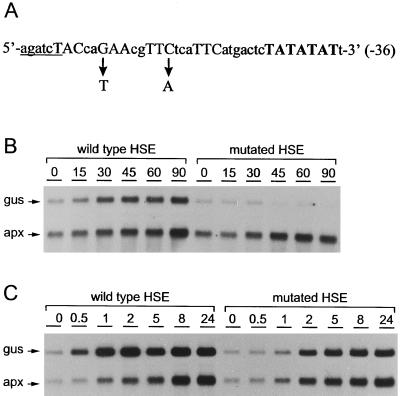
To prove that the putative HSE is involved in the regulation of apx1 gene expression in vivo, we designed an apx1 promoter-gus translational fusion at the ATG start codon. This construct contained 1.2 kb of apx1 promoter sequence and was used to introduce point mutations into the putative HSE. The HSE mutant version of the apx1 promoter was prepared by introducing two nucleotide replacements into the central inverted repeat aGAAcgTTCt, which then read aTAAcgTTAt (Fig. 5A). The replacement of the G base in the nGAAn basic HSE motif is known to have the most deleterious effect on the HSF binding in vitro (Fernandes et al., 1994) and dramatically affects heat-shock inducibility in vivo (Barros et al., 1992).
Figure 5.
Study of the stress responsiveness of the apx1 promoter mutated in HSE. A, Site-directed mutagenesis of the putative HSE. Sequences matching the nGAAn consensus are indicated in uppercase letters. The arrows point from the nucleotides that have been changed to the nucleotides introduced instead. The BglII site used for cloning is underlined and the TATA box is indicated in bold uppercase letters. B, Study of the apx1 promoter with mutated HSE in response to heat shock. In vitro-grown, 2-week-old transgenic Arabidopsis seedlings expressing apx1(HSEmut) and apx1(HSEwt) were shifted from 22°C to 37°C, and RNA was extracted from the whole seedlings at different times and analyzed by gel-blot hybridization using apx1- and gus-specific probes. The times in minutes are indicated at the top of the lanes. C, Effect of methyl viologen treatment on the apx1 promoter function with mutated HSE. The same plant material as in B was infiltrated with 10−5 m methyl viologen solution. RNA was extracted at different times after the treatment and analyzed by gel-blot hybridization using mixed apx1- and gus-specific probes. The times in hours are indicated at the top of the lanes. The difference between the endogenous apx1 and gus mRNA levels reflects the difference between specific radioactivities of the probes used rather than the levels of real mRNA. Infiltration of the plants with water was also used as a control and no increase in the mRNA levels was observed (data not shown).
The promoter-gus fusions were subcloned into a binary vector and stably integrated into the genome of Arabidopsis C24. The transgenic plants were monitored for their ability to express the apx1(HSEmut) mRNA under normal and stress conditions. Transgenic plants expressing the apx1(HSEwt) were used as a control. Because the size of the gus fusion mRNA was substantially higher than the apx1 messenger, we could easily distinguish both the fusion and endogenous apx1 expression on the same RNA gel-blot membrane by using corresponding mixed probes.
Under normal conditions the basal level of the apx1(HSEmut) and apx1(HSEwt) mRNAs seemed to be similar (Fig. 5B). However, a dramatic difference in expression was found after the heat-shock treatment. The levels of the endogenous apx1 and apx1(HSEwt) mRNAs clearly increased, as discussed above. In contrast, the apx1(HSEmut) was repressed, with mRNA being almost undetectable after 1.5 h of the onset of heat shock. These data confirm that the putative HSE is the functional HSE responsible for the heat-shock induction of the apx1 gene.
