Abstract
Duplex DNA containing an apurinic/apyrimidinic (AP) lesion undergoes cleavage significantly more rapidly in nucleosome core particles (NCPs) than it does when free. The mechanism of AP cleavage within NCPs was studied through independently generating lesions within them. AP mediated DNA cleavage within NCPs is initiated by DNA-protein crosslink (DPCun) formation followed by β-elimination to give DPCs containing cleaved DNA (DPCcl). Hydrolysis of DPCcl produces a DNA single strand break (SSB). C2-dideuteration of AP showed that deprotonation from this position is involved in the rate determining step. Experiments utilizing NCPs containing mutated histone H4 proteins indicated that lysine residues in the amino terminal tail are involved in both DPC formation and β-elimination steps. Lysines 16 and 20 seem to play a greater role in reacting with AP at superhelical location 1.5 but other amino acids (e.g. lysines 5, 8, and 12) compensate in their absence. The mechanism of rapid double strand breaks in bistranded, clustered AP lesions was studied by independently preparing reaction intermediates within model NCPs. A single strand break on one strand enhances the cleavage of a proximal AP on the opposite strand.
Introduction
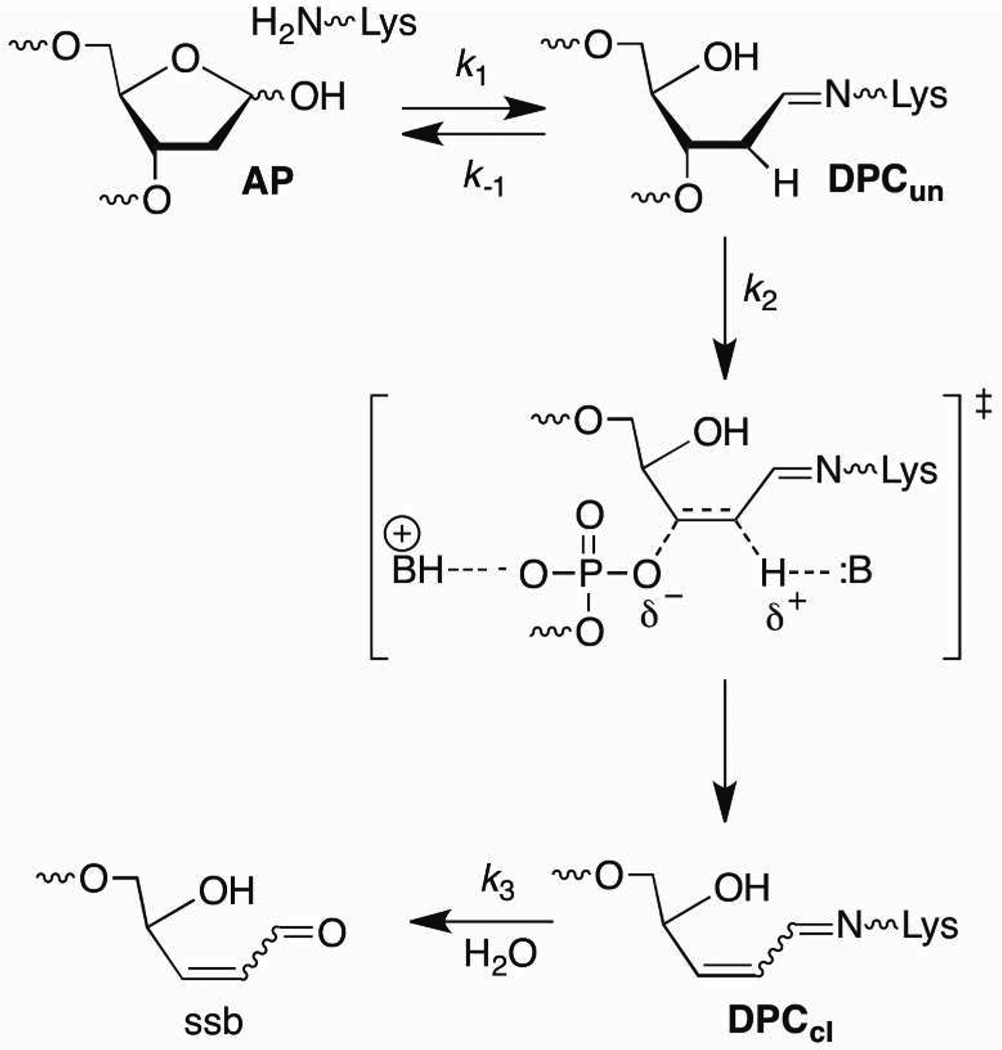
Apyrimidinic/apurinic sites (AP) are constantly produced in DNA due to spontaneous hydrolysis of the nucleotides' glycosidic bonds at rates sufficient to produce at least 10,000 of these lesions per cell per day.1 Nucleobase alkylation significantly increases the rate of hydrolysis of the glycosidic bond, and antitumor agents and carcinogens that alkylate nucleobases such as leinamycin and benzopyrene epoxides produce AP sites with varying ease following initial reaction with DNA.2,3 AP sites are also formed as intermediates during base excision repair by enzymes such as uracil DNA glycosylase.4 AP sites are alkali-labile lesions but undergo strand scission slowly in free DNA at physiological pH.5 Recently, we reported that the ~3 week half-life of an intact AP site in free DNA is decreased as much as 60-fold in a nucleosome core particle (NCP) composed of the α-satellite DNA sequence and that persistent DNA-protein cross-links (DPCs) are formed en route to strand breaks (Scheme 1).6 Herein we examine the mechanism by which the histone proteins in the nucleosome core particle catalyze strand scission from AP.
Scheme 1.
Lysine residues are critical in several processes involving AP cleavage. For instance, lysines are responsible for Schiff base formation during the β-elimination reactions characteristic of the lyase activity of enzymes involved in base excision repair, including Nth and DNA polymerase β.7–12 More recently, lyase activity has been ascribed to proteins not previously thought to play an active chemical role.13,14 Lysines are also components of small peptidic mimics of lyase enzymes.8,15–17 However, studies on the reactivity of AP sites with small molecules indicate that Schiff base formation is not limited to the lysine ε-amino group, nor is imine formation the only role for amines in β-elimination.17,18 Binding can contribute significantly to the lyase reactivity of a potential catalyst, as evidenced by
Lys•Trp•Lys tripeptide and an amino acridine (intercalator) equipped with an adenine to fill the void created by AP comparing the reactivity of two small molecules, the formation in duplex DNA.18 The latter is effective at cleaving DNA containing AP sites at 100,000-fold lower concentration than the Lys•Trp•Lys tripeptide.
