Abstract
Epigenetics is emerging as an attractive mechanism to explain the persistent genomic embedding of early-life experiences. Tightly linked to chromatin, which packages DNA into chromosomes, epigenetic marks primarily serve to regulate the activity of genes. DNA methylation is the most accessible and characterized component of the many chromatin marks that constitute the epigenome, making it an ideal target for epigenetic studies in human populations. Here, using peripheral blood mononuclear cells collected from a community-based cohort stratified for early-life socioeconomic status, we measured DNA methylation in the promoter regions of more than 14,000 human genes. Using this approach, we broadly assessed and characterized epigenetic variation, identified some of the factors that sculpt the epigenome, and determined its functional relation to gene expression. We found that the leukocyte composition of peripheral blood covaried with patterns of DNA methylation at many sites, as did demographic factors, such as sex, age, and ethnicity. Furthermore, psychosocial factors, such as perceived stress, and cortisol output were associated with DNA methylation, as was early-life socioeconomic status. Interestingly, we determined that DNA methylation was strongly correlated to the ex vivo inflammatory response of peripheral blood mononuclear cells to stimulation with microbial products that engage Toll-like receptors. In contrast, our work found limited effects of DNA methylation marks on the expression of associated genes across individuals, suggesting a more complex relationship than anticipated.
Keywords: population cohort, early-life environment, immune response
Epigenetic processes not only regulate developmental programming and cellular identity but might also mediate the interaction of the environment with the genome (1, 2). The regulation of epigenetic variation likely is complex, involving various factors, such as ethnicity and aging, environmental exposures, genetic allelic variation, and stochastic elements (3, 4). Along with chromatin proteins, DNA methylation constitutes a main component of the epigenome, and it is considered the most stable and accessible epigenetic mark for quantitative measurements in human populations. As such, the emerging interest in exploring associations of DNA methylation with disease and phenotypic variation has led to the development of principles for epigenome-wide association studies (EWASs) (5, 6). However, the underlying biology of epigenetics, along with technical and methodological issues, poses major challenges for the realization of EWASs in human populations (5, 7, 8).
In somatic cells, DNA methylation occurs almost exclusively on cytosine residues in the context of CpG dinucleotides, which are nonrandomly distributed across the human genome (9–12). Specifically, the density of CpGs in a particular genomic region varies, allowing a classification into low-density CpG (LC) regions, intermediate density CpG (IC) regions, and high-density CpG (HC) regions (13). In concert with other chromatin modifications, DNA methylation has the capacity to regulate gene expression (14). This is best understood for tumor suppressor genes in cancer, whose expression is negatively regulated by increased methylation of HC regions in their promoter (15). However, the relationship between promoter DNA methylation and gene expression might be less straightforward in nonmalignant somatic tissues, because recent work in established cell lines has failed to establish broad correlations across individuals (16, 17).
Human epigenetic population studies have natural limits rooted in tissue-specific differences of epigenetic marks, because only a very small number of all human cell types are easily accessible for interrogation. Specifically, these are buccal epithelial cells and peripheral blood leukocytes. Many epigenetic studies in human populations have used unfractionated leukocytes without accounting for possible interindividual differences in subset composition and the distinct epigenomes likely associated with each cell type, an issue that has long been recognized in analogous gene expression studies (18–20). Despite these limitations, tremendous interest has arisen in relation to identifying biological and environmental factors that interact with the epigenome, and the functional consequences thereof (21). In the context of epigenetic population studies, it is important to appreciate that the intimate linkage between epigenetic marks and tissue specification results in between-tissue variation in DNA methylation that greatly exceeds between-individual differences within any one tissue (18, 22, 23).
In regard to developmental origins of adult human phenotypes, epigenetics has emerged as an attractive candidate responsible for persistent biological embedding of experiences during development or early life (24, 25). Specifically, epigenetic marks have been linked in previous research to diverse environmental exposures, including nutrition and maternal mood during pregnancy, early-life socioeconomic status (SES), abuse, and parental stress (26–31). Although often supported by analogous findings in animal models, the extant research in humans derives from a small group of correlational studies that generally used relatively small sample sizes and diverse technological platforms to interrogate the epigenome (8). Furthermore, rigorous statistical approaches for the analysis of genome-wide measurements of DNA methylation have not always been applied, or yet agreed on, by the community, thus hampering careful assessment of reported associations and comparisons between different studies (5, 8, 32).
Here, we aimed to characterize variation in human DNA methylation in an accessible tissue, to identify some of the factors that sculpt the epigenome, and to determine its functional relation to gene expression. We used a diverse suite of assessments ranging from blood composition to demographic and psychosocial factors to test for correlation to DNA methylation. By combining statistical approaches aimed at minimizing errors due to multiple testing and at deriving principal components governing coordinated variation in DNA methylation, we were able to begin developing a comprehensive framework and resource of factors predicting DNA methylation in a normative human population.
Results
Study Cohort and DNA Methylation Measurements.
To begin exploring epigenetic variation and its predictors in human populations, we purified peripheral blood mononuclear cells (PBMCs) from a community cohort of 92 individuals in the Vancouver lower mainland area (33). Individuals ranged in age from 24 to 45 y (median = 33.04, SD = 5.03) and were 62% female (n = 57) vs. 38% male (n = 35). Although this cohort broadly resembled the community average, it was stratified according to early-life SES. To measure DNA methylation in genomic DNA derived from PBMCs, we used the Infinium HumanMethylation27 array platform (Illumina), which enables the simultaneous quantitative assessment of 27,578 CpG loci at single-nucleotide resolution in the promoters or first exons of ∼14,475 genes. For each CpG site, a β-value is derived, which approximately corresponds to the percentage of methylated DNA molecules in a given sample. After filtering to remove technically unreliable or sex-biased probes on the X and Y chromosomes, we included a total of 22,922 CpGs in our analysis.
Variable DNA Methylation Loci Existed in Peripheral Blood.
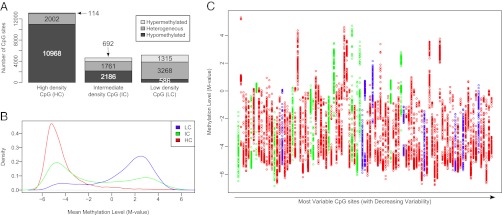
We decided to focus this work on PBMCs, because these are clinically relevant cells commonly used in immunological assays and are devoid of multinucleate granulocytes. The correlation between individuals was R2 = 0.986, which was lower than the correlation for technical replicates (R2 = 0.994), with the overall DNA methylation distribution having a highly bimodal pattern similar to the one observed in deep PBMC methylome sequencing from one individual (10) (Fig. S1A). Next, we divided CpG loci based on mean DNA methylation levels into hypomethylated (less than 20% β-values), heterogeneously methylated (β-value levels between 20% and 80%), and hypermethylated (more than 80% β-values) categories, according to published criteria (10, 22). Not unexpectedly, we found distinct contextual CpG density-dependent differences with HC region loci primarily present in the hypomethylated category, whereas LC region loci were primarily present in the heterogeneous and hypermethylated categories (10) (Fig. 1A).
