Abstract
Early life experience is associated with long-term effects on behavior and epigenetic programming of the NR3C1 (GLUCOCORTICOID RECEPTOR) gene in the hippocampus of both rats and humans. However, it is unlikely that such effects completely capture the evolutionarily conserved epigenetic mechanisms of early adaptation to environment. Here we present DNA methylation profiles spanning 6.5 million base pairs centered at the NR3C1 gene in the hippocampus of humans who experienced abuse as children and nonabused controls. We compare these profiles to corresponding DNA methylation profiles in rats that received differential levels of maternal care. The profiles of both species reveal hundreds of DNA methylation differences associated with early life experience distributed across the entire region in nonrandom patterns. For instance, methylation differences tend to cluster by genomic location, forming clusters covering as many as 1 million bases. Even more surprisingly, these differences seem to specifically target regulatory regions such as gene promoters, particularly those of the protocadherin α, β, and γ gene families. Beyond these high-level similarities, more detailed analyses reveal methylation differences likely stemming from the significant biological and environmental differences between species. These results provide support for an analogous cross-species epigenetic regulatory response at the level of the genomic region to early life experience.
Keywords: conservation, neuronal plasticity
Variation in early life experience is associated with differences in life-long health and behavioral trajectories in animals as well as humans. For example, differences in maternal care in rats during the first week of life are associated with long-term effects on behavior and brain function that persist into adulthood, including alterations in the stress response (1). In humans, similar effects are observed. For instance, childhood maltreatment associates with development of both externalizing and internalizing personality traits and psychopathology in adulthood (2). The association in both rats and humans of stable developmental phenotypes with early life experience suggests that molecular mechanisms may serve as a memory of these early life experiences in both species. In fact, there is evidence that these long-term effects are, at least in part, mediated by epigenetic alterations in the brain. In particular, recent studies have found aberrant DNA methylation in the NR3C1 (GLUCOCORTICOID RECEPTOR) gene promoter of the hippocampi of both rats and humans associated with differential early life experience (3, 4). Exposure of infant rats to stressed caretakers displaying abusive behavior produced persisting changes in methylation of the BDNF gene promoter in the adult prefrontal cortex (5). Early life stress in mice caused sustained DNA hypomethylation of an important regulatory region of the AVP gene (6).
Although explanations involving a single site are appealing, it is unlikely that the broad systemic response to early life experience would be associated with a few site-specific epigenetic changes. Indeed, we have previously shown that several hundred genes are differentially expressed in the hippocampi of adult rat offspring that received low compared with high maternal licking and grooming (LG) (7). Moreover, in the hippocampi of humans with documented childhood abuse, we have recently discovered methylation differences in the rRNA gene promoters that are scattered across the genome (8). Furthermore, recent evidence suggests that epigenetic regulation is not restricted to the few thousand bases around the transcription start sites of genes. Epigenetic changes associated with transcriptional changes can appear within the body of a gene (9) or even at high frequency across megabase-sized domains simultaneously deactivating dozens of neighboring genes (10, 11). These results led us to hypothesize that the epigenetic response to early life experience is not limited to a single gene promoter but that NR3C1, along with neighboring genes, might belong to a domain under coordinated control. To test this hypothesis, we recently investigated DNA methylation, H3K9 acetylation, and transcriptional profiles in a region encompassing 6.5 million base pairs centered at NR3C1 in the hippocampus of adult rat offspring of high and low LG (12). We confirmed our hypothesis by identifying hundreds of robust DNA methylation differences between the offspring of high and low LG that were scattered across this large region.
Considering the parallel behavioral and epigenetic responses in humans and rats to early life environments described above, it is reasonable to assume that at least part of the broad epigenetic responses observed in rats to early life experiences may be evolutionarily conserved in humans. Therefore, in this study we investigated the extent of this conservation in humans by generating epigenetic profiles of the analogous region in humans, the 6.5 million base pair region centered at NR3C1 (heretofore referred as the NR3C1 locus). Such a cross-species investigation is further supported by the fact that there is an ∼35% sequence homology between the rat and human NR3C1 loci, and that 80% of the genes in the human region have orthologs in the rat region.