The octameric core of histone proteins in nucleosome core particles utilizes a large number of positively charged, basic residues to bind the ~146 base pairs of DNA that complete ~1.7 turns as it wraps around the protein complex.19,20 In addition, the histone proteins contain lysine and arginine rich, flexible amino terminal tails that are potentially able to interact with DNA at a variety of nucleotide positions. The ε-amino groups of lysine residues in these tails are often post-translationally modified via acetylation or methylation.21–24 This has significant biochemical consequences, but also alters the amines' chemical properties. Complexation of AP containing DNA within NCPs provides a high effective molarity of lysine and arginine amino acids that could catalyze strand scission by β-elimination via general base catalysis, neutralization of the phosphate monoester leaving group, and/or the formation of Schiff bases (Scheme 1). The rapid cleavage of DNA containing an oxidized abasic site produced by an antitumor agent within chromatin provided the first glimpse into the nucleosome's ability to catalyze strand scission.25 We have taken advantage of the ability to independently generate lesions at defined sites in DNA, and technology for selectively mutating the histone proteins that make up NCPs to investigate this chemistry.6,26
Results and Discussion
Design and preparation of nucleosome core particles containing AP
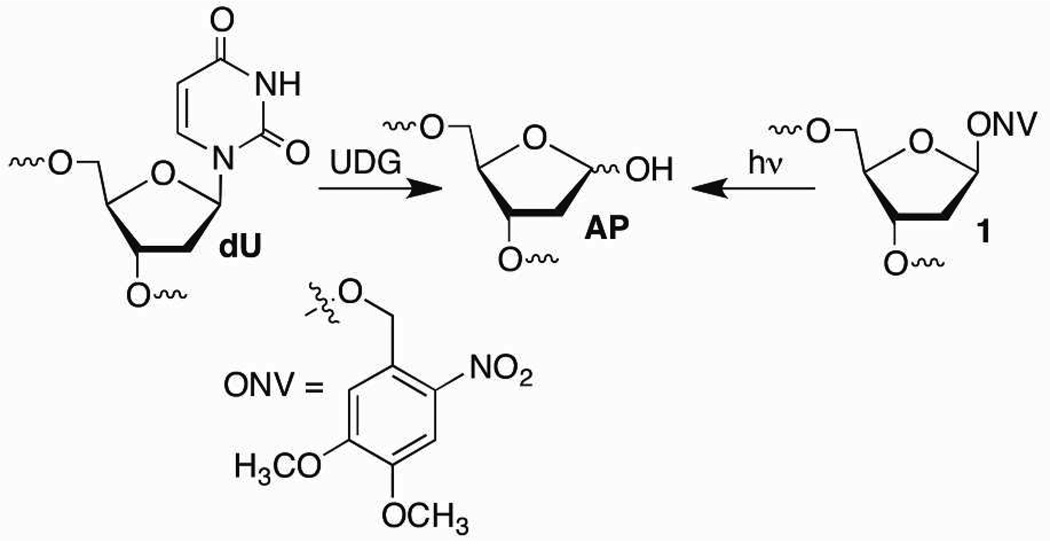
AP reactivity was examined in two 145–146 bp sequences, the α-satellite and "601" DNA.19,27 X-ray crystal structures of NCPs composed of either DNA sequence have been reported.19,20 The DNA sequences are positioned remarkably similarly in the core particles, but the 601 sequence was chosen for its strong binding.27 The DNA molecules were prepared by enzymatic ligation of chemically synthesized oligonucleotides and purified by denaturing polyacrylamide gel electrophoresis (PAGE).6,26,28 The AP site was generated after nucleosome core particle reconstitution from either 1 (photolysis) or dU (uracil DNA glycosylase, UDG) (Scheme 2). The photolytic precursor was employed in all kinetic experiments in order to eliminate enzyme activity as a variable.29,30 UDG treatment on dU was used when determining which histone protein(s) formed cross-links with AP because of the necessity to radiolabel the 5'-phosphate of the lesion.6,28
Scheme 2.
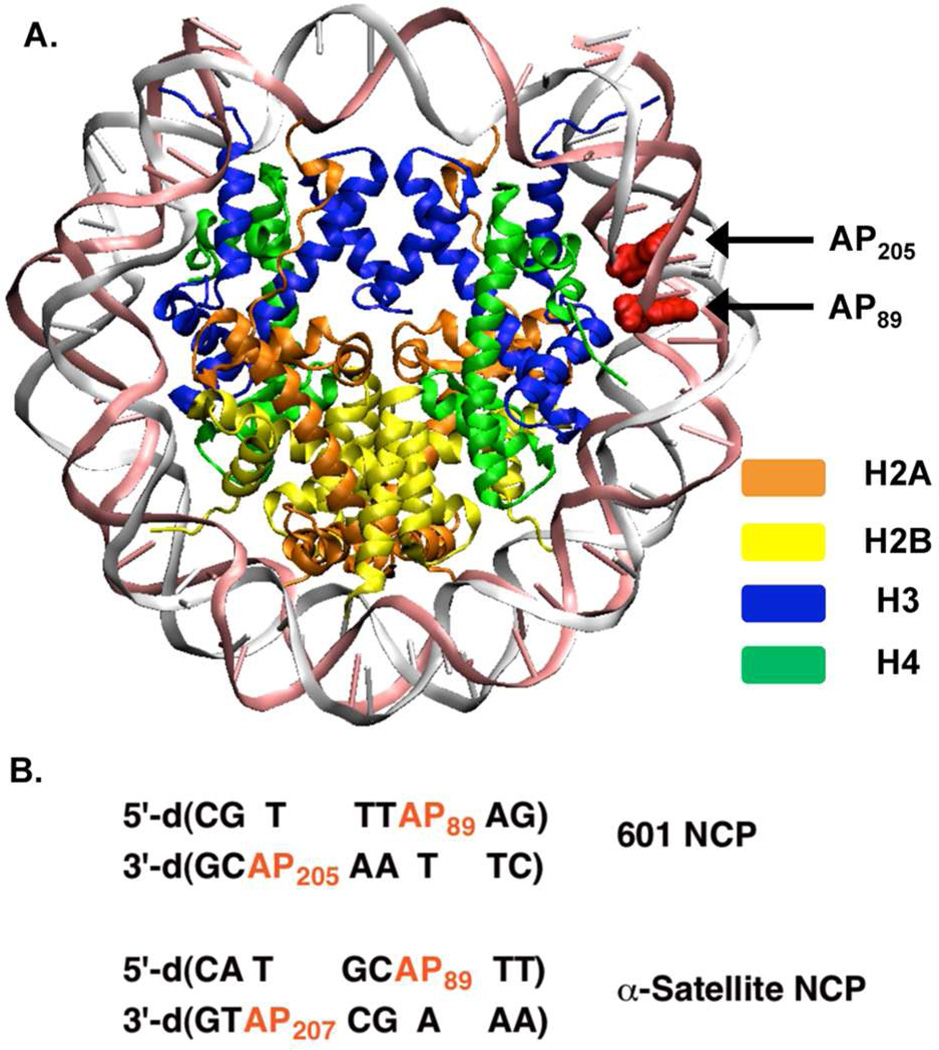
The histone octamer composed of a tetramer of dimers was assembled from purified Xenopus proteins (H2A, H2B, H3, and H4) that were over expressed in E. coli following literature protocols.31 Purified mutant forms of the H4 protein were obtained using Quickchange® as previously described.26 Nucleosome core particles composed of octamers consisting of wild type (wt) or mutated H4 proteins were reconstituted using reported methods.31 Abasic sites were generated at two positions in the region that is approximately 1.5 helical turns from the dyad axis of the DNA (superhelical location (SHL) 1.5), a known hot spot for DNA binding/damaging molecules.32,33 In each nucleosomal core particle, AP was produced individually at position 89 on one strand and 3 nucleotides away on the opposite strand (AP207 when using the 146 bp α-satellite DNA or AP205 in the 145 bp 601 DNA) (Figure 1).28 X-ray crystal structures of nucleosomes containing α-satellite or 601 DNA with native nucleotides at the positions where AP is introduced indicate that all of the lesions should face towards the octameric core. Nucleosome core particles containing AP on both strands (bistranded lesions) were employed in experiments focused on double strand break (DSB) formation. DNase I digestion demonstrated that the rotational positioning of the nucleosome core particles was unaffected by the modifications.28
Figure 1.
Independent generation of AP lesions in nucleosome core particles. (A) X-Ray crystal structure of the 601 NCP showing nucleotide positions modified. From: PDB: 3LZ0 (B) Local sequences in 601 and α-satellite NCPs in which AP sites are incorporated. See Supporting Information for entire DNA sequences and crystal structure of α-satellite NCP.