Fig. 1.
Variable DNA methylation in PBMCs derived from a human cohort. (A) Distinct distribution of mean DNA methylation levels dependent on the context of CpG site. All CpG sites were classified into LC regions, IC regions, and HC regions. CpG sites were further divided based on mean DNA methylation levels into hypomethylated (less than 20% β-values, dark gray), heterogeneously methylated (β-value levels between 20% and 80%, gray), and hypermethylated (more than 80% β-values, light gray). (B) Mean DNA methylation levels are represented by M-values and divided according to CpG density categories. M-values are log transformations of methylated intensity over unmethylated intensity and resulted in a much more uniform variability. A M-value of 0 is equivalent to a β-value of 0.5, with negative M-values indicating less than and positive M-values indicating more than a β-value of 0.5. (C) Population distribution of the 99 CpG loci with a mean SD > 1, with CpG density category indicated by colors as in B. Each column depicts DNA methylation levels at one specific CpG site, with every individual in our study graphically represented by one dot. The most variable CpG site is shown in the leftmost column, with CpG sites arranged in columns of decreasing variability going from left to right.
Having established the general pattern of promoter methylation in our cohort, we next asked whether there was appreciable interindividual variation for each CpG site by comparing its SD with its mean methylation, akin to an approach used in mouse liver (34). To avoid the uneven variability across the methylation range when using β-values that causes the extremes to have much lower variability, we transformed the data into M-values (35). These represent the log of methylated intensity over unmethylated intensity and resulted in a much more uniform variability, consistent with published data (Fig. S1 B and C). Using M-values did not alter the general distribution of CpGs in HC, IC, or LC regions (Fig. 1B and Fig. S1A). Displaying SD distributions from M-values suggested that HC region loci were most variable, followed by IC and LC region loci, although the differences were small (Fig. S1D). This was statistically substantiated by ANOVA, which revealed significant differences when comparing the SD of HC, IC, and LC sites (P = 4.25E-14). This was attributed to HC sites, which were significantly different from IC (P = 2.75E-6) and LC (P = 2.20E-16) sites as determined by Tukey “honestly significant difference” comparison. Using an SD cutoff of 0.5, ∼7.8% of all CpG sites (1,779 of 22,922) were variable, whereas an SD cutoff of 1 resulted in 99 variable CpG sites (0.43%). The variation across the methylation range and individuals for these 99 CpG sites generally showed a continuous pattern (Fig. 1C). Although all our subsequent analyses were done using the M-value–transformed data, we retransformed them after a given analysis back to β-values because these are easier to understand.
Blood Composition Was Associated with DNA Methylation.
Although lacking granulocytes, PBMCs are still a somewhat heterogeneous mixture of white blood cells. Primarily, these are lymphocytes and monocytes, which, on average, comprised 31.56% (SD = 8.37) and 6.74% (SD = 2.1) of the total circulating leukocytes in our cohort, respectively. (The other 63% of cells in the pool were granulocytes, which we removed before DNA extraction by means of density-gradient centrifugation). We wanted to determine whether the amount of lymphocytes and monocytes present in an individual would affect the DNA methylation patterns derived from PBMCs. To reduce the number of tests, we eliminated all CpGs with less than 5% or more than 95% β-values across all samples, corresponding to sites that were almost uniformly unmethylated and uniformly methylated in our cohort, thus leaving 17,870 CpGs for all subsequent analyses. Using Spearman correlation and correction of the false discovery rate (FDR) at a q value <5% and further filtering for a minimum methylation difference >5%, we identified high-confidence associations of 264 CpG sites with lymphocyte percentage and 248 CpG sites with monocyte percentage, as determined by whole blood cell count. As expected, there was substantial overlap between these two sets of CpG loci (n = 119), which resulted in a sum total of 393 subtype-associated high-confidence CpG loci, representing 2.1% of the 17,870 CpGs included in this analysis. Using more relaxed criteria at an FDR at a q value <25% and no filtering for absolute methylation difference, we found 1,323 CpGs associated with lymphocyte percentage and 2,182 CpGs associated with monocyte percentage, respectively. Accounting for the overlap (n = 463), the remaining subtype-associated 3,042 CpG sites corresponded to 17.0% of the 17,870 CpGs assessed. The association of PBMC DNA methylation with lymphocyte percentage derived from whole blood was further substantiated by P-value distributions (Fig. S2A) and quantile-quantile (Q-Q) plots (Fig. S2B) that deviated clearly from what would be expected by chance alone. Similar results were found in the analogous representations for monocyte percentage (Fig. S2 C and D).
Using a second smaller cohort, we compared the DNA methylation profile of PBMCs with that of CD14+ monocytes and CD3+ T cells purified by immunomagnetic selection. Based on within-person comparisons, we found 1,208 CpG sites that were specific for one of the three classes as determined by ANOVA, primarily driven by the differences between monocytes and T cells/PBMCs (details are provided in SI Results and Fig. S3). Reassuringly, this set included 93.2% (246 of 264) of the high-confidence CpGs associated with lymphocyte percentage and 90.7% (225 of 248) of the CpGs associated with monocyte percentage in the community cohort analysis, suggesting that our statistical approach to determine correlations was very well-supported by experimental data.
These findings illustrated that at a large number of CpG loci, methylation readouts are influenced by the cellular composition of a blood sample. Purification of the desired cell type population is the most reliable approach for eliminating this unwanted interindividual variability. However, it is not always possible to perform such purifications due to the volume of blood and specialized technology required to obtain sufficient amounts of all salient cell types. To circumvent this problem, we developed a multiple regression approach that can be applied to methylation data post hoc, assuming a complete blood cell count was performed simultaneously (Materials and Methods). After applying this approach to our original dataset, the previously observed correlations between subset percentages and DNA methylation readouts were eliminated. Thus, statistical corrections may be able to minimize confounding caused by differences in individuals’ leukocyte subtype composition in future methylation studies.
Demographics Were Associated with Variable DNA Methylation.