Results
Methylation Profiles in the NR3C1 Locus of Rat and Human Hippocampi.
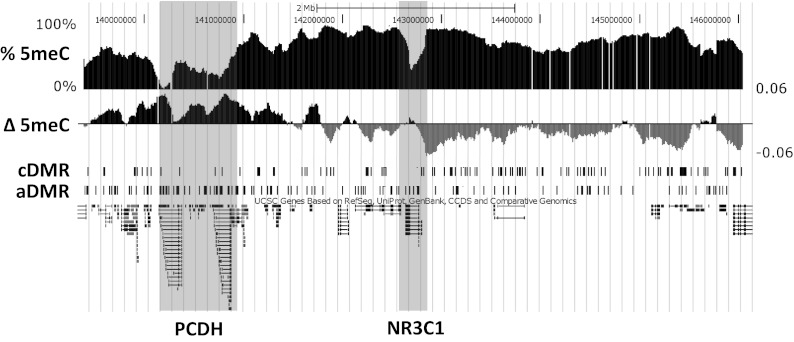
We generated DNA methylation profiles in hippocampal samples obtained from the Quebec Suicide Brain Bank of 12 suicide completers with a history of severe childhood abuse and 12 nonabused controls. Profiles covered the genomic region from 3.25 Mb upstream to 3.25 Mb downstream of the NR3C1 gene at 100-bp spacing and were created by using the method of methylated DNA immunoprecipitation (meDIP) followed by hybridization to a custom-designed Agilent 44K tiling microarray. Fig. 1 depicts the locus tiled with probes including the locations of genes along with estimated methylation levels and differences between the abuse group and controls. Previously published rat methylation profiles were generated using identical methods from the hippocampi of adult rat offspring of high and low LG and covering the synteneic region from 3.25 Mb upstream to 3.25 Mb downstream of the NR3C1 gene at 100-bp spacing (12).
Fig. 1.
Associations of human DNA methylation with early life abuse in the 6.5-Mb NR3C1 locus. Track images obtained from the University of California, Santa Cruz genome browser (human genome assembly hg18) show % 5meC: average methylation levels across all samples estimated from microarray probe intensities; Δ 5meC: mean log2 fold differences between abused and control sample probe intensities, where positive values are shown in black and indicate higher methylation in abused samples, and gray values indicate higher methylation in control samples; cDMR: locations of cDMRs (significantly higher methylation in control samples); and aDMR: locations of aDMRs (significantly higher methylation in abuse samples). The locations of the protocadherin families of genes and NR3C1 are identified by shading.
Conservation of the NR3C1 Locus Gene Architecture.
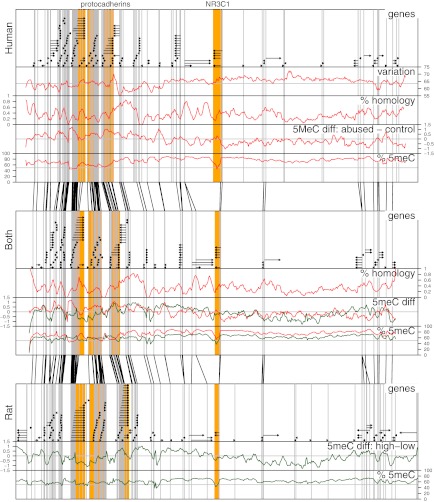
Overall organization of the NR3C1 locus is significantly conserved, as shown by the almost identical order of orthologous genes across the locus (Fig. 2). Fig. 2, Top shows the positions of genes in human, and Fig. 2, Bottom shows their positions in rat. Fig. 2, Middle shows the rat–human “hybrid” created by assigning orthologous genes to positions similar to their relative locations in the human and rat genomes. Gray vertical lines in each panel coincide with transcription start sites. Black lines between adjacent panels link transcription start sites of orthologous genes in neighboring panels.