Isolated AP reactivity in nucleosome core particles composed of wild type histone proteins
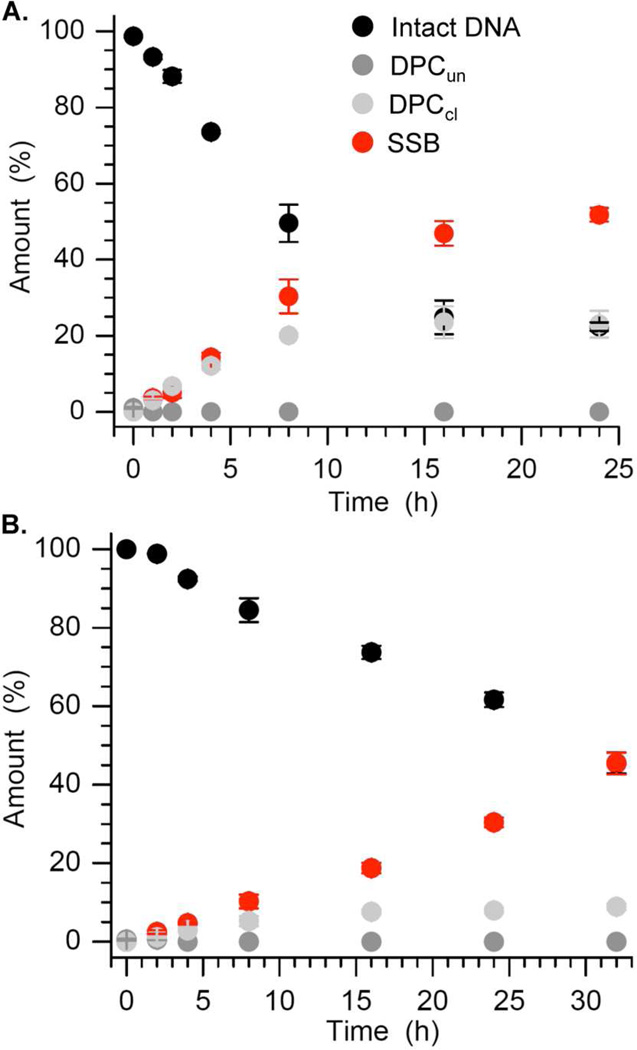
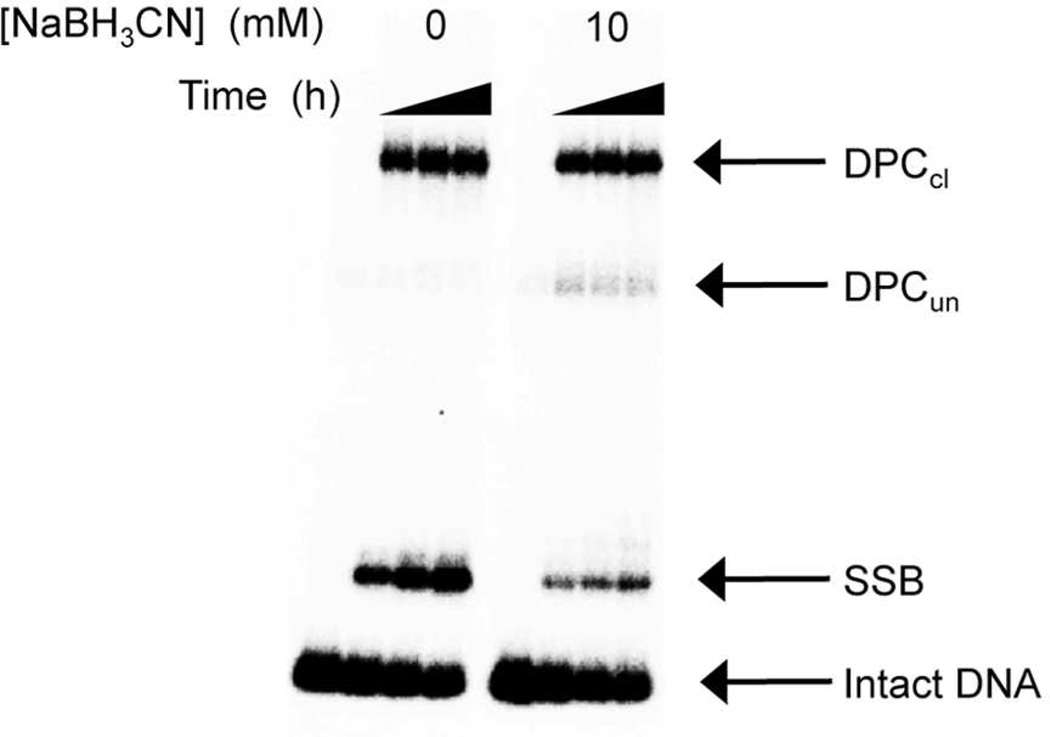
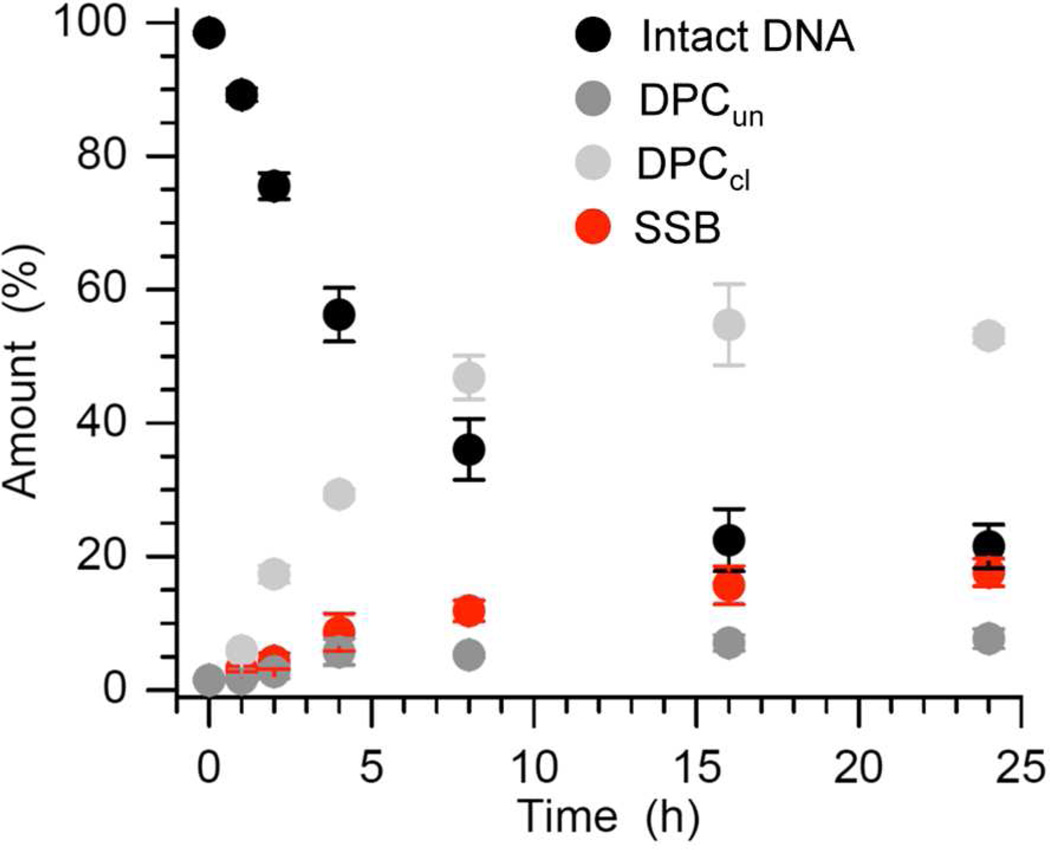
Using NaBH3CN as a trapping agent, we previously established that AP cleavage in NCPs proceeds through DNA-protein cross-links.6 The disappearance of AP89 and AP205 in NCPs containing the 601 DNA sequence was followed over 24 hours and analyzed by sodium dodecyl sulfate (SDS) PAGE (Figure 2, Table 1).28 AP site disappearance fit well to first order kinetics (Table 1) and 3 products were observed. Two products migrated considerably more slowly in the SDS PAGE gel, in which proteins are denatured and noncovalently bound DNA dissociates from the proteins. The slower migrating products are identified as DNA-protein cross-links based upon their transformation into faster migrating products upon proteinase K treatment. The products (Figure 3) were characterized following their isolation from the corresponding bands from the gel by the crush and soak method. The desalted materials were treated with proteinase K, and reanalyzed by denaturing PAGE analysis.28 The migration of the proteinase K treated material in a denaturing PAGE gel was compared to independently prepared DNA containing intact AP and material that was cleaved by treatment with NaOH. The DNA from the more slowly migrating DNA-protein cross-link comigrated with cleaved DNA in the denaturing PAGE gel, while the DNA from the faster moving product comigrated with the intact AP containing DNA.28 These DNA-protein cross-links were concluded to contain cleaved (DPCcl) and uncleaved DNA (DPCun), respectively. Similar analysis of the product that migrates slightly more slowly than starting material in the SDS PAGE gel indicated that these were single strand breaks (SSBs, Figure 3).
Figure 2.
Reaction of (A) AP89 and (B) AP205 in a 601 nucleosome core particle as a function of time.
Table 1.
AP reactivity as a function of position in 601 nucleosome core particles and free DNA at 37 °C.
| Nucleosome Core Particle | Free DNA | |||
|---|---|---|---|---|
| Position | NaBH3CN | kDis (10−6 s−1)a | t1/2 (h) | t1/2 (h)b |
| AP89 | - | 21.8 ± 2.0 | 8.9 ± 0.8 | 986 |
| AP89 | + | 20.7 ± 4.5 | 9.3 ± 2.0 | - |
| AP205 | - | 6.5 ± 1.1 | 29.6 ± 5.2 | 1008 |
| AP205 | + | 11.3 ± 0.6 | 17.1 ± 0.9 | - |
Rate constants are averages ± std. dev. of at least 5 experiments, each consisting of 3 independent reactions.
Data were for a single experiment consisting of 3 independent reactions in 100 mM NaCl, 1 mM EDTA, 10 mM HEPES (pH 7.5).
