Abstract
The Protein Information Management System (PiMS) is a laboratory information management system (LIMS) designed for use with the production of proteins in a research environment. The software is distributed under the CCP4 licence, and so is available free of charge to academic laboratories. Like most LIMS, the underlying PiMS data model originally had no support for protein–protein complexes. To support the SPINE2-Complexes project the developers have extended PiMS to meet these requirements. The modifications to PiMS, described here, include data model changes, additional protocols, some user interface changes and functionality to detect when an experiment may have formed a complex. Example data are shown for the production of a crystal of a protein complex. Integration with SPINE2-Complexes Target Tracker application is also described.
Keywords: Laboratory Information Management System (LIMS), Informatics, Data management, Data model, Protein complex
1. Introduction
Recent years have seen the development of many laboratory information management systems (LIMS), The majority of such LIMS are developed commercially and/or devoted to relatively simple process management, for examples see http://limsource.com/products/vproduct.html (2010). Academically developed LIMS related to structural biology include LISA (Haebel et al., 2001), XTRACK (Harris and Jones, 2002), SESAME (Zolnai et al., 2003), CLIMS (Fulton et al., 2004), HALX (Prilusky et al., 2005) and MOLE (Morris et al., 2005). The Protein Information Management System (PiMS) is an academically funded LIMS focused on the management of data describing the production and crystallization of proteins in a research environment (Morris et al., 2011). PiMS was originally developed as part of the UK BBSRC’s SPoRT initiative and is now principally supported by the CCP4 project (Collaborative Computational Project, Number 4, 1994). PiMS is distributed under the CCP4 licence and, therefore, is free to any laboratory (commercial or academic) with a CCP4 licence. Details of licensing and downloading PiMS can be found on the project web site, http://www.pims-lims.org/.
PiMS is a fully featured and flexible LIMS, but it is built around a small number of central concepts that are already familiar to practitioners in the field: Target, Construct, Sample, Experiment and Protocol.
A Target represents a single biological macromolecule (usually a protein) whose study is the purpose of some research. In structural genomics, the biological entities of interest have been the open reading frames (ORFs) of organisms and the proteins that each of them encodes. This two-sequence model of a Target has been incorporated into PiMS.
A Construct in PiMS is a record of what the scientist intends to express, including sequence mutations, truncations, fusions and affinity tags. Scientists usually design multiple constructs in their attempts to express each Target. PiMS facilitates the design and recording of Constructs by recommending PCR primer sequences and maintaining lists of standard primer extensions.
A Sample is intended to represent any physical sample. It may contain molecules related to a Target or Construct (in the form of DNA or protein), or it may represent a reagent that has been produced in (or brought into) the laboratory.
An Experiment describes how Samples were produced and acted on, and relates to a particular Construct. Samples are the optional inputs to and outputs from Experiments, thus allowing complicated workflows to be built up.
A Protocol is a reusable template from which Experiments can be created. A set of Experiments based on a single Protocol can be grouped together, most usefully to represent plate-based experiments.
Many proteins are biologically active only in a complex with other proteins (e.g. Gavin et al., 2006) and so many research projects, including the SPINE2-Complexes project, have research objectives which are not adequately represented by a single ORF product. To record work on complexes some modifications to PiMS are required. However, the representation of complexes presents some design challenges:
-
•
Is a complex something that is hypothesised to exist naturally in biology or any set of expressed constructs that interact?
-
•
Should one distinguish between, for example, two proteins in a complex and the same two proteins merely co-existing in a solution?
-
•
Is it valuable to distinguish between monomers and homodimers for a single protein species?
-
•
How should LIMS deal with the programmatic and user-interface complexity introduced by the many-to-many relationships required to describe complexes?
-
•
How are complexes “related” to each other for searching and comparison purposes?
In dealing with these issues the PiMS developers were guided by the SPINE2-Complexes Target Tracker (see below) which, in turn, drew on and extended the model developed for the 3D-Repertoire project (Romier et al., 2006). This technical note describes the data model design decisions that were taken as well as outlining the other changes within PiMS that were required to manage this extra richness: the definition of new Protocols specific to creating complexes, how the user interface changes were kept to a minimum so as not to impact on non-complex work and the development of new functionality to recognise when experiments were intended to form potential complexes. Finally, these changes are illustrated by considering the use of PiMS to record the creation of a real protein complex crystal.