We also analyzed the oxidative stress response of apx1(HSEmut). Results of the induction with methyl viologen in the same system are shown in Figure 5C. The promoter containing the mutant HSE was still induced, suggesting that this type of apx1 induction depends mainly on some other cis element(s) to be identified further. However, quantitative analysis of the intensity of the bands using phosphor imaging revealed that the induction by methyl viologen of the apx1(HSEmut) was weaker, ranging from 70% of the apx1(HSEwt) level during the first 2 h of the treatment to 30% of the apx1(HSEwt) level by the end of the treatment. The high reproducibility of the induction pattern of the endogenous apx1 in the transgenic plants expressing both the apx1(HSEmut) and apx1(HSEwt) ruled out the possibility that the observed differences were due to variability in the methyl viologen treatments.
In Vivo Footprinting of the apx1 Promoter
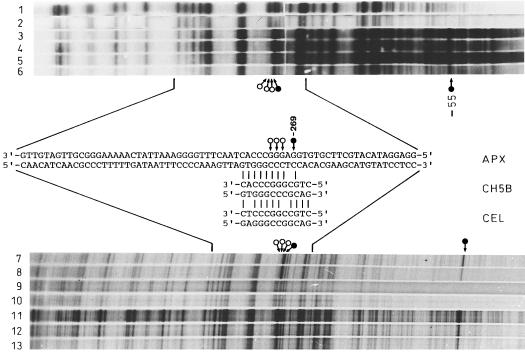
As a second approach to gaining insight into the promoter of the apx1 gene, we analyzed DNA-protein interactions in the proximal part of the apx1 promoter using ligation-mediated PCR-DMS in vivo footprinting (Mueller and Wold, 1989; Pfeifer et al., 1989). The G ladder was visualized using two different modifications of this method, blotting with subsequent DNA hybridization (Pfeifer et al., 1989) and extension of 32P-labeled primer (Mueller and Wold, 1989). Figure 6 shows the results obtained with DNA extracted from DMS-treated Arabidopsis leaf protoplasts compared with DMS-treated naked DNA. As confirmed by the two methods and several independent experiments, G at −273, −272, and −271 on the noncoding strand were protected from DMS modification and G at −269 was hypersensitive to DMS. Similar G/C-rich sequences were found in several plant promoters that had homology to the ethylene-inducible bean chalcone synthase and avocado cellulase promoters (Fig. 6). A strong DMS hypersensitivity at G −55 was detected close to the TATA box in the HSE sequence, confirming our results on the functionality of HSE in the apx1 promoter.
Figure 6.
In vivo footprinting analysis of the proximal part (approximately 400 bp) of the apx1 promoter. Lanes 1, 2, 9, and 10 are DMS-treated naked genomic Arabidopsis DNA from two independent samples. Reactions were performed with DNA samples from the DMS-treated protoplasts. The samples loaded in lanes 3 to 6, 7 and 8, and 10 to 13 were derived from two independent preparations and were loaded in all other lines. Protoplasts were treated with 0.2% DMS for 5 min (lanes 3, 5, 7, and 8) and 10 min (lanes 4, 6, 11, 12, and 13). The footprinting was done by the hybridization approach (Pfeifer et al., 1989) or by extension of a labeled primer (top and bottom autographs, respectively; Muller and Wold, 1989). Arrows with open circles indicate complete protection from the DMS modification, and arrows with filled circles indicate the G-269 and G-55 hypersensitive to DMS. Between the autoradiographs, the corresponding sequences of the apx1 gene promoter are shown and the protected bases on the noncoding strand are pointed out as above. Below, an apx1 promoter sequence, part of the promoter sequences of the bean chalcone synthase gene CH5B (Broglie et al., 1989), and the avocado cellulase gene CEL (Cass et al., 1990) are shown. The vertical bars show the similarity between these sequences.
DISCUSSION
APXs are thought to play an essential role in protecting plants from oxidative stress. It was shown previously that steady-state transcript levels of the pea apx1 gene strongly increase after treatment of plants with ethephon, methyl viologen, heat shock, and drought stress (Mittler and Zilinskas, 1992, 1994). In agreement with this observation, we found that the steady-state mRNA levels of the apx1 from Arabidopsis are up-regulated by methyl viologen, ethephon, and heat shock. The heat-shock response is very fast; a significant increase in transcript levels is seen already after 15 min. The observed changes in apx1 mRNA levels after various environmental stimuli indicate that the transcriptional activation of the apx1 gene might be an important, although not the sole (as shown for the pea apx1 [Mittler and Zilinskas, 1992, 1994]) control step leading to higher APX activity in plants under oxidative stress.