Figure 3.
Representative SDS PAGE gels (with and without NaBH3CN) of products produced from AP89 in the 601 NCP.
The rate constants for AP disappearance in the 601 NCP (Table 1) are within experimental error of those reported at the comparable positions containing α-satellite DNA.6 The acceleration in the NCPs compared to the free DNA is approximately 3-fold greater in the 601 sequence than it was in the α-satellite NCP. For instance, reactivity at AP89 is ~110-times greater in the 601 NCP than in the same sequence of free DNA. However, the greater acceleration in the 601 core particle compared to that containing α- satellite DNA is almost completely due to slower rate constants for AP cleavage in the free DNA, whose half-life is as long as 6 weeks.
Although the kinetics of AP disappearance were very similar, the product distributions in the 601 (Figure 2) and α-satellite6 NCPs were different from one another. For example, the total yield of DNA-protein cross-links (DPCs) was lower in the 601 nucleosome core particles at AP89 and AP205, and was compensated for by higher single strand break (SSB) yields. DPCs containing uncleaved DNA (DPCun) were not observed after 1 h, which is slightly sooner than in the α-satellite NCP. The DPCcl reach their maximum yield at 16 h and then decrease, which is also faster than in α-satellite NCP. The DPCs never rose above 10% at AP205; whereas they were the major products in the α-satellite DNA containing nucleosome. This indicates the hydrolysis of the putative Schiff base in DPCcl (k3, Scheme 1), especially those involving AP205 is faster in the 601 NCP.
NaBH3CN was used in situ to trap DPC intermediates by reducing the C=N to C-N. Incubating the 601 NCPs in the presence of NaBH3CN (10 mM) significantly increases the yield of DNA-protein cross-links at the expense of SSBs, but the majority of DPCs trapped still consist of cleaved DNA (DPCcl) (Figure 4). Higher concentrations of reducing agent result in higher yields of DPCun at the further expense of single strand break and DPCcl levels. Despite the changes in product distribution, the rate constants for AP89 disappearance in the NCP is unaffected by incubating with NaBH3CN, whereas the half-life of AP205 is reduced by more than 40% in the presence of the reducing agent (Table 1). These data affirm the previous proposal that AP cleavage is catalyzed by the histone protein(s) within the nucleosome core particle and proceeds through DNA-protein crosslinks.6 However, the different effects of NaBH3CN on the rate constant of AP disappearance suggest that the equilibrium corresponding to the initially formed Schiff base (DPCun, Scheme 1) is more favorable for AP89 than for AP205. Incubation of NCP core particle in the presence of NaBH3CN traps DPCun and prevents its reversion to starting material.
Figure 4.
Reaction of AP89 in a 601 nucleosome core particle as a function of time in the presence of NaBH3CN (10 mM).
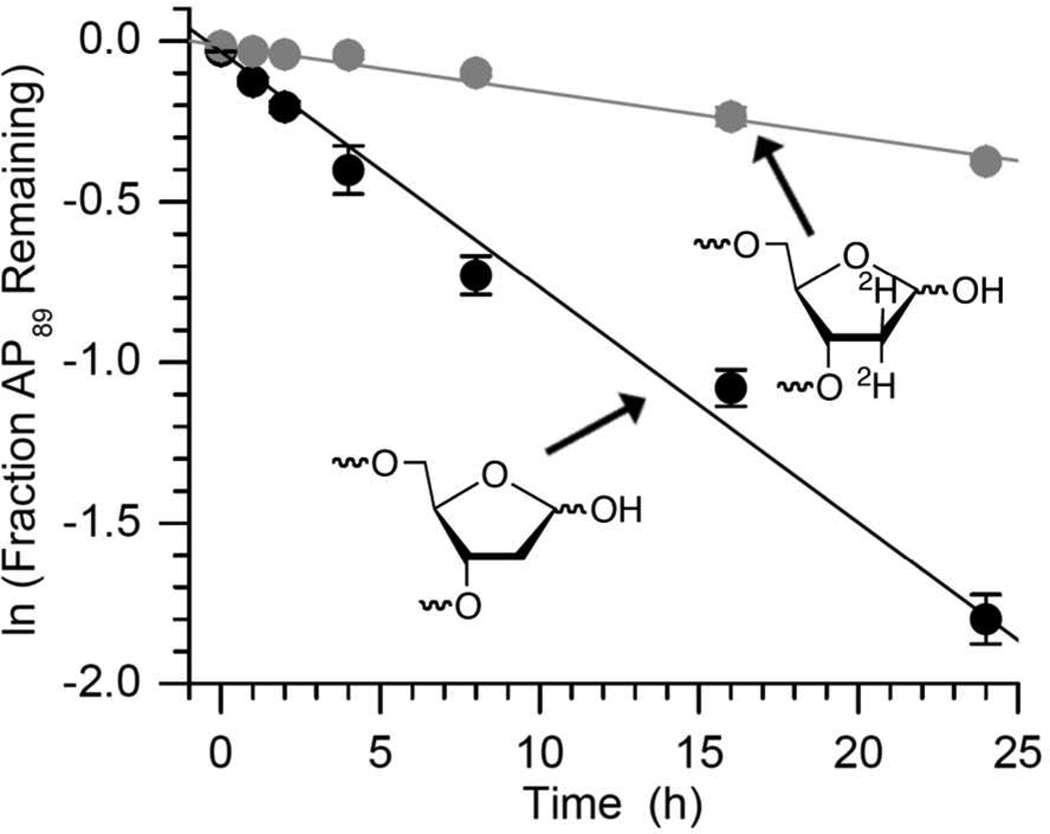
The effect of C2-dideuteration on the rate of disappearance of the lesion was determined for AP89 (Figure 5). A significant kinetic isotope effect (5.2 ± 0.5) was measured. As in the case of 2-deoxyribonolactone (L) cleavage, this indicated that C2-deprotonation is involved in the rate determining step for AP89 disappearance within NCPs.26 C2-Dideuteration also reinforced the notion that AP89 Schiff base hydrolysis (k−1, Scheme 1) is rapid because despite reducing the rate constant for elimination from DPCun only 3% of this cross-linked intermediate builds up over 24 h.28 Overall, the observed rate constant for disappearance of the AP site in the NCP (kDis) is expressed in the context of a pre-equilibrium mechanism (Eqn. 1, Scheme 1).
| (1) |
Figure 5.
Effect of C2-dideuteraton on the rate of disappearance of AP89 in a 601 nucleosome core particle.
The role of lysines in the histone H4 tail on AP reactivity
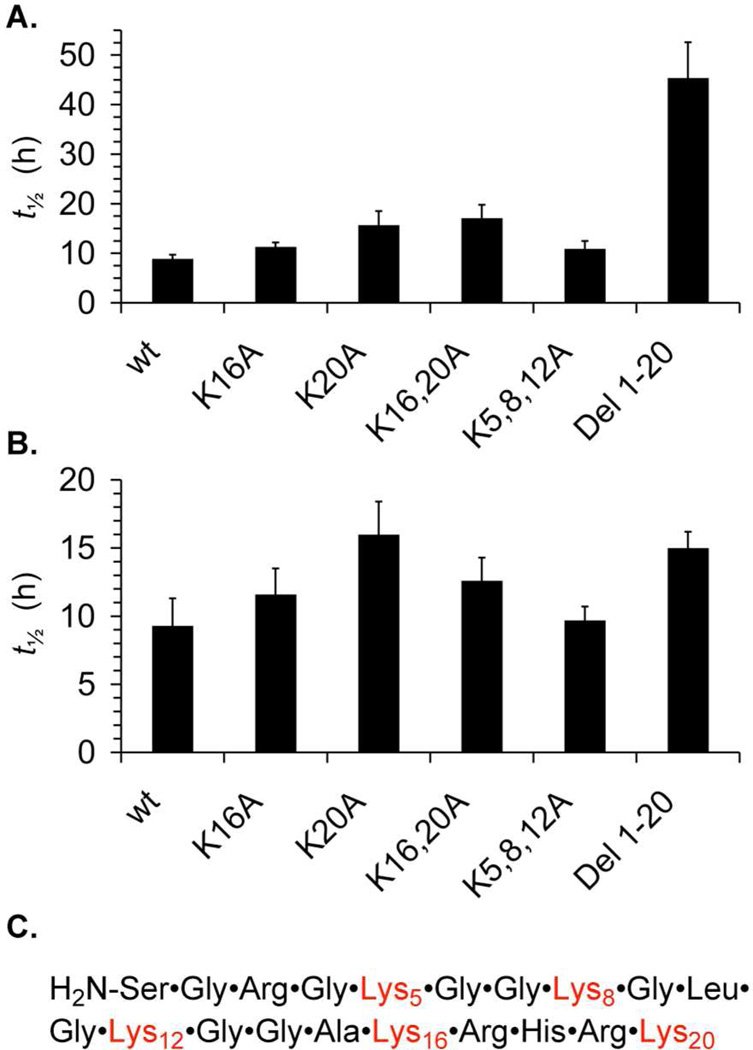
Histone H4 is responsible for more than 95% and 88% of the DNA-protein cross-links at AP89 and AP207, respectfully in α-satellite DNA containing nucleosome core particles.6 Removing the 19 N-terminal amino acids from H4 reduces the rate constant for AP disappearance 2–3 fold in the respective NCPs but has no effect on the contribution of cross-linking by histone H4 to AP89 relative to other proteins. However, the amount of cross-linking between AP207 and histone H3 increases to 28% at the expense of the H4 protein (62%). Four lysines are present in the first 19 amino terminal amino acids of the H4 protein, and a fifth is located at position 20 (Figure 6C). It is possible that either the α- and/or ε-amine of Lys20 in the 19 amino acid deletion mutant of histone H4 is responsible for Schiff base formation. However, we were unable to identify the amino acid(s) responsible for Schiff base formation by mass spectrometry.