2. Methods
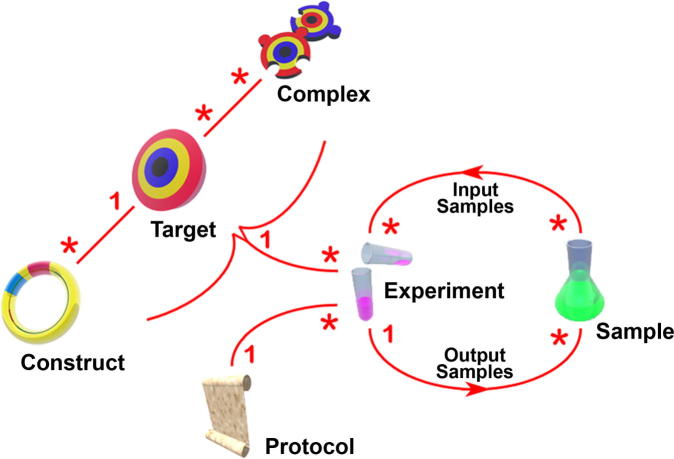
The PiMS data model was changed to allow a collection of macromolecules to be recorded as the target of research. This new concept is called the PiMS Complex and it is intended to represent a biologically relevant complex, i.e. between two or more naturally occurring proteins. Thus, a group of Targets describing individual proteins (and any small molecule ligands) defines a Complex. If a complex is studied by making multiple constructs for each component protein then all complexes between expressed constructs are linked to the same Complex. This reduces the number of complexes that have to be described and helps in detecting relationships between complexes (i.e. common components in multiple complexes). This implementation minimises the impact on existing PiMS use: the individual proteins of a complex are often research targets in their own right and producing a protein complex often involves producing the individual proteins prior to combination. A second data model change was also required: to allow an Experiment to relate to a Complex rather than to a Construct when appropriate. This second change was more difficult to implement as it required significant changes to the internal organisation of PiMS, however these changes were hidden from the user. The central PiMS concepts and their revised relationships are shown schematically in Fig. 1.
Fig.1.
A schematic showing the relationships between the core PiMS concepts relevant to the recording of work on complexes. The “1”s and “*”s on the red lines indicate one-to-many and many-to-many relationships, respectively. For example, a Construct belongs to a single Target but a Target may be a component of many Complexes. The icons are used throughout the PiMS user interface to indicate the relevant object types. An additional many-to-many relationship exists between Sample and Construct but is not relevant to the work described here.
New user-interface components that allow users to create and search Complexes have been added under the “Target” menu in PiMS. There are only two required pieces of information for a new Complex, its Name and a description. Targets can be added to, and removed from, a Complex through a simple search-and-select interface, with recently viewed Targets shown at the top of the list for convenience.
Extra links have been provided on the PiMS Experiment and Sample pages to allow users to navigate easily back to the parent Complex(es). All laboratory activities are recorded as Experiments. Each Experiment has some general information (its status, who ran it, when it was run, whether it worked, which Construct, Target and now Complex it relates to, etc.) and some specific information (what Samples went in, what Samples were produced and runtime parameters/results such as PCR annealing temperature). A Sample produced by one Experiment goes on to be used as an input to another Experiment and the production history of any Sample can be obtained by retracing this chain. Since a PiMS Sample can represent any experimental product – including a single protein or a protein complex – recording an Experiment involving a complex is no different to recording any other Experiment.
A PiMS Protocol, a reusable Experiment template, records the categories of Samples that are allowed as inputs and produced as outputs amongst other things. Importantly, PiMS avoids the use of special categories for dealing with protein complexes: a PiMS Plasmid describes a plasmid whether it is bi-cistronic or not; a PiMS Soluble Protein could equally well refer to a single protein (purified or not), multiple proteins, a well-defined protein–protein complex or some combination of these species. In this way PiMS avoids a difficult (and potentially artificial) distinction: whether mixing two protein components gives a mixture or a complex. Indeed, even with a single protein species there is ambiguity since its oligomeric state may vary over time or as a function of concentration.