As an initial step in the analysis of the apx1 promoter, we studied a putative HSE in more detail for at least two reasons. First, the putative HSE is the sole conserved sequence between promoters of pea and Arabidopsis apx1, implying its importance for the promoter activity. Moreover, the promoter of the other recently cloned apx gene from Arabidopsis, apx2, also contains the two putative HSEs (Santos et al., 1996; our data). Second, the heat-shock response is known to be induced not only by heat but also by a number of other environmental stimuli including oxidative stress. For example, it was shown that hydrogen peroxide treatment results in the induction of heat-shock proteins in Drosophila melanogaster (Courgeon et al., 1988); and Salmonella typhimurium cells (Morgan et al., 1986).
Moreover, oxidative stress imposed by menadione induced HSF phosphorylation and an HSF-dependent transcriptional activation of the yeast metallothionein cup1 gene (Liu and Thiele, 1996). Direct evidence has also been presented that proteins containing nonnative disulfate bonds as a result of oxidative stress can serve as a signal for the activation of the heat-shock response (McDuffee et al., 1997). Conversely, oxidative stress has been shown to play a major role in heat-induced cell death in yeast (Davidson et al., 1996). In parsley a small heat-shock protein was found to be induced by ozone and heat shock (Eckey-Kaltenbach et al., 1997). These few examples suggest that there is a considerable overlap in cellular processes induced by heat shock and oxidative stress and in the subsets of genes reacting to these stimuli. Therefore, we addressed the question of whether the HSE may play a key role in the regulation of apx1 gene expression.
In vitro analysis of the interaction between recombinant tomato HSF and the apx1 promoter confirmed that the apx1 HSE represents a functional HSF-binding site. Furthermore, the apx1 promoter with a mutated HSE loses inducibility and even becomes repressed under the heat-shock treatment. Nevertheless, the inducibility by methyl viologen is retained. A careful analysis of the phosphor images, however, demonstrated that the strength of apx1(HSEmut) transcription was lower than apx1(HSEwt) after the methyl viologen treatment. This finding may not be surprising in view of the indiscriminate chemical reactivity of AOS, which probably leads to the elicitation of a number of redundant and/or overlapping signaling pathways ending on different cis-acting elements in the apx1 promoter.
We carried out an additional in vivo analysis of the apx1 promoter aiming to find other putative cis-acting elements. For several “stress-related” plant genes it has been shown that relevant cis elements are located close to the TATA box, usually about 200 to 400 bp upstream from the transcription start. To study this sequence in detail, in vivo footprinting analysis of the apx1 promoter was started, which particularly focused on the first 300 bp. We found strong differences in DMS reactivity of G residues in a sequence (GTGGGCCCTCC) located at approximately −270 bp from the transcription start. This sequence is highly similar to the G/C-rich boxes found in the chlorophyll a/b-binding protein and alcohol dehydrogenase genes, although further experiments are necessary to show whether it is recognized either by the GCBP-1 (Olive et al., 1991) or the GC-1 (Schindler and Cashmore, 1990) transcription factors. A similar element is present in a bean chalcone synthase promoter, where it is essential for ethylene induction (Broglie et al., 1989), and a homologous element is also found in promoters of ethylene-regulated cellulase genes in avocado (Cass et al., 1990).
ACKNOWLEDGMENTS
We thank Alfredo Herrera-Estrella, Ranjan Perera, Wim Van Camp, Didier Hérouart, Marc Van den Bulcke, and Vladimir Mironov for many stimulating discussions and helpful comments in the course of this work. We are grateful to Wilson Ardiles, Raimundo Villarroel, and Jan Gielen for help with the DNA sequencing and computer sequence analysis, to Enric Belles-Boix for technical assistance, to Martine De Cock for preparing the manuscript, and to Vera Vermaercke, Rebecca Verbanck, and Karel Spruyt for excellent artwork.