We next tested whether demographic, psychosocial, and environmental factors predicted DNA methylation, because we had ascertained a comprehensive set of 15 variables belonging to these categories in our cohort (Table S1). We categorized associated loci into either the high-confidence group as above (5% or less FDR) or a medium-confidence group (25% or less FDR), similar to an approach we used previously (31). The latter is more liberal in cognizance of our small sample size and, in some cases, the more distant relationship between variables and PBMC DNA methylation. Details of all correlation analyses, including the number of CpG loci belonging to each category and the fraction of those having a methylation change of more than 5%, are presented (Table S1).
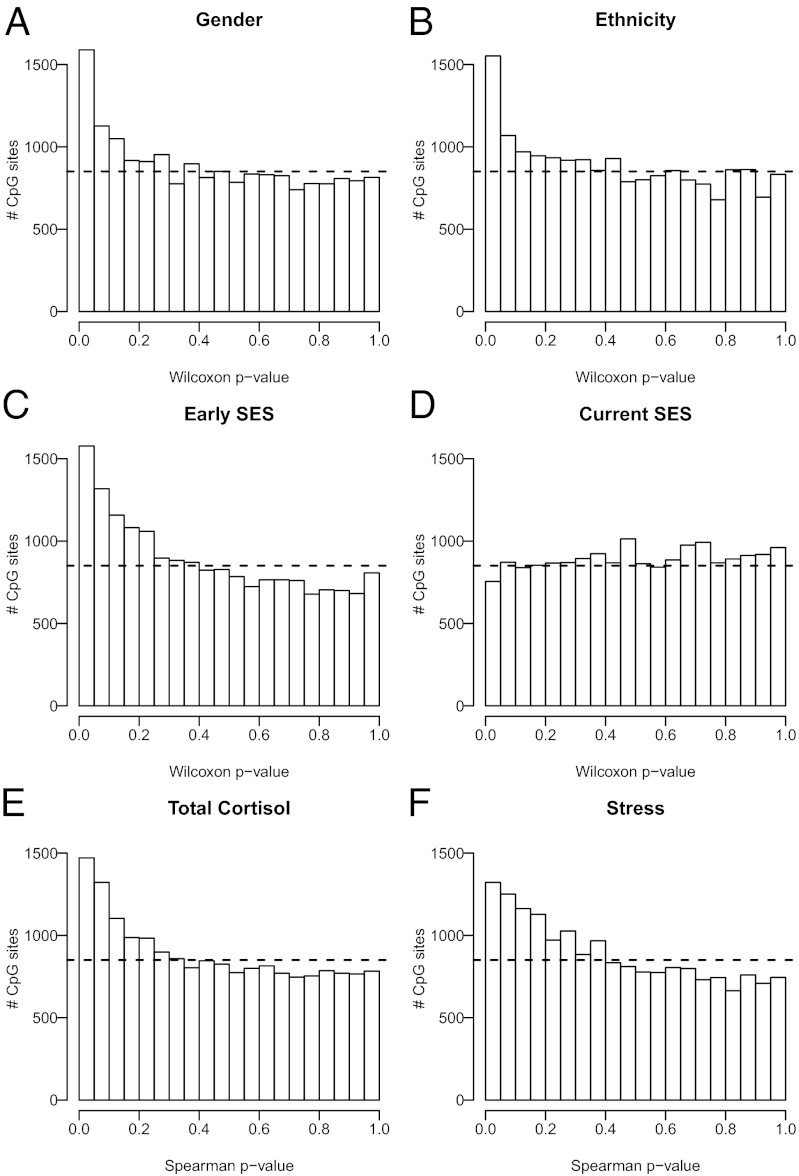
Because our cohort included both males and females, we first tested whether DNA methylation was associated with sex. We identified 487 autosomal CpG loci associated with sex, of which 123 belonged to the high-confidence class. However, the majority of differences were subtle, with only 27 CpGs having more than 5% methylation change between the genders. In addition to the FDR method, these findings were further substantiated by a skewed P-value distribution (Fig. 2A) and a Q-Q plot that deviated from the randomly expected values (Fig. S4A). Age is another demographic characteristic that has been associated with DNA methylation. Despite the narrow age range present in our cohort, we identified 2 high-confidence and 13 medium-confidence CpG sites that were associated with aging, with 6 of them having more than 5% methylation change (Table S1).
Fig. 2.
Demographic and psychosocial factors were associated with DNA methylation. Graphical representations of P-value distributions. In each case, the dashed line represents the uniform distribution that was expected by chance. The skewed distributions with an enrichment of CpG sites having small P values suggested that sex (A), ethnicity (B), and early-life SES (C) were correlated with DNA methylation. (D) This contrasted with the lack of correlation suggested by the uniform P-value distribution of current SES. Furthermore, cortisol output (E) and perceived stress (F) were both correlated with DNA methylation. Testing for correlations was done using either Wilcoxon tests (A–D) or Spearman ρ statistics (E and F).
Given that most community cohorts will include participants from a variety of racial/ethnic backgrounds, we next tested whether this was associated with variation in DNA methylation. Due to the small sample size, we divided the cohort into Caucasian (n = 63) and non-Caucasian (n = 29) subjects, with the latter being primarily of Asian or mixed descent. Even with this admittedly rough grouping, we found 299 medium-confidence CpG sites associated with ethnicity, of which 21 had more than 5% methylation change (Table S1). Consistent with DNA methylation being associated with ethnicity, both the P-value distribution (Fig. 2B) and the Q-Q plot were skewed (Fig. S4B).
Early-Life Poverty and Adult Stress Were Correlated with DNA Methylation.
Our cohort was assembled to test for influences of early-life SES lasting into adulthood irrespective of current SES (33). Thus, half of the cohort grew up in low-SES households and the remainder in high-SES households, as defined by the commonly used occupational prestige of the subjects’ parents. Occupational prestige reflects the long-term outcome of a person’s educational background and provides a rough approximation of his/her earning potential. It takes into account social context in the sense that status rankings are based on values that are likely to be somewhat culturally bound (33). Each group was further stratified into low-SES and high-SES categories based on the subject’s current SES. We identified three medium-confidence CpG sites associated with early-life SES, although none had more than 5% change in DNA methylation (Table S1). Perhaps more importantly, we observed the characteristic nonrandom P-value distribution (Fig. 2C) and Q-Q plot skewing (Fig. S4C). This contrasted with current SES, in which both the P-value distribution (Fig. 2D) and the Q-Q plot (Fig. S4D) resembled what would be expected by chance. Interestingly, SES analysis also provided a clear example of the importance of accounting for differential blood cell counts between individuals. Specifically, when using noncorrected DNA methylation data, current SES actually had a more suggestive P-value distribution pattern, which strongly decreased after applying our regression algorithm described above (Fig. S5A). In contrast, this method made no difference for early-life SES (Fig. S5B).