Fig. 2.
Associations of human and rat DNA methylation with early life abuse in the 6.5-Mb NR3C1 locus. Top, Middle, and Bottom: Each panel shows the 6.5-Mb NR3C1 locus. Top: Human locus. Bottom: Rat locus. Middle: Human–rat “hybrid” panel created by assigning orthologous genes to positions similar to their relative locations in the human and rat genomes. Each panel is divided into five labeled parts: genes: black horizontal arrows denote genes and the direction of mRNA synthesis; variation: graph indicates regions of high and low methylation variation across all human subjects; % homology: graph shows the percentage of bases in the human genome that were mapped by the lastz alignment tool to the rat genome; 5meC diff: graph shows mean log2 fold differences between sample groups (i.e., between abused and control humans and between high- and low-LG rats); and % 5meC: graph shows methylation levels estimated from microarray probe intensities. Across each panel, gray vertical lines demark transcription start sites. Black lines between panels link the positions of transcription start sites of orthologous genes. The lack of crossings between these lines illustrates conservation of gene architecture around NR3C1 between rats and humans.
Expected DNA Methylation Patterns Confirmed.
Methylation levels were estimated from microarray meDIP profiles by deconvoluting individual CpG methylation levels from the intensities of nearby probes (13). Estimates of these levels across the locus in human and rat are shown in Fig. 2 and are compared directly in the middle panel showing the human–rat hybrid. Overall, methylation levels seem to rise and fall in unison. Indeed, they do have a small but statistically significant correlation (P < 0.0013; R = 0.048). Given the regulatory role of DNA methylation, these patterns are unlikely to be random. For example, dips should correspond to active transcription start sites, CpG islands, and the 3′ ends of genes (14, 15). As shown for the rat profiles, we observed lower methylation levels around transcription start sites (P ≤ 1.68 × 10−272, Wilcoxon rank sum test), at the 3′ ends of genes (P ≤ 9.9 × 10−28), and inside CpG islands (P ≤ 10−300). In contrast to rat methylation levels, human methylation levels were lower near methylation-sensitive transcription factor binding sinks (P ≤ 0.06). As the name suggests, methylation-sensitive transcription factor binding sinks are regions enriched for methylation-sensitive transcription factor binding sites as predicted by binding motifs. The lack of methylation decrease in these regions in rats is likely due to the fact that most transcription factor binding motifs have been derived from human studies rather than rat studies.
Conservation of a Widespread Methylation Response.
Both the rat and human profiles revealed hundreds of differentially methylated regions (DMRs) associated with early life experience scattered unevenly across the NR3C1 locus (Fig. 1). In total, there were 281 human DMRs, of which 126 had increased methylation in controls (cDMRs), and 155 had increased methylation in the individuals with histories of childhood abuse (aDMRs). Real-time PCR of meDIP samples was used to validate selected DMRs. We investigated 11 of these differences inside gene promoters located across the locus (Fig. 3). The rat profiles revealed more than twice as many DMRs (723), of which 373 were more methylated in high-LG offspring (hDMRs) and 350 were more methylated in low-LG offspring (lDMRs). This larger number in rat is possibly due to the greater genetic similarity and less environmental variability leading to increased power to detect differences within rat groups compared with human groups.
Fig. 3.
Validation of microarray calls. Real-time PCR validation of microarray meDIP data is shown. Eleven of the 28 promoters identified as being differentially methylated by microarray (Table S1) were subjected to real-time PCR quantification of enrichment. The y axis represents concentration values generated by methylation-enriched and input DNA. Left: Concentration levels in the abuse group. Right: Concentration levels in the control group. Each real-time PCR was performed in triplicate. Error bars indicate SEM.
As observed in the rat profiles, the placement of the DMRs across the locus is nonuniform, resulting in large regions enriched with DMRs and others almost completely depleted of any DMRs (Fig. 1). In the sections below we explore these patterns in more detail.
Conservation of Long-Range Methylation Dependencies.