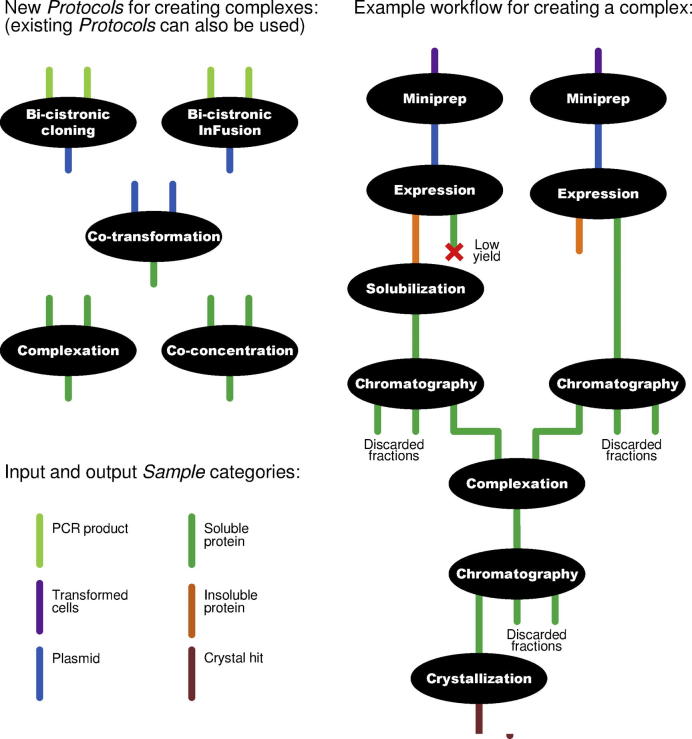
Existing Protocols are just as valid for complexes as for single proteins. However, as part of the work described here, the standard set of PiMS Protocols was augmented by five new Protocols which describe Experiments uniquely relevant to work on complexes: two for bi-cistronic expression of two products from a single vector; one for co-expression of two products by co-transformation of two plasmids into one cell; one for mixing of two separately expressed products (“Complexation”); and one for co-concentration of two separately expressed products. A characteristic of these Protocols is that they specify two or more input Samples with the same category. For example, the Complexation Protocol specifies two input Samples belonging to category Soluble Protein (Fig. 2).
Fig.2.
A schematic showing the new standard PiMS Protocols for working with complexes (left-hand side) and an example of how Experiments based on these Protocols fit together to build up a workflow (right-hand side). Protocols specify the categories of Samples that are allowed as inputs and produced as outputs (colour-coded in the diagram). Experiments “snap together” by matching one output category to the next input category. By specifying identical input categories, a Protocol allows unrelated Samples of the same type to be combined, shown by the “Complexation” step in the example workflow.
The flexibility of the Protocol system combined with the dynamic generation of production history means that no further changes are required for PiMS to be used to record the production of complexes. PiMS has a user interface that allows Protocols to be created, copied, edited and deleted making it straightforward to create new Protocols describing novel methods of complex formation.
3. Results and discussion
Although significant in design terms, recording protein complexes is only a small part of PiMS usage. Therefore, it was essential that introducing this functionality should not make PiMS any harder to use. This goal was achieved primarily due to the flexible nature of the PiMS Protocol system and its approach to Sample typing. Furthermore, these features make PiMS just as easy to use for recording and reporting work on complexes as single proteins, as illustrated by the following examples.
3.1. Production of a Hedgehog/Hedgehog Interacting Protein Complex
Hedgehog (Hh) family proteins are ubiquitous in tissue growth, patterning and morphogenesis. Dysregulation of Hh signalling can have severe pathological consequences and is an intensely active field of research. Several cell surface receptors, for example Hedgehog Interacting Protein (HIP), transduce and/or regulate Hh signals. The Hh–HIP interaction has been implicated in neuronal pathway development as well as stem cell maintenance and cancer. Bishop et al. (2009) undertook a structural study of the Hh–HIP complex as part of the SPINE2-Complexes project and their work was recorded in PiMS using the new functionality for complexes. The Hh–HIP complex was produced by separate expression and purification of the components followed by mixing to form the complex and a further purification step prior to crystallisation trials.