We next tested whether psychosocial stress was associated with DNA methylation. Both subjects’ self-reports of total cortisol levels and perceived stress, based on 18 saliva samples collected over 3 d of monitoring, had P values (Fig. 2 E and F) and distribution Q-Q plots (Fig. S4 E and F) that were significantly different from patterns expected by chance, suggesting that these factors might be correlated with PBMC DNA methylation. This was further substantiated by the identification of five medium-confidence CpG sites that were correlated to total cortisol levels, one of which had a change of >5% in DNA methylation levels (Table S1).
DNA Methylation Predicted ex Vivo Stimulation Response of PBMCs.
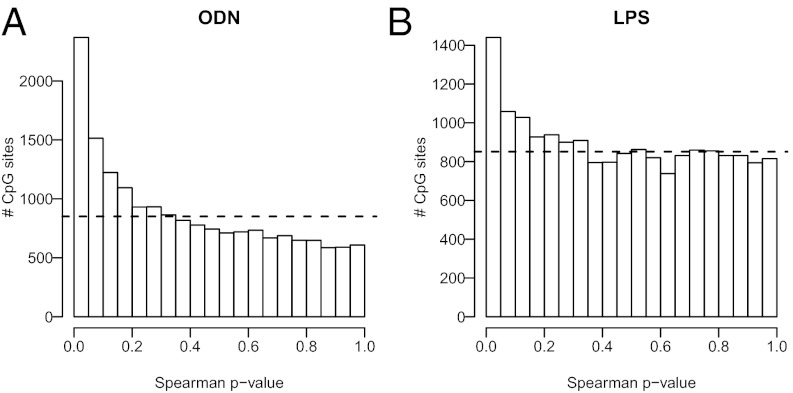
A portion of the blood collected for methylation studies was also used to quantify the functional capacity of PBMCs as determined by their capacity to engage distinct Toll-like receptor (TLR) signaling pathways. TLRs are a family of receptors expressed by cells of the innate immune system and other tissues involved in host defense. On recognizing common microbial threats, TLRs activate signaling cascades that orchestrate an early-stage immune response involving inflammation. TLRs are considered the interface between the microbial world and the immune system. Importantly, we have previously shown that PBMCs from low early-life SES show greater stimulated production of the proinflammatory cytokine IL-6 after ex vivo stimulation with TLR ligands (36). After they had been separated through centrifugation, PBMCs were cultured for 6 h with a variety of microbial products known to act through distinct TLR signaling pathways. Responses were quantified by production of the proinflammatory IL-6, which was measured in culture supernatants using highly sensitive immunoassays. Interestingly, we found strong associations between DNA methylation and PBMC responsivity to several TLR ligands (Table S2). In particular, 2,408 medium-confidence CpG sites were associated with IL-6 responses to oligodeoxynucleotide (ODN; unmethylated DNA), a TLR-9 agonist, with 364 of them having a methylation change >5%. In addition, DNA methylation at 52 medium-confidence CpG sites correlated with IL-6 responses to LPS (a molecule on the surface of Gram-negative bacteria), a TLR-4 agonist, and 8 of them survived filtering for methylation changes >5% (Table S2). For ex vivo IL-6 responses to ODN and LPS, the P-value distributions (Fig. 3 A and B) and Q-Q plots (Fig. S6 A and B) clearly differed from a random pattern. Reassuringly, many biologically plausible candidates related to immune and inflammatory response were included in the sets of most strongly associated genes, including chemokine ligand 11 (ρ = 0.3007, q value = 0.10, methylation change = −9.0%) for the TLR-9 stimulation and IL-1β (ρ = −0.4191, q value = 0.12, methylation change = 5.6%) for the TLR-4 stimulation. We also identified a small number of medium-confidence CpG sites associated with IL-6 responses to peptidoglycan (a bacterial cell wall component that functions as a TLR-2 agonist) and IL-1β (a proinflammatory cytokine used to activate PBMCs nonspecifically (Table S2), despite the fact that their P-value distributions barely deviated from random distributions (Fig. S6 C and D).
Fig. 3.
DNA methylation was predictive of PBMC ex vivo response. As judged by the skewed P-value distributions, IL-6 production in PBMCs was associated with DNA methylation on ex vivo stimulation for 6 h by ODN (A) and LPS (B). Dashed lines represent the uniform distribution expected by chance. Testing for correlations was done using Spearman ρ statistics.
Principal Component Analysis of DNA Methylation Revealed Correlated Patterns Among Individuals.