Figure 6.
Effect on histone H4 protein structure on half-life for disappearance of AP89 in a 601 nucleosome core particle in the (A) absence or (B) presence of NaBH3CN. (C) Amino terminal sequence of histone H4 protein. Legend: wild type, wt; lysine, K; alanine, A.
Our inability to identify specific amino acids involved in Schiff base formation using mass spectrometry led us to probe the roles of individual lysines in the H4 protein by examining the reactivity of AP89 in 601 nucleosome core particles containing various mutated forms of this protein (Figure 6). Lysines were targeted for mutation because they are most typically associated with Schiff base formation. Histone H4 was primarily responsible for DPC formation with AP89 in 601 NCPs containing wild type H4 and all mutant proteins studied (vide infra).28 Substituting alanines for lysines 5, 8, and 12 had no effect on the half-life of AP89 and replacing Lys16 decreased the reactivity only slightly (Figure 6A). The Lys16,20Ala double mutant showed the largest effect of any of the substitution mutants, but AP89 reactivity was still only ~2-fold slower than in the NCP composed of wild type H4. Deleting the 20 N-terminal amino acid H4 protein tail containing 5 lysine residues had the largest effect, increasing the half-life for AP89 disappearance ~5-fold. However, even this increase in the AP89 half-life accounts for only a portion of the accelerated reactivity (>100-fold) of the abasic site within the nucleosome core particle compared to free DNA (Table 1).
The mutant forms of histone H4 were also used to probe Schiff base formation by trapping these intermediates with NaBH3CN (Figure 6B). The half-life for AP89 disappearance increases less than 2-fold in any NCP containing mutant forms of H4, including the 20 amino acid deletion. One interpretation of these experiments is that different lysines in the H4 tails have similar potency for reacting with AP89 to form Schiff bases. Alternatively, AP89 would disappear with approximately the same rate constant in the core particles containing mutant H4 proteins because reaction with NaBH3CN is the rate determining step. We cannot rule this out.
The protein tail may also be involved in the β-elimination step that transforms DPCun to DPCcl (Scheme 1). When the NCP reconstituted using the H4 deletion mutant was incubated in the presence of NaBH3CN SSBs are largely replaced by DNA-protein cross-links containing uncleaved DNA (DPCun, Scheme 1, Table 2).28 Elimination cannot occur after reduction by NaBH3CN. Recall, that DPCcl was the major product trapped from NCP containing wild type proteins incubated with NaBH3CN (Figure 4, Table 2). DPCcl was also the major product when the Lys5,8,12Ala H4 mutant was incubated with NaBH3CN. However, larger DPCun:DPCcl ratios were observed when NCPs containing Lys16Ala and/or Lys20Ala H4 mutants were incubated in the presence of reducing agent (Table 2). These data are consistent with the hypothesis that lysines 16 and 20 are more effective at inducing β-elimination at AP89 than are lysines 5, 8, and 12, which is why NaBH3CN is better able to compete with elimination when the former are mutated to alanine.
Table 2.
Effect of mutations in histone H4 protein on the distribution of DNA-protein cross-links at AP89 in a 601 nucleosome core particle in the presence of NaBH3CN.
| histone H4 protein | DPCun : DPCcl |
|---|---|
| Wild type | 0.15 |
| Lys5,8,12Ala | 0.12 |
| Lys16Ala | 0.26 |
| Lys20Ala | 0.64 |
| Lys16,20Ala | 1.22 |
| 1–20 Deletion | 10.3 |
Despite the effect of various mutated proteins on AP89 reactivity, histone H4 remained the major species responsible for Schiff base formation. Analysis of DPCs isolated from core particles incubated in the presence of NaBH3CN in which the 5'-phosphate of AP89 was 32P-labeled revealed that histone H4 accounted for more than 95% of the crosslinked material in the 601 NCP composed of wild type proteins, alanine mutants, and even in the NCP composed of the 20 amino acid deletion.6,28 The α-amino group of the Nterminal amine,34 which based upon the crystal structure of the 601 NCP containing undamaged DNA, is located near position 89 is postulated to be responsible for cross-linking in the deletion mutant.20
Based upon these observations we propose that lysines are involved in the rate determining elimination step, as well as in Schiff base formation. Lysines 16 and 20 seem to play a greater role in reacting with AP89 but other amino acids (e.g. Lys5, 8, and 12) compensate in their absence. The flexibility of the protein tail presumably enables multiple lysine residues to react with AP89. The experiments utilizing the H4 deletion protein suggest that other amino acids (e.g. arginines) and/or the H3 histone tail may also contribute to the rate limiting elimination step, especially when H4 is modified (Scheme 1), but this has yet to be investigated.
Double strand break formation from bistranded lesions
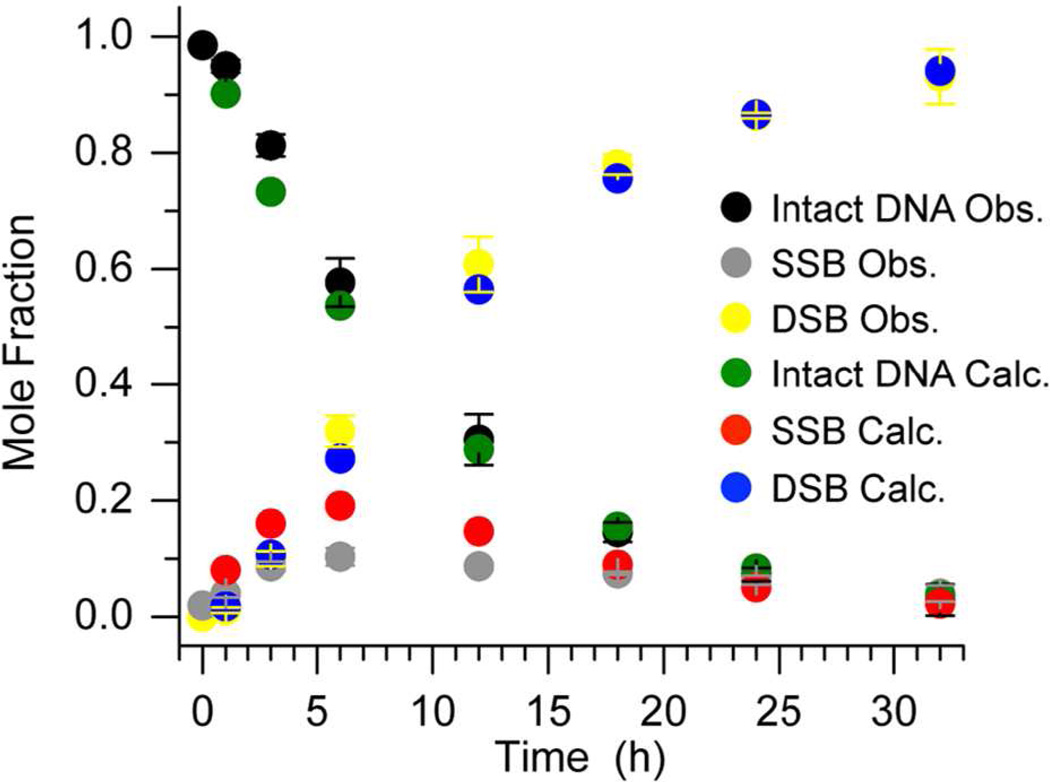
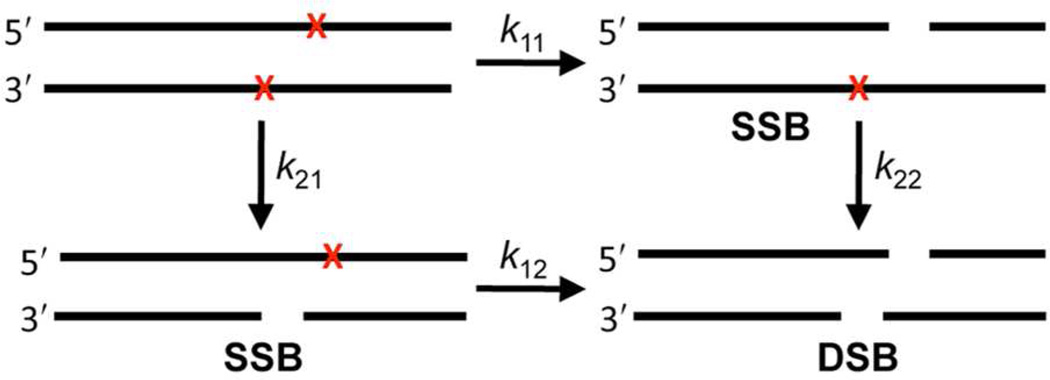
Bistranded lesions contain two damage sites within ~1.5 helical turns, are produced by γ-radiolysis, and are believed to be especially deleterious to cells because they are repaired more slowly than individual modifications or strand breaks.35–41 The chemical reactivity of bistranded lesions in nucleosome core particles had not been described prior to our study involving α-satellite containing NCPs.6 Double strand breaks resulted from DNA containing AP sites at positions 89 and 207 with no apparent induction period (Figure 7), even though the overall rate constant for core particle disappearance was at most slightly greater than when only a single lesion at AP89 was present.