The project was used as a test case for the new features of PiMS and the new Protocols were created as part of this process. The “Complexation” Protocol was developed for the key complex-formation step and has two input Samples and one output Sample, all of type Soluble Protein (Fig. 2). A total of 25 different Experiments were recorded in PiMS to account for the whole process from construct design to crystal growth. After minimal training, a research student was able to record the work (and even contribute to defining the new Protocols) within PiMS. For navigating across, and keeping track of, the complicated multi-threaded workflow that was generated, the interactive diagram features of PiMS (Morris et al., 2011) were found to be particularly useful.
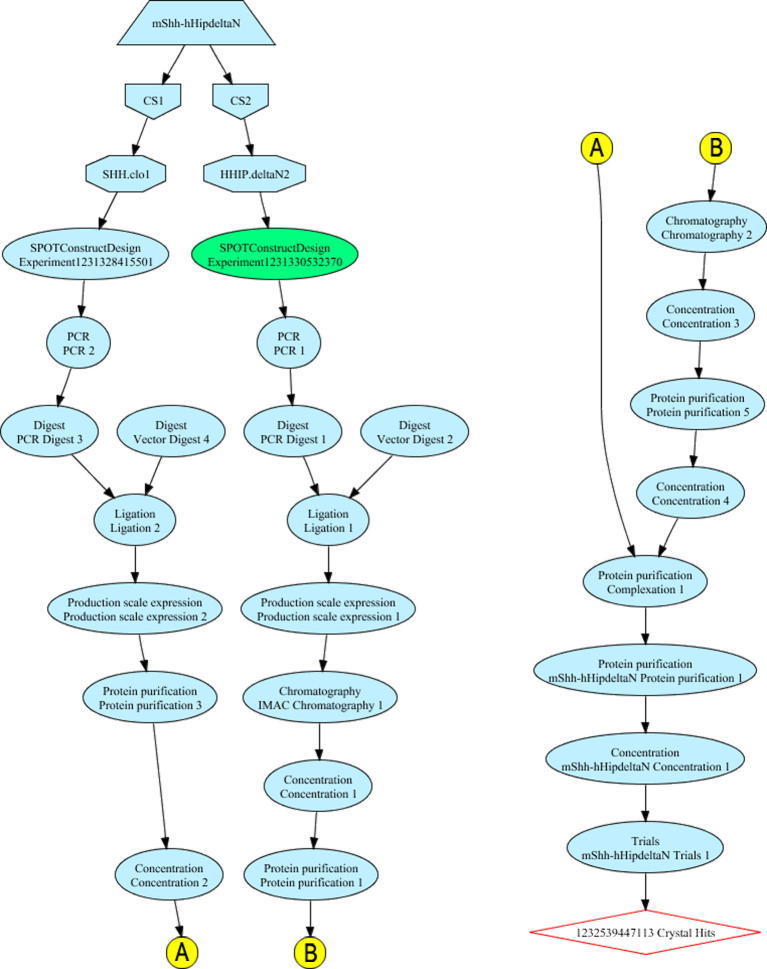
The whole project history can be summarised clicking on the PiMS “Sample History Report” button, which appears near the top of every Sample page (Morris et al., 2011), for the resulting crystal hits. This one-page report details the full production history that led to that Sample. It can include all the Complexes, Targets, Constructs, Samples, Experiments and Results involved in the workflow and present these both in tabular form and as a workflow diagram. For the Hh–HIP example, the summary workflow diagram (i.e. just the Complexes, Targets, Constructs and Experiments) is shown in Fig. 3. Such reports assist in the write-up of the work, giving a clear view of the successful workflow from amongst many possible dead ends and failures. Indeed, the long-term goal is for such a report to generate the “Materials and methods” for publication directly, although at the present time the exported PDF version of the report is probably more suited to inclusion as Supplementary information. The URL of the report can be directly shared with others, subject to PiMS’ access control, thus providing an efficient progress report for colleagues, collaborators and principal investigators.
Fig.3.