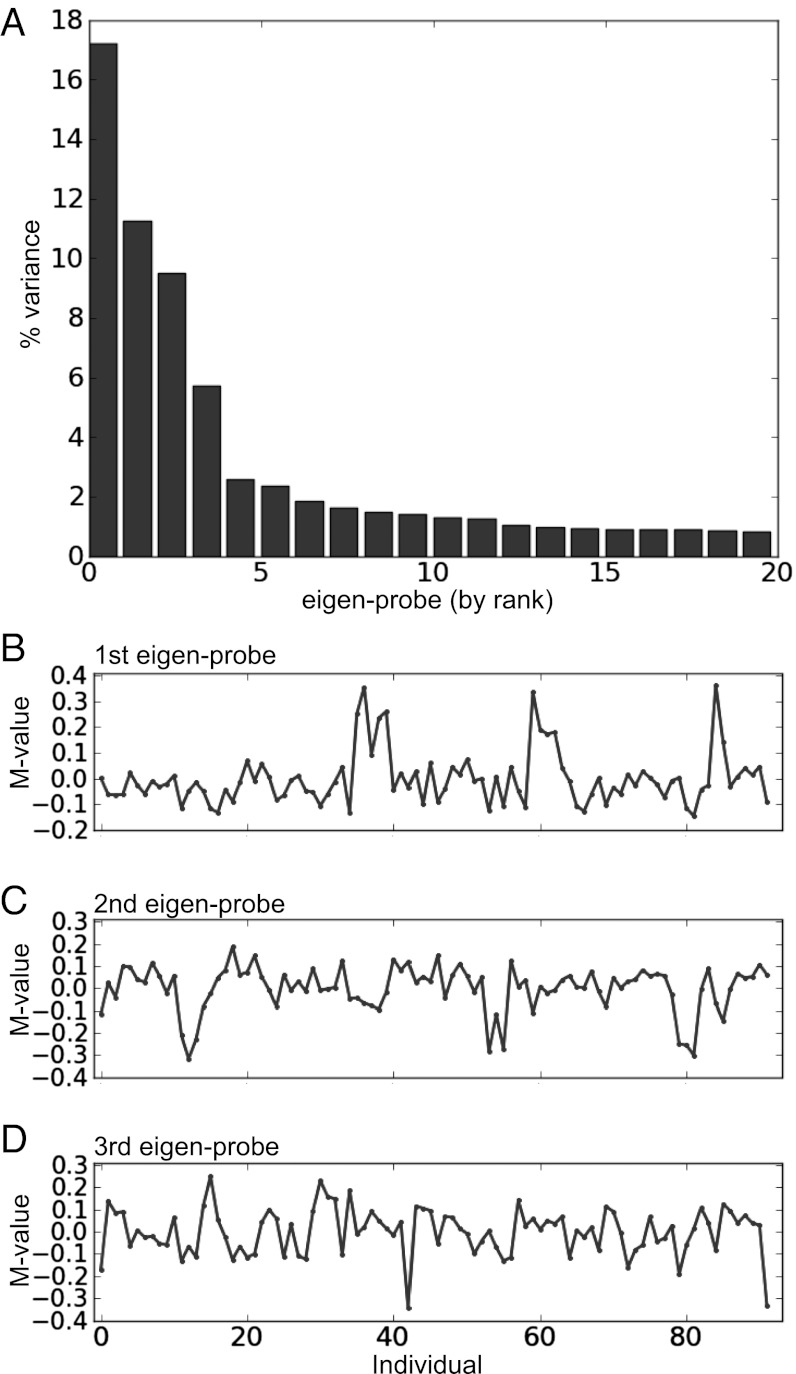
The above analysis shows that it was possible to identify probes that correlate with specific phenotypic traits. Next, we applied a different approach to identify common patterns of methylation variation across the population, identifying individuals within the population that show correlations in DNA methylation. Using principal component analysis (PCA) allows us to identify those individuals who have CpG loci that show the most covariation across the population. These population variances can then be correlated and tested for statistical enrichment for any particular trait. It is possible that the identified patterns of methylation variation across the population may possess multiple statistically significant traits. Using a covariance matrix between individuals, we calculated the eigenvectors of this matrix to determine the principal components. We called these “eigen-probes” because they revealed the dominant type of probe patterns across the population, in analogy to the “eigen-genes” identified in gene expression microarrays (37).
The top/zeroth eigen-probe associated with the largest eigenvalue merely reflected a difference in the population mean probe intensity from one probe to the next and was not considered in what follows because it is not informative for variations across individuals. The remaining eigen-probes revealed variation in DNA methylation across the population. For example, the first, second, and third eigen-probes accounted for 17%, 11%, and 9% of the variance, respectively (Fig. 4A). Closer examination showed that there were a few dominant patterns of variation, suggesting that there were CpG probes in the data whose methylation either strongly rose or decreased for these individuals compared with the rest of the individuals in the population. The first eigen-probe revealed a group of ∼10 individuals who showed correlations in their methylation, reflected as positive peaks (Fig. 4B). The second and third eigen-probes also showed sets of individuals whose methylation strongly covaried compared with others (Fig. 4 C and D).
Fig. 4.
PCA revealed covariant DNA methylation patterns in a population. (A) Percentage of variation in DNA methylation accounted for by the top 20 eigen-probes. (B–D) DNA methylation signatures for the top 3 eigen-probes over the population of individuals. Using a methylation variation (M-value) cutoff of ±0.1 allowed us to create groups of correlated individuals for each eigen-probe that we could then test for enrichment for particular traits (main text).
We correlated the eigen-probes with the variation of traits (e.g., age, body mass index, IL-6 response to ODN) across the population (Materials and Methods) and found 9 eigen-probes of the 92 that correlated strongly with one of the traits after Bonferroni correction for multiple testing (P values < 0.001). As previously found on a probe-by-probe basis, traits that showed correlation with a pattern of methylation across the population were ODN level, ethnicity, sex, and stress. Only one of the eigen-probes showed correlation with multiple traits (depression and stress). At this level of stringency, only the second eigen-probe (Fig. 4C) of the top three eigen-probes correlated with any particular trait over the whole population (in this case, IL-6 response to ODN; P = 0.001).
The above analysis was based on assessing correlations between patterns of methylation with quantitative traits over the entire population. Given that the top eigen-probes revealed that there were distinct groups of individuals who strongly covaried in their methylation patterns compared with others in the population, we tested these subgroups for enrichment in a given trait. For the first eigen-probe (Fig. 4B), we identified a group of 10 subjects that was enriched for low early-life SES (8 of 10 individuals, Bonferroni-corrected P = 0.006.). The groups formed from the second eigen-probe (Fig. 4C) using the above criterion showed no strong enrichment for any trait. The other top eigen-probes showed groups enriched for ethnicity and exercise. Thus, analysis of the top eigen-probes showed that it was possible to group individuals based on common variation in methylation and that these groups possessed enriched traits that may not have been revealed when considering how the trait correlates with the pattern of methylation over the whole population.
Variable DNA Methylation Across Individuals Was Not Closely Linked to Gene Expression.