Figure 7.
Observed and calculated (Kintecus)42 reactivity of bistranded AP in α-satellite DNA containing NCP.
We modeled single and double strand break growth in the bistranded α-satellite DNA containing NCP (Figure 7), that were previously measured by SDS gel electrophoresis following proteinase K digestion, using a simplified kinetic scheme (Scheme 3) that ignores the persistence of DNA-protein cross-links following strand scission (DPCcl, Scheme 1).6,42 By assuming that the rate constants for disappearance (cleavage) of the first AP site in the bistranded core particle (k11 = 2.3 × 10−5 s−1, k21 = 6.1 × 10−6 s−1) were the same as in core particles in which a single lesion was present, the program Kintecus42 did an excellent job fitting DSB growth but the low SSB yields as a function of time were less well reproduced (Figure 7). With these caveats, the predicted rate constants for double strand break formation from the SSBs (k12 = 3.9 × 10−4 s−1, k22 = 6.5 × 10−5 s−1) were 10–17 times faster than those for initial single strand cleavage.
Scheme 3.
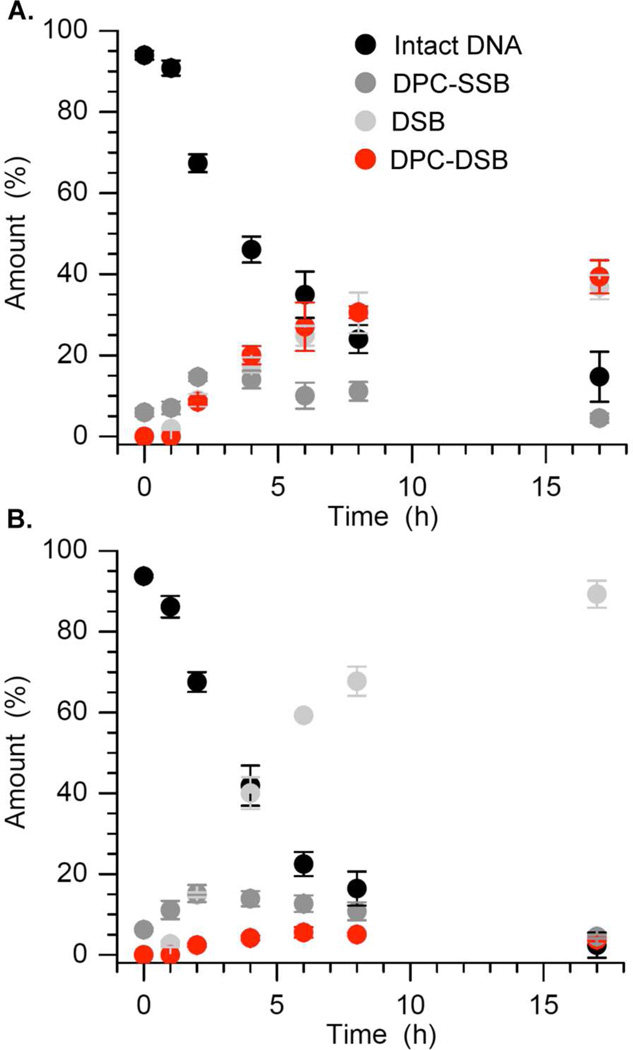
The 601 NCP containing abasic sites at positions 89 and 205 also yields double strand breaks rapidly with little if any induction period (Figure 8). In contrast to the NCP containing α-satellite DNA, the overall rate constant for disappearance of the 601 NCP containing the bistranded lesion (strand containing AP89 32P-labeled: kDis = 7.1 ± 2.0 × 10−5 s−1, t½ = 2.7 ± 0.8 h; strand containing AP205 32P-labeled: kDis = 6.1 ± 0.9 × 10−5 s−1, t½ = 3.2 ± 0.5 h) is more than twice as fast as the core particle containing a single lesion at AP89.28 Single strand breaks are not detected by SDS PAGE, regardless of which strand is 32P-labeled (Figure 8). The DNA-protein cross-links containing cleaved DNA (DPC-SSB, Scheme 4) yield (~15%) reaches a maximum at ~2 h and then declines to ~4% over the course of the reaction. Proteins cross-linked to the labeled strand are detectable in DSBs. The yields of protein cross-linked DSBs (DPC-DSB) correlate with what is observed in core particles containing a single abasic site (Figure 2). For instance, the DPC-DSB yield is greater in the core particle in which the strand containing AP89 is 32P-labeled (~40%, Figure 8A), whereas the comparable product never reached 6% when the strand containing AP205 is 32P-labeled (Figure 8B).
Figure 8.
Reaction of bistranded abasic sites within a nucleosome core particle as a function of time in which (A) the strand containing AP89 is 32P-labeled and (B) the strand containing AP205 is 32P-labeled.
Scheme 4.