Screenshot of the Sample History Report diagram for the production history of Hh–HIP complex crystal (Bishop et al., 2009). The trapezoid at the top represents the Complex; pentagons represent Targets; octagons represent Constructs; ellipsoids represent Experiments and the red diamond at the end represents the Sample whose production history is shown. All other Samples are represented by the lines between Experiments. Within PiMS the diagram is interactive: clicking on one of the shapes will take you to the relevant page in PiMS. The diagram has been reformatted for publication by splitting in half and introducing the yellow A and B continuation markers.
3.2. Recording and representing data for many complexes: SPINE2-Complexes Target Tracker
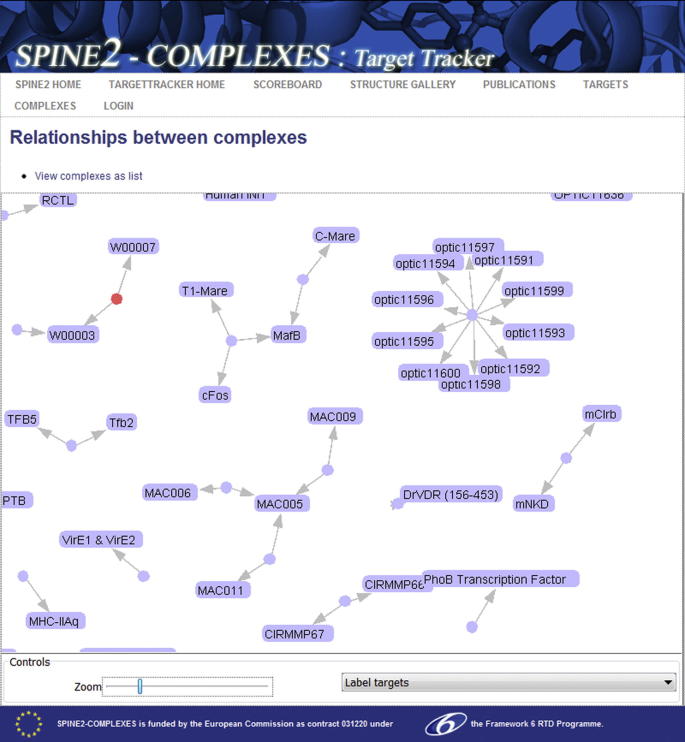
Structural genomics and proteomics initiatives have, for some time, coordinated and publicised their work by reporting to a central registry, TargetDB (Chen et al., 2004; http://targetdb.pdb.org/, 2010). TargetDB uses an ORF-based definition of a target which is not well suited to recording protein complexes and so the SPINE2-Complexes project developed its own registry, called “Target Tracker” (http://www.spine2.eu/SPINE2TT/, 2010). The Target Tracker enables project members to enter the list of complexes on which they were working, to report progress toward production and characterisation of those complexes (and their components) and to compare related complexes across the project. While the majority of data were entered manually, Target Tracker includes an interface that allows for bulk upload of data from an XML document and a specification of the document’s schema. One goal of Target Tracker development was to provide a graphical interface for showing the project’s status (Fig. 4; http://www.spine2.eu/SPINE2TT/ComplexMap.jsf, 2011). This view is created on the fly by a dynamic Java applet and can be scaled and manipulated by user. Complexes and their components are tied together by arrows and the layout is controlled by a pseudo-force field derived from sequence similarity.
Fig.4.
Screenshot from the SPINE2-Complexes Target Tracker graphical view of complexes (http://www.spine2.eu/SPINE2TT/ComplexMap.jsf, 2011). Circles represent complex targets and rectangles show the names of the component proteins. Note that proteins such as MAC005 and MafB are components of multiple, hence related, complexes. The view can be manually rearranged using drag-and-drop functionality and clicking on a complex or target reports the progress toward that component.
Support for Target Tracker was added to PiMS as part of the SPINE2-Complexes project. The Sample History Report already allowed the export of a PDF report, and this functionality was extended to enable the export of a Target Tracker-compliant XML document. An additional button (labelled “Export to Spine2”) is present on the Sample History Reports for those Samples which are linked to Complexes. While PiMS is able to infer the potential existence of an unrecorded Complex from a Sample’s production history, the decision was taken to require manual declaration of Complexes prior to exporting data to Target Tracker. Therefore, additional checking is required in PiMS to ensure that all production paths share a common predefined Complex.