Abbreviations:
- AOS
active oxygen species
- APX
ascorbate peroxidase
- apx1(HSEmut)
apx1 promoter-gus fusion mutated in HSE
- apx1(HSEwt)
wild-type apx1 promoter-gus fusion
- DMS
dimethyl sulfate
- hHSF
hexahistidine-HSF fusion
- HSE
heat-shock cis element
- HSF
heat-shock factor
- tHSF
T7 immunotag-HSF fusion
Footnotes
This research was supported by grants from the Belgian Program on Interuniversity Poles of Attraction (Prime Minister's Office, Science Policy Programming, no. 38) and the Vlaams Actieprogramma Biotechnologie (no. ETC 002), by the International Human Frontier Science Program (no. RG-43494M), and by the European Community Biotech Program as part of the Project of Technological Priority (1993–1996). D.I. is a Research Director of the Institut National de la Recherche Agronomique (France).
LITERATURE CITED
- Altmann T, Damm B, Halfter U, Willmitzer L, Morris P-C (1992) Protoplast transformation and methods to create specific mutants in Arabidopsis thaliana. In C Koncz, N-H Chua, J Schell, eds, Methods in Arabidopsis Research. World Scientific Publishing, Singapore, pp 310–330
- Asada K. Production and action of active oxygen species in photosynthetic tissues. In: Foyer CH, Mullineaux PM, editors. Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants. Boca Raton, FL: CRC Press; 1994. pp. 77–104. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1993) Current Protocols in Molecular Biology, Vol 1. Current Protocols, New York
- Barros MD, Czarnecka E, Gurley WB. Mutational analysis of a plant heat shock element. Plant Mol Biol. 1992;19:665–675. doi: 10.1007/BF00026792. [DOI] [PubMed] [Google Scholar]
- Benfey PN, Chua N-H. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science. 1990;250:959–966. doi: 10.1126/science.250.4983.959. [DOI] [PubMed] [Google Scholar]
- Bowler C, Van Montagu M, Inzé D. Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:83–116. [Google Scholar]
- Broglie KE, Biddle P, Cressman R, Broglie R. Functional analysis of DNA sequences responsible for ethylene regulation of a bean chitinase gene in transgenic tobacco. Plant Cell. 1989;1:599–607. doi: 10.1105/tpc.1.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I, Marschner H. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiol. 1992;98:1222–1227. doi: 10.1104/pp.98.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass LG, Kirven KA, Christoffersen RE. Isolation and characterization of a cellulase gene family member expressed during avocado fruit ripening. Mol Gen Genet. 1990;223:76–86. doi: 10.1007/BF00315799. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Last RL. Differential accumulation of antioxidant mRNAs in Arabidopsis thaliana exposed to ozone. Plant Physiol. 1995;109:203–212. doi: 10.1104/pp.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgeon A-M, Rollet E, Becker J, Maisonhaute C, Best-Belpomme M. Hydrogen peroxide (H2O2) induces actin and some heat-shock proteins in Drosophila cells. Eur J Biochem. 1988;171:163–170. doi: 10.1111/j.1432-1033.1988.tb13772.x. [DOI] [PubMed] [Google Scholar]
- Davidson JF, Whyte B, Bissinger PH, Schiestl RH. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5116–5121. doi: 10.1073/pnas.93.10.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block M, Botterman J, Vandewiele M, Dockx J, Thoen C, Gosselé V, Movva R, Thompson C, Van Montagu M, Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987;6:2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckey-Kaltenbach H, Kiefer E, Grosskopf E, Ernst D, Sandermann H., Jr Differential transcript induction of parsley pathogenesis-related proteins and of a small heat shock protein by ozone and heat shock. Plant Mol Biol. 1997;33:343–350. doi: 10.1023/a:1005786317975. [DOI] [PubMed] [Google Scholar]