Kintecus analysis of the time dependence for SSBs and DSBs in the 601 NCP containing AP89 and AP205 does not yield as good a fit for product formation as was observed for the core particle containing α-satellite DNA (Figure 7).28 Using the rate constants for disappearance of AP89 or AP205 in core particles containing isolated lesions may be a source of the poor fitting because the bistranded lesion disappears more rapidly than do the isolated abasic sites in the 601 NCP. Nonetheless, the rate constants for cleavage of the second AP site are also predicted to be much faster than the first.28
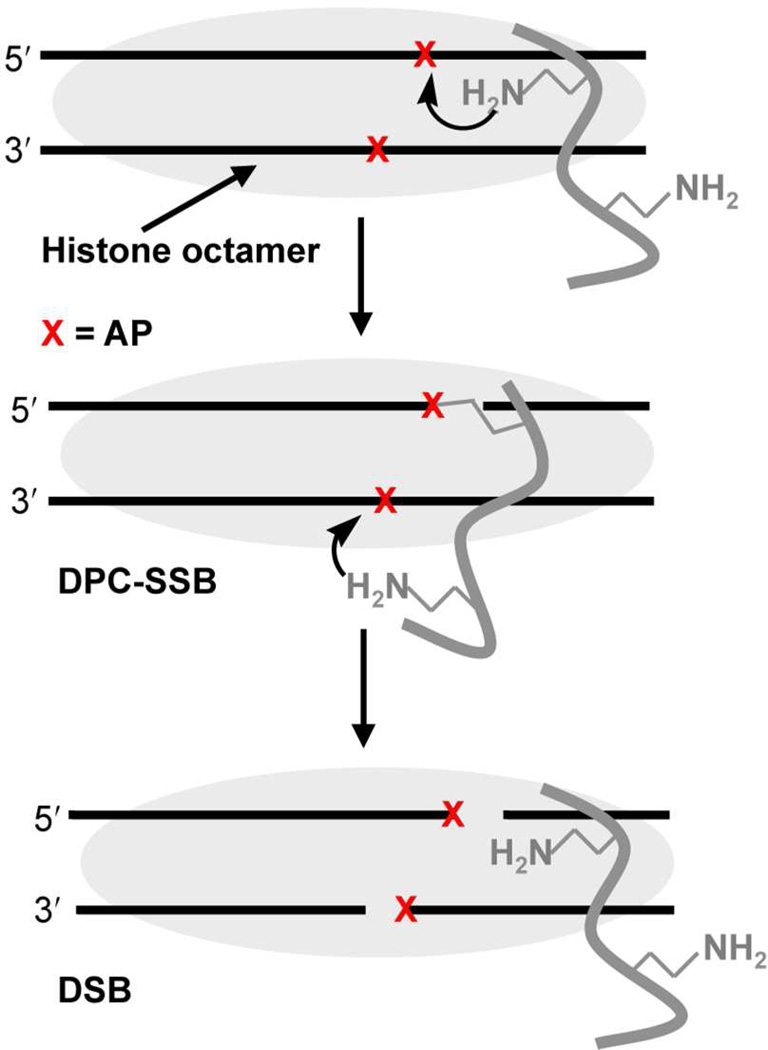
We suggested a template mechanism6 for the faster cleavage of the second AP site in bistranded abasic sites. In this mechanism (Scheme 4), DNA-protein cross-link formation at one abasic site increases the effective molarity of other lysines with respect to a neighboring lesion, resulting in more rapid DSB formation than expected (k12 >k11 and k22 > k21, Scheme 3).
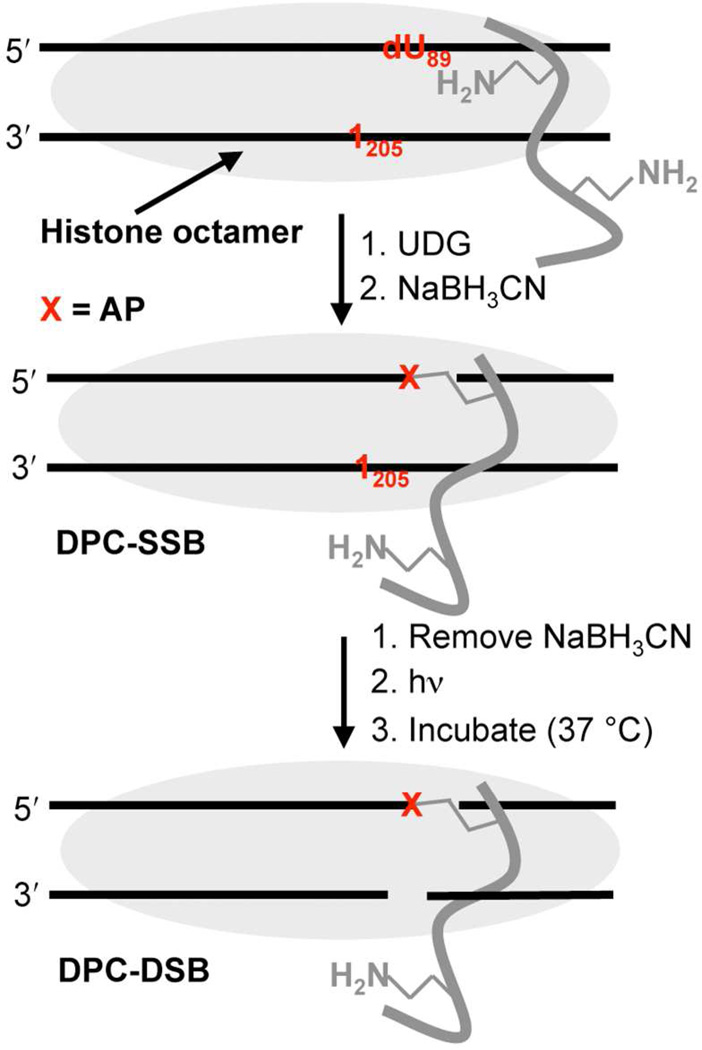
To test the viability of the proposed template mechanism, the kinetic effect of a DNA-protein cross-link on the reactivity of a proximal AP site was independently examined by preparing a model nucleosome core particle. We took advantage of the ability to generate AP sites from two orthogonal precursors (Scheme 2). Two 601 NCPs containing dU89 and 1205 or 189 and dU205 on opposite strands were prepared and treated with UDG, followed by incubation in the presence of NaBH3CN to trap the DNA-protein crosslinks (Scheme 5). Using this method DPC-SSBs (Scheme 5) were formed in 50–70% yield after 24 h. Following removal of the reducing agent, the AP site on the opposite strand was generated photochemically. The reactivity of the photochemically generated AP site in the presence of the DPCcl was monitored as a function of time.28 A marked acceleration in AP reactivity was observed in both NCPs. The half-life for AP89 decreased from ~9 h (Table 1) to less than 2 h (t1/2 = 113 ± 32 min, k = 1.1 ± 0.4 × 10−4 s−1) when cleaved AP205 was trapped by a histone protein (DPCcl, Scheme 1). The effect of DPC-SSB involving AP89 on AP205 reactivity was even more striking. The half-life for AP205 in a NCP decreased from almost 30 h (Table 1) to less than 1 h (t1/2 = 52 ± 9 min, k = 2.3 ± 0.4 × 10−4 s−1).
Scheme 5.
Experiments using the independently synthesized DNA-protein cross-link suggest that the template mechanism can result in faster cleavage of the second AP in bistranded basic sites. However, it does not rule out an even simpler explanation. For instance, given that the majority of the templated proteins would contain a strand break (DPCSSB, Scheme 5), we considered whether double strand break formation is accelerated due to the presence of a SSB. Such an effect could be rationalized by thinking of the AP site in the vicinity of a strand break as being in a more single strand like environment. Abasic lesions typically undergo strand scission in single stranded DNA more readily than those in double stranded DNA.43–45
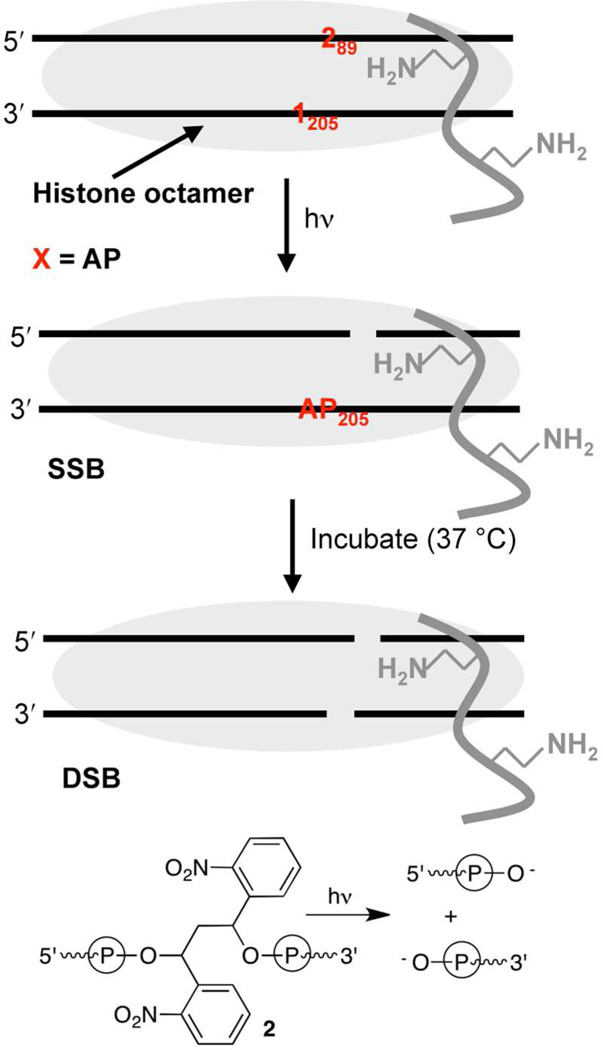
In order to test the effect of a strand break on AP reactivity core particles containing 1 and a photolabile precursor (2, Scheme 6) to a direct strand break developed by Taylor were independently synthesized.46 Substituting a direct strand break for AP205 increased the reactivity of AP89 (t1/2 = 267 h) by 3.7-fold in naked 601 DNA. Similarly, AP205 was 2.8-fold more reactive (t1/2 = 364 h) when a strand break was present at position 89. Greater AP reactivity in the presence of a strand break was also observed in the nucleosome core particles. The half-life for AP89 (t1/2 = 156 ± 19 min, k = 7.4 ± 0.9 × 10−5 s−1) decreased more than 3-fold in the vicinity of a strand break compared to when it was an isolated lesion within a NCP (Table 1). The reactivity of AP205 (t1/2 = 96 ± 8 min, k = 1.2 ± 0.1 × 10−4 s−1) increased even more when part of a bistranded lesion containing a strand break on the opposite strand at position 89. AP205 reacted 20-times faster in a NCP containing a proximal strand break on the opposite strand compared to when it was the only lesion present.