While the exported XML is in Target Tracker’s format, the schema was largely derived as an extension to and expansion of that of TargetDB at the time (version 1, http://targetdb.sbkb.org/target.dtd, 2011). The XML could be post-processed to match the current TargetDB schema (version 2, http://targetdb.sbkb.org/TargetDB/documentation/targetdb.v2.dtd, 2011) for uploading both the complex and its individual components to TargetDB.
4. Conclusion
Few LIMS have specific features for recording protein complexes and dealing with the extra data model richness they introduce. To the best of our knowledge, the extensions of the PiMS LIMS described here make it uniquely suited to this task and the way these extensions have been implemented has not introduced undue complexity for users. In PiMS, it is as easy to work with complexes as it is to work with single protein species. We have demonstrated the utility of this new functionality in recording the production of a protein complex from the field of cell signalling. Furthermore, by showing how PiMS can export data to the SPINE2-Complexes Target Tracker we have provided a model for data exchange with other applications. PiMS is available from the project website (http://www.pims-lims.org/, 2010) and is distributed under the CCP4 licence (http://www.ccp4.ac.uk/ccp4license.php, 2010).
Acknowledgments
This work was done on behalf of the EC-funded project #31220 (SPINE2Complexes). The PiMS developers were supported by the CCP4 project and by the BBSRC as part of Grants BBC5121291, BBC5121371 and BBC5121451. The support of the MRC for the OPPF and the Wellcome Trust (Core Award 075491/Z/04) is also acknowledged.
References
- Bishop B., Aricescu A.R., Harlos K., O’Callaghan C.A., Jones E.Y., Siebold C. Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat. Struct. Mol. Biol. 2009;16(7):698–703. doi: 10.1038/nsmb.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Oughtred R., Berman H.M., Westbrook J. TargetDB: a target registration database for structural genomics projects. Bioinformatics. 2004;20(16):2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4, 1994. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D50, 760–763. [DOI] [PubMed]
- Fulton K.F., Ervine S., Faux N.G., Forster R., Jodun R.A. CLIMS: crystallography laboratory information management system. Acta Crystallogr. 2004;D60:1691–1693. doi: 10.1107/S0907444904017615. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Haebel P.W., Arcus V.L., Baker E.N., Metcalf P. LISA: an intranet-based flexible database for protein crystallography project management. Acta Crystallogr. 2001;D57:1341–1343. doi: 10.1107/s0907444901009295. [DOI] [PubMed] [Google Scholar]
- Harris M., Jones T.A. Xtrack – a web-based crystallographic notebook. Acta Crystallogr. 2002;D58:1889–1891. doi: 10.1107/s0907444902012696. [DOI] [PubMed] [Google Scholar]
- Morris C., Wood P., Griffiths S.L., Wilson K.S., Ashton A.W. MOLE: a data management application based on a protein production data model. Proteins. 2005;58:285–289. doi: 10.1002/prot.20291. [DOI] [PubMed] [Google Scholar]
- Morris C., Pajon A., Griffiths S.L., Daniel E., Savitsky M. The Protein Information Management System (PiMS): a generic tool for any structural biology research laboratory. Acta Crystallogr. 2011;D67:249–260. doi: 10.1107/S0907444911007943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J., Oueillet E., Ulryck N., Pajon A., Bernauer J. HalX: an open-source LIMS (Laboratory Information Management System) for small- to large-scale laboratories. Acta Crystallogr. 2005;D61:671–678. doi: 10.1107/S0907444905001290. [DOI] [PubMed] [Google Scholar]
- Romier C., Ben Jelloul M., Albeck S., Buchwald G., Busso D. Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr. 2006;D62:1232–1242. doi: 10.1107/S0907444906031003. [DOI] [PubMed] [Google Scholar]
- Zolnai Z., Lee P.T., Li J., Chapman M.R., Newman C.S. Project management system for structural and functional proteomics: sesame. J. Struct. Funct. Genomics. 2003;4:11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]