- Fernandes M, Xiao H, Lis JT. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor–heat shock element interactions. Nucleic Acids Res. 1994;22:167–173. doi: 10.1093/nar/22.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick KE, Gonzalez L, Jr, Harrison CJ, Nelson HCM. Yeast heat shock transcription factor contains a flexible linker between the DNA-binding and trimerization domains. Implications for DNA binding by trimeric proteins. J Biol Chem. 1994;269:12475–12481. [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford, UK: Clarendon Press; 1989. [Google Scholar]
- Hu MC-T, Davidson N. Mapping transcription start points on cloned genomic DNA with T4 DNA polymerase: a precise and convenient technique. Gene. 1986;42:21–29. doi: 10.1016/0378-1119(86)90146-0. [DOI] [PubMed] [Google Scholar]
- Jespersen HM, Kjærsgård IVH, Østergaard L, Welinder KG. From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase. Biochem J. 1997;326:305–310. doi: 10.1042/bj3260305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, del Río LA, Sevilla F. Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 1997;114:275–284. doi: 10.1104/pp.114.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger PE, Sarge KD, Morimoto RI. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol Cell Biol. 1993;13:3370–3383. doi: 10.1128/mcb.13.6.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Saji H, Tanaka K, Kondo N. Genomic DNA structure of a gene encoding cytosolic ascorbate peroxidase from Arabidopsis thaliana. FEBS Lett. 1993;315:313–317. doi: 10.1016/0014-5793(93)81185-3. [DOI] [PubMed] [Google Scholar]
- Kubo A, Saji H, Tanaka K, Kondo N. Expression of Arabidopsis cytosolic ascorbate peroxidase gene in response to ozone or sulfur dioxide. Plant Mol Biol. 1995;29:479–489. doi: 10.1007/BF00020979. [DOI] [PubMed] [Google Scholar]
- Kubo A, Saji H, Tanaka K, Kondo N. Cloning and sequencing of a cDNA encoding ascorbate peroxidase from Arabidopsis thaliana. Plant Mol Biol. 1992;18:691–701. doi: 10.1007/BF00020011. [DOI] [PubMed] [Google Scholar]
- Lis J, Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- Liu X-d, Thiele DJ. Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996;10:592–603. doi: 10.1101/gad.10.5.592. [DOI] [PubMed] [Google Scholar]
- Loewus FA, Loewus MW. Biosynthesis and metabolism of ascorbic acid in plants. CRC Crit Rev Plant Sci. 1987;5:101–119. [Google Scholar]
- Lopez F, Vansuyt G, Casse-Delbart F, Fourcroy P. Ascorbate peroxidase activity, not the mRNA level, is enhanced in salt-stressed Raphanus sativus plants. Physiol Plant. 1996;97:13–20. [Google Scholar]
- Mathews MC, Summers CB, Felton GW. Ascorbate peroxidase: a novel antioxidant enzyme in insects. Arch Insect Biochem Physiol. 1997;34:57–68. [Google Scholar]
- McDuffee AT, Senisterra G, Huntley S, Lepock JR, Sekhar KR, Meredith MJ, Borrelli MJ, Morrow JD, Freeman ML. Proteins containing non-native disulfide bonds generated by oxidative stress can act as signals for the induction of the heat shock response. J Cell Physiol. 1997;171:143–151. doi: 10.1002/(SICI)1097-4652(199705)171:2<143::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Mehlhorn H, Cottam DA, Lucas PW, Wellburn AR. Induction of ascorbate peroxidase and glutathione reductase activities by interactions of mixtures of air pollutants. Free Radical Res Commun. 1987;3:193–197. doi: 10.3109/10715768709069784. [DOI] [PubMed] [Google Scholar]
- Mittler R, Zilinskas BA. Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. J Biol Chem. 1992;267:21802–21807. [PubMed] [Google Scholar]