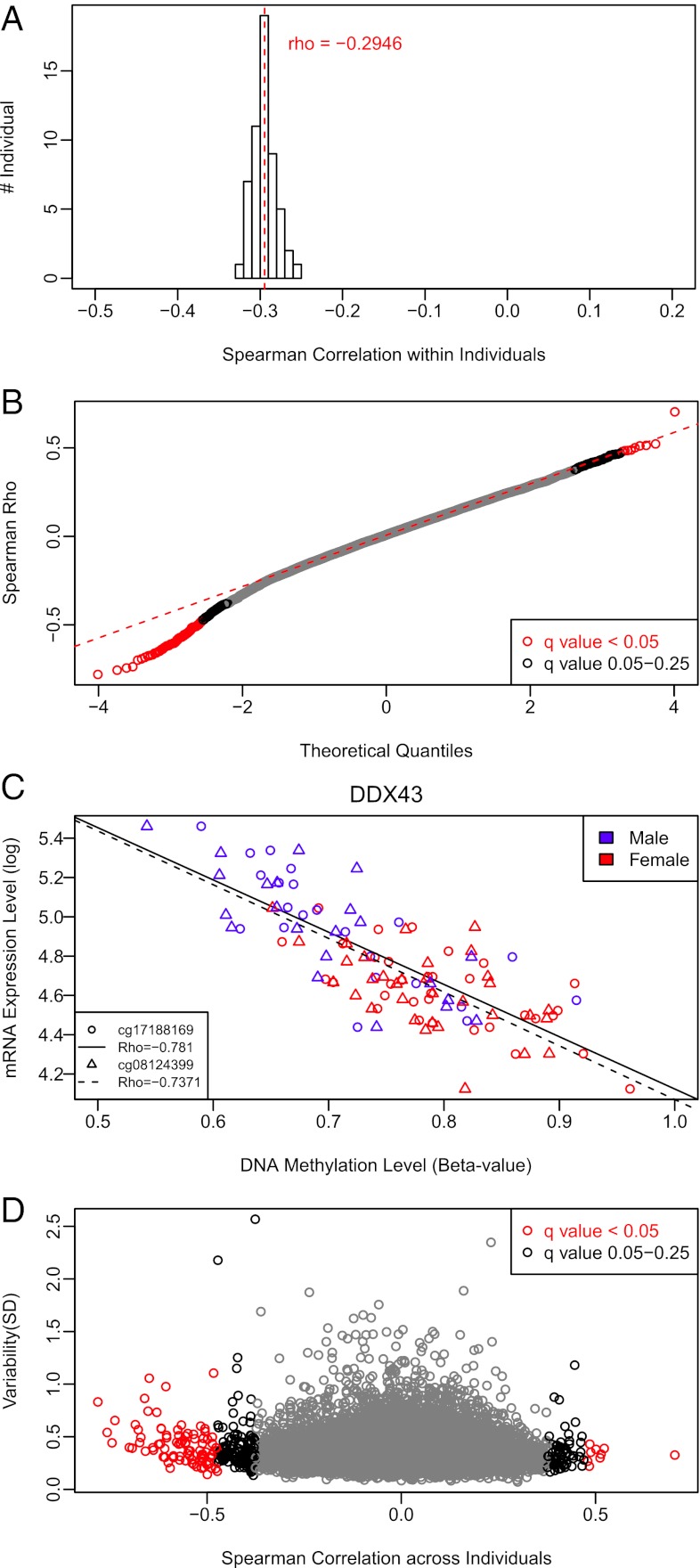
For 55 of our subjects, we had previously published gene expression studies with mRNA levels in the very same PBMC fraction used to extract genomic DNA for methylation studies (33, 38). This allowed for a direct assessment of the linkage between DNA methylation and gene expression. As expected, the overall correlation between DNA methylation and expression of the associated gene within an individual was negative, with a mean Spearman correlation of ρ = −0.29 (Fig. 5A). This means that highly methylated genes were expressed at low levels, and vice versa. We note that although this was generally true, a substantial proportion of genes behaved differently from this pattern. For example, some lowly methylated promoters were associated with low expression levels and some highly methylated promoters were associated with high expression levels (Fig. S7). However, rather than focusing on individual correlations, it is much more relevant for epidemiological studies to compare the methylation state of a given gene’s promoter and its expression level across individuals. If DNA methylation and gene expression were tightly linked, we would expect that changes in DNA methylation between individuals would correspond to changes in expression levels between individuals. Using this approach, our analysis showed that both Q-Q plots (Fig. 5B) and P-value distribution (Fig. S8) were indeed consistent with a nonrandom correlation between DNA methylation and gene expression. However, using FDR correction, we found that only 97 of the 16,419 CpG sites tested (0.6%) had high-confidence correlations (FDR < 5%) to expression of their associated genes, with an additional 213 CpG sites (1.3%) being correlated when medium-confidence FDR correction was used (Table S3). This relationship was largely driven by the genomic context of a given CpG, because LC region loci were overrepresented with an odds ratio of 3.139 (P = 3.544E-8) and HC region loci were underrepresented with an odds ratio of 0.2713 (P = 8.564E-9). Although the majority of our high- and medium-confidence CpGs had the expected negative correlation to gene expression, there clearly were CpGs with a positive correlation (Fig. 5B). Specifically, among 97 of the high-confidence CpG sites, 88 (90.7%) had a negative correlation, whereas 9 (9.3%) had a positive correlation. However, we noticed that the magnitude of change in DNA methylation even for high-confidence CpG loci was often below 5% (Table S3). The most striking example of a robust association was the negative correlation between DDX43 gene expression and two CpGs in its promoter (Fig. 5C). DDX43 encodes an ATP-dependent RNA helicase in the DEAD-box family that is overexpressed in various solid cancers and hematological malignancies and, coincidentally, was also one of the genes identified as having sex-specific DNA methylation. Next, we examined whether highly variable DNA methylation was associated with concurrent expression changes. Surprisingly, many CpG loci that had substantial variation were not correlated with gene expression, suggesting that variable DNA methylation was not obligatorily transmitted to the functional level of mRNA production (Fig. 5D).
Fig. 5.
DNA methylation and gene expression were not tightly linked across individuals. (A) Histogram of correlations between DNA methylation and expression of the associated genes within each individual across all 16,419 CpG sites demonstrated the canonical negative correlation between DNA methylation and expression. Average correlation is shown in red. (B) Q-Q plot shows the association of DNA methylation and mRNA expression of associated genes across individuals. Although the majority of significant correlations were negative, a substantial fraction was unexpectedly positive. (C) Representative example of two CpG loci in the promoter of the DDX43 gene that had a strong negative correlation with expression across individuals. These sites were also differentially methylated between males and females. Lines of least square and Spearman correlation between DNA methylation at each site and mRNA expression of DDX43 gene are shown on the graph. (D) Only a minority of variable CpG sites had a significant correlation with mRNA expression across individuals. The extent of variation in DNA methylation is shown as SD on the y axis, whereas the correlation between DNA methylation and expression of the associated gene is shown on the x axis. Each circle indicates one CpG site. Testing for correlations was done using Spearman ρ statistics, and the red circles in B and D indicate CpG sites that survive FDR correction at a q value <5%, whereas the black circles indicate CpG sites with q values between 5% and 25%.