Scheme 6.
Conclusions
Histone cross-linking to one abasic site accelerates AP cleavage on the opposite strand 5 to 30-fold. However, a proximal single strand break also enhances AP cleavage on the opposite strand 3–20 times within the NCP. In the case of bistranded, clustered lesions containing two AP sites, DNA-protein cross-link (DPCcl, Scheme 1) formation on one AP site may catalyze the opposite strand cleavage through a template mechanism (Scheme 4). However, mechanistic economy (Ockham's razor) requires that the increased rate of double-strand break formation be ascribed to the formation of a single strand break, which is common to both model systems examined above (Schemes 5 and 6).47,48
Whether this is generally the case throughout nucleosome core particles is uncertain and AP reactivity needs to be examined at other superhelical locations. In addition, these experiments do not account for the entire acceleration of abasic site cleavage that is observed in nucleosomes compared to naked DNA. Further studies are warranted to reveal the factors responsible for this acceleration.
Regardless of the source of the large acceleration in abasic site cleavage within nucleosome core particles, this study further validates that the octameric histone core is more than a structural scaffold. Histone catalyzed cleavage of (oxidized) abasic site cleavage suggests that bistranded lesions containing them are de facto double strand breaks. This chemistry may help explain the high cytotoxicity of DNA damaging agents that produce bistranded lesions.26,49,50
Supplementary Material
ACKNOWLEDGMENT
We are grateful for generous financial support from the National Institute of General Medical Sciences (GM-063028). We thank Professor Jack Norton for helpful discussion with the kinetic analysis and the reviewers for helpful comments.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental procedures for all experiments, complete sequences of all DNAs used to prepare nucleosome core particles, representative autoradiograms and kinetic plots, NMR and mass spectra of key compounds, and mass spectra of oligonucleotides containing 1 and 2. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Lindahl T. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Viswesh V, Gates K, Sun D. Chem. Res. Toxicol. 2010;23:99–107. doi: 10.1021/tx900301r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gates KS, Nooner T, Dutta S. Chem. Res. Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 4.Stivers JT, Jiang YL. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama H, Fujiwara T, Ura A, Tashiro T, Yamamoto K, Kawanishi S, Saito I. Chem. Res. Toxicol. 1994;7:673–683. doi: 10.1021/tx00041a013. [DOI] [PubMed] [Google Scholar]
- 6.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kow YW, Wallace SS. Biochemistry. 1987;26:8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- 8.Mazumder A, Gerlt JA, Absalon MJ, Stubbe J, Cunningham RP, Withka J, Bolton PH. Biochemistry. 1991;30:1119–1126. doi: 10.1021/bi00218a033. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto Y, Kim K. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 10.Feng J-A, Crasto CJ, Matsumoto Y. Biochemistry. 1998;37:9605–9611. doi: 10.1021/bi9808619. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto Y, Kim K, Katz DS, Feng J-A. Biochemistry. 1998;37:6456–6464. doi: 10.1021/bi9727545. [DOI] [PubMed] [Google Scholar]
- 12.David SS, Williams SD. Chem. Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 13.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strande N, Roberts SA, Oh S, Hendrickson EA, Ramsden DA. J. Biol. Chem. 2012;287:13686–13693. doi: 10.1074/jbc.M111.329730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behmoaras T, Toulme J-J, Helene C. Proc. Nat. Acad. Sci. USA. 1981;78:926–930. doi: 10.1073/pnas.78.2.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behmoaras T, Toulme J-J, Helene C. Nature. 1981;292:858–859. doi: 10.1038/292858a0. [DOI] [PubMed] [Google Scholar]
- 17.Lhomme J, Constant JF, Demeunynck M. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Fkyerat A, Demeunynck M, Constant JF, Michon P, Lhomme J. J. Am. Chem. Soc. 1993;115:9952–9959. [Google Scholar]
- 19.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 20.Vasudevan D, Chua EYD, Davey CA. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Carey M, Workman JL. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Suganuma T, Workman JL. Annu. Rev. Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 23.Kulaeva OI, Zheng G, Polikanov YS, Colasanti AV, Clauvelin N, Mukhopadhyay S, Sengupta AM, Studitsky VM, Olson WK. J. Biol. Chem. 2012;287:20248–20257. doi: 10.1074/jbc.M111.333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura T, Liu Y-J, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- 25.Bennett RAO, Swerdlow PS, Povirk LF. Biochemistry. 1993;32:3188–3195. doi: 10.1021/bi00063a034. [DOI] [PubMed] [Google Scholar]
- 26.Zhou C, Greenberg MM. J. Am. Chem. Soc. 2012;134:8090–8093. doi: 10.1021/ja302993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowary PT, Widom J. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 28.See Supporting Information.
- 29.Cole HA, Tabor-Godwin JM, Hayes JJ. J. Biol. Chem. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinz JM, Rodriguez Y, Smerdon MJ. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luger K, Rechsteiner TJ, Flaus AJ, Waye MMY, Richmond TJ. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 32.Davey G, Wu B, Dong Y, Surana U, Davey CA. Nucl. Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuduvalli PN, Townsend CA, Tullius TD. Biochemistry. 1995;34:3899–3906. doi: 10.1021/bi00012a005. [DOI] [PubMed] [Google Scholar]
- 34.Raindlova V, Pohl R, Hocek M. Chem. Eur. J. 2012;18:4080–4087. doi: 10.1002/chem.201103270. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhry MA, Weinfeld M. J. Biol. Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 36.Mckenzie JA, Strauss PR. Biochemistry. 2001;40:13254–13261. doi: 10.1021/bi015587o. [DOI] [PubMed] [Google Scholar]
- 37.Georgakilas AG, Bennett PV, Sutherland BM. Nucleic Acids Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgakilas AG, Bennett PV, Wilson DM, Sutherland BM. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill P, Wardman P. Int. J. Radiat. Biol. 2009;85:9–25. doi: 10.1080/09553000802640401. [DOI] [PubMed] [Google Scholar]
- 40.Sage E, Harrison L. Mutat. Res. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asaithamby A, Hu B, Chen DJ. Proc. Nat. Acad. Sci. USA. 2011;108:8293–8298. doi: 10.1073/pnas.1016045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ianni JC. Kintecus, Windows Version 2.80. 2002 www.kintecus.com. [Google Scholar]
- 43.Zheng Y, Sheppard TL. Chem. Res. Toxicol. 2004;17:197–207. doi: 10.1021/tx034197v. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Gil JM, Greenberg MM. Angew. Chem. Int. Ed. 2003;42:5882–5885. doi: 10.1002/anie.200352102. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Stubbe J. Biochemistry. 2004;43:5278–5286. doi: 10.1021/bi0495376. [DOI] [PubMed] [Google Scholar]
- 46.Zhang K, Taylor J-S. Biochemistry. 2001;40:153–159. doi: 10.1021/bi001781j. [DOI] [PubMed] [Google Scholar]
- 47.Berson JA. In: Rearrangements in Ground and Excited States. de Mayo P, editor. Vol. 1. New York: Academic Press; 1980. [Google Scholar]
- 48.Hoffmann R, Minkin VI, Carpenter BK. Bull. Soc. Chim. France. 1996;133:117–130. [Google Scholar]
- 49.Kappen LS, Goldberg IH. Biochemistry. 1989;28:1027–1032. doi: 10.1021/bi00429a016. [DOI] [PubMed] [Google Scholar]
- 50.Xi Z, Goldberg IH. In: Comprehensive Natural Products Chemistry. Kool ET, editor. Vol. 7. Amsterdam: Elsevier; 1999. pp. 553–592. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.