- Mittler R, Zilinskas BA. Regulation of pea cytosolic ascorbate peroxidase and other antioxidant enzymes during the progression of drought stress and following recovery from drought. Plant J. 1994;5:397–405. doi: 10.1111/j.1365-313x.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- Morgan RW, Christman MF, Jacobson FS, Storz G, Ames BN. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci USA. 1986;83:8059–8063. doi: 10.1073/pnas.83.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PR, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Nover L. Heat Shock Response. Boca Raton, FL: CRC Press; 1991. [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB. The HSF world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive MR, Peacock WJ, Dennis ES. The anaerobic responsive element contains two GC-rich sequences essential for binding a nuclear protein and hypoxic activation of the maize Adh1 promoter. Nucleic Acids Res. 1991;19:7053–7060. doi: 10.1093/nar/19.25.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inzé D. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell. 1989;1:81–93. doi: 10.1105/tpc.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O, Xiao H, Lis JT. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-x. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Steigerwald SD, Mueller PR, Wold B, Riggs AD. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989;246:810–815. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Rao MV, Paliyath G, Ormrod DP. Ultraviolet-B- and ozone-induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol. 1996;110:125–136. doi: 10.1104/pp.110.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M, Gousseau H, Lister C, Foyer C, Creissen G, Mullineaux P. Cytosolic ascorbate peroxidase from Arabidopsis thaliana L. is encoded by a small multigene family. Planta. 1996;198:64–69. doi: 10.1007/BF00197587. [DOI] [PubMed] [Google Scholar]
- Scharf K-D, Rose S, Zott W, Schöff F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Cashmore AR. Photoregulated gene expression may involve ubiquitous DNA binding proteins. EMBO J. 1990;9:3415–3427. doi: 10.1002/j.1460-2075.1990.tb07549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöner S, Krause GH. Protective systems against active oxygen species in spinach: response to cold acclimation in excess light. Planta. 1990;180:383–389. doi: 10.1007/BF00198790. [DOI] [PubMed] [Google Scholar]
- Shirzadegan M, Christie P, Seemann JR. An efficient method for isolation of RNA from tissue cultured plant cells. Nucleic Acids Res. 1991;19:6055. doi: 10.1093/nar/19.21.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Dennis ES, Ellis JG, Llewellyn DJ, Tokuhisa JG, Wahleithner JA, Peacock WJ. OCSBF-1, a maize ocs enhancer binding factor: isolation and expression during development. Plant Cell. 1990;2:891–903. doi: 10.1105/tpc.2.9.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Colombé SV. Drought influences the activity of enzymes of the chloroplast hydrogen peroxide scavenging system. J Exp Bot. 1988;39:1097–1108. [Google Scholar]
- Tanaka K, Masuda R, Sugimoto T, Omasa K, Sakaki T. Water deficiency-induced changes in the contents of defensive substances against active oxygen in spinach leaves. Agric Biol Chem. 1990;54:2629–2634. [Google Scholar]
- Tanaka K, Suda Y, Kondo N, Sugahara K. O3 tolerance and the ascorbate-dependent hydrogen peroxide decomposing system in chloroplasts. Plant Cell Physiol. 1985;26:1425–1431. [Google Scholar]
- Thomsen B, Drumm-Herrel H, Mohr H. Control of the appearance of ascorbate peroxidase (EC 1.11.1.11) in mustard seedling cotyledons by phytochrome and photooxidative treatments. Planta. 1992;186:600–608. doi: 10.1007/BF00198042. [DOI] [PubMed] [Google Scholar]
- Troutt AB, McHeyzer-Williams MG, Pulendran B, Nossal GJV. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc Natl Acad Sci USA. 1992;89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansuyt G, Lopez F, Inzé D, Briat JF, Fourcroy P. Iron triggers a rapid induction of ascorbate peroxidase gene expression in Brassica napus. FEBS Lett. 1997;410:195–200. doi: 10.1016/s0014-5793(97)00587-5. [DOI] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]