Discussion
This study broadly assessed variation of DNA methylation in one of the few accessible tissues in a human community cohort, thereby providing a reference framework for factors that are associated with epigenetic variation. Having established the presence of epigenetic variation in the cohort, we showed that the leukocyte composition of peripheral blood covaried with patterns of DNA methylation at many sites. Demographic factors, such as sex, age, and ethnicity, were also associated with variable DNA methylation, as were stress and cortisol output. Consistent with epigenetic marks constituting a mechanism for biological embedding of early-life experiences, we found an association between DNA methylation and low early-life SES. Interestingly, we found cases in which DNA methylation was strongly related to the ex vivo inflammatory response of PBMCs to stimulation with microbial products that engage TLRs. The general validity of these correlations was further supported by the identification of similar DNA methylation patterns between individuals using PCA, which suggested the coordinated effects of several individual factors giving rise to commonalities in epigenetic marks. Lastly, although DNA methylation within an individual had the expected negative correlation with gene expression, we found limited effects of DNA methylation marks on the expression of their associated genes across individuals. This suggested that in human populations, the relationship between DNA methylation and gene expression was more complex than might have been expected from research in model systems.
The number of sites with variable DNA methylation was relatively small, yet the extent of variation at a given CpG locus could be substantial. When examining correlates of this variation, we found leukocyte composition to be the strongest quantitative and qualitative correlate of variable DNA methylation. Specifically, a large number of CpG sites were associated with the relative amount of either lymphocytes or monocytes present in the leukocyte fraction. Reassuringly, the vast majority of these sites were distinctly methylated in CD3+ T cells or CD14+ monocytes that had been isolated from the larger PBMC population through immunomagnetic separation. Thus, our data suggest that careful measurements of leukocyte composition differences between individuals must be made, because these might affect associations between environmental variables and DNA methylation. Perhaps not surprisingly, this is reminiscent of similar findings in gene expression studies, and as such, it has profound implications for the interpretation of DNA methylation studies using DNA derived from whole blood (19, 20). This problem can be mitigated by applying a statistical model that incorporates differential cell counts and by using these “corrected” methylation values for subsequent analyses, as we have done here. Although not always “doable” in practice, an even better approach would be to perform the epigenetic profiling after immunomagnetic purification of the specific blood cell subtypes of interest for a particular biological question.
We were surprised by the relative scarcity of statistically significant associations between PBMC DNA methylation at any single CpG site and the 12 environmental and psychosocial factors assessed here. However, the skewed distribution patterns of P values and the deviations from random in the Q-Q plots supported a general trend toward correlation between DNA methylation and certain variables, such as early-life SES, perceived stress, and cortisol output. Although this likely indicated small effects, their widespread prevalence is a unique finding and was not inconsistent with the lack of any single CpG sites surviving an FDR of 5%, given the stringent multiple-testing correction necessary in any analysis involving thousands of statistical tests. It is tempting to speculate that larger cohorts will help to substantiate the statistical significance of these general correlations. Regardless, our findings might provide the opportunity to serve as a starting point for performing power calculations relevant for epigenetic epidemiology (5, 8).
In part, the lack of strong statistical associations might also be linked to the characteristics of our cohort. Specifically, for some variables of interest, such as smoking and alcohol consumption, we did not have the range necessary for proper statistical assessments. In addition, with the exception of early-life SES, our cohort did not include individuals at the edges of the phenotypic spectrum. For example, given that our participants did not have chronically high stress levels, inclusion of such individuals might result in stronger correlations than the one already indicated by the nonrandom P value distribution. We note that similar array-based approaches have uncovered associations of DNA methylation with disparate variables and exposures not interrogated here, including hair dye use, smoking, sun exposure, exercise in sedentary people, posttraumatic stress disorder, parental stress, and institutionalized children (31, 39–45). In each case, the number of significant CpG loci is rather small and comparable to our findings, as is the limited magnitude of change in DNA methylation, which rarely exceeds 10%. A similarly limited extent of epigenetic alterations is found in nonmalignant complex diseases, such as diabetes mellitus and systemic lupus erythematosus, even when the affected tissue was interrogated (46–48). Although this might be reflective, in part, of the limited number of CpGs present on the array, it is certainly consistent with our findings.
One of the most remarkable and surprising findings reported here was the association of DNA methylation with PBMC inflammatory responses to TLR ex vivo stimulation. Specifically, the number of correlated CpG sites and the magnitude of methylation change were only slightly lower than the numbers we found for leukocyte cell composition, although the latter was generally more statistically significant. The underlying biology for the expansive correlation of DNA methylation with IL-6 production in response to ex vivo stimulation with ODN and LPS remains to be determined, as do the reasons for this effect apparently being limited to certain stimuli.
Reassuringly, the general trend emerging from PCA largely corroborated the findings from individual analyses. PCA revealed the common patterns of covariation in methylation across the population, and a small number of eigen-probes showed correlation with a single quantitative trait. The predictive traits were consistent with those found on the probe-by-probe analysis. By examining the eigen-probes, it was possible to identify groups of individuals who shared covarying methylation patterns on these subsets of probes. We have found that these individuals were indeed enriched for particular traits or combinations thereof, as nicely exemplified by the enrichments of subjects with low early-life SES. The groupings of individuals that the first eigen-probes revealed also showed that correlating methylation patterns over the whole population with a particular trait can potentially miss enrichments that exist within subgroups. This and other potential interactions will provide the basis for further exploration of the specific DNA methylation sites underlying these groupings, and their relation to each other.
One issue of particular interest relates to the role of epigenetics in the embedding of early-life experiences. Although our data from both the correlation analysis and the PCA clearly support an association of early-life SES with adult DNA methylation, the small number of statistically significant correlated CpG loci appears to be at odds with existing data from a comparable but smaller 1958 British cohort study reporting DNA methylation changes in 1,252 promoters correlating to early-life SES (26). However, this result is based on an analysis treating neighboring microarray probes as independent data points to identify significantly differentially methylated regions. Because the probes have an average spacing of ∼100 bp, which is smaller than the size of DNA fragments hybridized to the array, neighboring probes are not truly independent, and treating them as independent would be expected to inflate the significance of any small differences between groups. In addition, details of FDR calculations and data normalization not adequately described in the paper could further add to these inflated statistics. Lastly, it is possible that the differences are rooted, in part, in the use of unfractionated leukocytes in the 1958 cohort study. Regardless, in principle, the relationship between early-life but not current SES and DNA methylation reported here and in the 1958 cohort paper mirrors the association of gene expression profiles with SES that we published earlier using the same cohort (33).
Our experimental approach of measuring DNA methylation and mRNA expression from the exact same primary PBMC samples derived from our cohort resulted in several intriguing findings. Although we did confirm the well-established general relationship between DNA methylation levels and gene expression across genes within an individual, we found that across individuals, only a small percentage of all DNA methylation loci had a significant relation to expression of their associated gene. As expected, the majority of the latter correlations were negative, although there were a substantial number of genes for which there was a positive correlation between DNA methylation and gene expression. In the context of epigenetic epidemiology, it was perhaps most striking that CpG loci with variable DNA methylation did not have an obvious association with gene expression. This suggested that variation in DNA methylation does not necessarily relate to variation in gene expression in primary PBMCs, and hence that the relationship between gene expression and promoter CpG methylation is more complex than previously appreciated. The reason for this is unclear, but it might be related to multiple additional layers of epigenetic modifications cooperating with DNA methylation to regulate gene expression. It is also possible that variable DNA methylation serves to poise its associated gene for expression in response to future events, such as a challenge to the immune system. Our findings with respect to the ex vivo stimulation hinted at this possibility, although we did not explicitly test mRNA expression after engaging TLRs. Regardless, our data were consistent with published work that either failed to establish strong connections between differential DNA methylation and gene expression or found small but significant sets of genes with positive correlations between promoter methylation and gene expression (16, 22, 46, 49). Given these complexities, it is obvious that the simple formula of decreased methylation equaling elevated expression might not always be suitable for human cohort studies, even though this picture has often been conveyed in the nascent field of social epigenetics.
The data presented here provide important insight into factors that need to be considered when evaluating epigenetic variation in human populations. For example, the identification of autosomal loci associated with demographic factors, such as sex, ethnicity, and age, suggests that these relationships need to be taken into account when testing for effects of the environment on the epigenome. In addition, it is important to consider that variation of DNA methylation can be linked to allelic variation at nearby SNPs, although this was not assessed in our study (16, 17, 50, 51). Indeed, when appropriately incorporating the concepts introduced here and addressing published concerns integral to human epigenetic epidemiology, there is no doubt that the early promise of social epigenetics will mature to yield important contributions to our understanding of the causes and effects of phenotypic variation in a developmental context (5, 8, 21, 25).
Materials and Methods
Detailed information on materials and methods used in this study is provided in SI Materials and Methods. Briefly, the Illumina HumanMethylation27 array platform was used to measure DNA methylation at 27,578 CpG sites in PBMC genomic DNA obtained from a published community cohort constituting 92 individuals of mixed sex and ethnicity (33). DNA methylation was correlated to a variety of biological, demographic, and psychosocial variables carefully ascertained in each individual using either the Wilcoxon rank sum test or Spearman correlation. Multiple testing corrections were done using the FDR method to determine a q value (52).
Supplementary Material
Acknowledgments
We thank E. Magda Price for generously sharing array annotations, Sarah Mah for suggestions, and Tanya Erb for editorial assistance. M.S.K. is a Scholar of the Canadian Institute for Advanced Research (CIFAR) in the Experience-Based Brain and Biological Development program, and a Scholar of the Mowafaghian Foundation. E.E. is also a CIFAR Scholar. H.B.F. is an Alfred P. Sloan Fellow and a Pew Scholar in the Biomedical Sciences. This work was primarily supported by grants to G.E.M., E.C., and M.S.K. from the National Institute of Child Health and Human Development (Grant HD058502), the British Columbia Ministry of Child and Family Development via the Human Early Learning Partnership, and AllerGen Networks of Centres of Excellence. Additional funding was provided by Grant R24-MH081797 (to M.S.K.) from the National Institute of Mental Health. Research in E.E.’s laboratory is supported by a grant from the National Sciences and Engineering Council of Canada.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Biological Embedding of Early Social Adversity: From Fruit Flies to Kindergartners,” held December 9–10, 2011, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/biological-embedding.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE37008).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121249109/-/DCSupplemental.
References
- 1.Mohn F, Schübeler D. Genetics and epigenetics: Stability and plasticity during cellular differentiation. Trends Genet. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feil R, Fraga MF. Epigenetics and the environment: Emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 4.Pujadas E, Feinberg AP. Regulated noise in the epigenetic landscape of development and disease. Cell. 2012;148:1123–1131. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Hatchwell E, Greally JM. The potential role of epigenomic dysregulation in complex human disease. Trends Genet. 2007;23:588–595. doi: 10.1016/j.tig.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41:74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illingworth RS, Bird AP. CpG islands—‘A rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 14.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 16.Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell JT, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byun HM, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18:4808–4817. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobb JP, et al. Inflammation and Host Response to Injury Large-Scale Collaborative Research Program Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci USA. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meaney MJ. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010;81:41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- 22.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies MN, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochberg Z, et al. Child health, developmental plasticity, and epigenetic programming. Endocr Rev. 2011;32:159–224. doi: 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hertzman C, Boyce T (2010) How experience gets under the skin to create gradients in developmental health. Annu Rev Public Health 31: 329–347, 3p following 347. [DOI] [PubMed]
- 26.Borghol N, et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41:62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heijmans BT, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberlander TF, et al. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 29.Schroeder JW, et al. DNA methylation in neonates born to women receiving psychiatric care. Epigenetics. 2012;7:409–414. doi: 10.4161/epi.19551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Essex MJ, et al. Epigenetic vestiges of early developmental adversity: Childhood stress exposure and DNA methylation in adolescence. Child Dev. 2011 doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 33.Miller GE, et al. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci USA. 2009;106:14716–14721. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du P, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirschfeld AF, et al. Prevalence of Toll-like receptor signalling defects in apparently healthy children who developed invasive pneumococcal infection. Clin Immunol. 2007;122:271–278. doi: 10.1016/j.clim.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Alter O, Brown PO, Botstein D. Generalized singular value decomposition for comparative analysis of genome-scale expression data sets of two different organisms. Proc Natl Acad Sci USA. 2003;100:3351–3356. doi: 10.1073/pnas.0530258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen E, Miller GE, Kobor MS, Cole SW. Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol Psychiatry. 2011;16:729–737. doi: 10.1038/mp.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langevin SM, et al. The influence of aging, environmental exposures and local sequence features on the variation of DNA methylation in blood. Epigenetics. 2011;6:908–919. doi: 10.4161/epi.6.7.16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan ES, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21:3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grönniger E, et al. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010;6:e1000971. doi: 10.1371/journal.pgen.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrès R, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Ressler KJ, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naumova OY, et al. Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Dev Psychopathol. 2012;24:143–155. doi: 10.1017/S0954579411000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkmar M, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakyan VK, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Javierre BM, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–419. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibbs JR, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leek JT, Storey JD. A general framework for multiple testing dependence. Proc Natl Acad Sci USA. 2008;105:18718–18723. doi: 10.1073/pnas.0808709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.