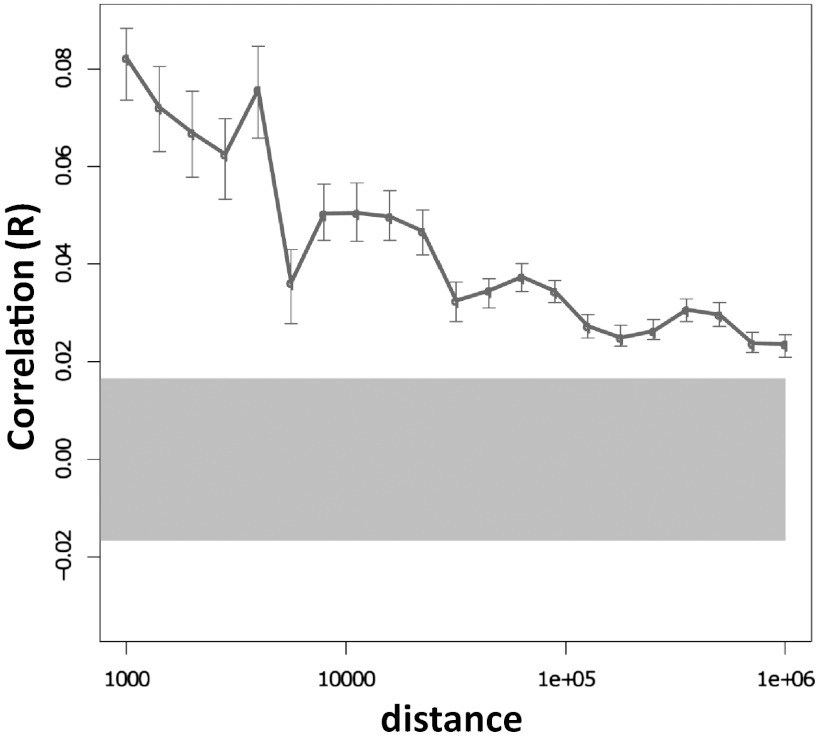
In both species, DMRs showing the same direction of change according to environmental experience seem to form clusters covering large genomic regions, supporting a high-level organization linking distant sites. In general, there seem to be consistent dependencies between methylation differences as far apart from NR3C1 as 1 million base pairs in both species (Fig. 4; figure 3a in ref. 12). As an example of this long-range clustering, observe in Fig. 1 that if the human NR3C1 locus is partitioned into two parts, one part left and the other part right of NR3C1, aDMRs are enriched in the left part (P ≤ 2.2 × 10−32; hypergeometric), and cDMRs are enriched in the right part (P ≤ 2.2 × 10−32). Partitioning the rat NR3C1 locus in the same way, hDMRs are enriched in the left part (P ≤ 0.031), and lDMRs are enriched in the right part (P ≤ 0.013). To determine whether any of the rat DMRs were conserved in human, hDMR and lDMR sequences were mapped to the human genome using BLAT. In total, 111 of these sequences mapped successfully to the human locus; however, none of them overlapped with human DMRs. Interestingly though, just as hDMRs are enriched to the left and lDMRs are enriched to the right of NR3C1 in rat, the mapped hDMRs are also enriched to the left and the mapped lDMRs are enriched to the right of NR3C1 in the human locus (Fig. S1). This suggests that change according to environment is conserved across species at a high level, although details about those changes differ between species.
Fig. 4.
Correlation of human DNA methylation associations with early life abuse in the 6.5-Mb NR3C1 locus. Pearson correlations of DNA methylation differences between the subject groups at various genomic distances. Error bars show 95% confidence intervals for the correlation values. The gray highlight shows the expected 95% confidence interval if there is no correlation between methylation differences at different genomic sites. This confidence interval does not overlap with the error bars associated with distances less than 1 Mb, suggesting the existence of systematic dependencies between methylation differences at distances up to 1 Mb.
Conservation of Enriched Methylation Response in Suspected Regulatory Sites.
Given the regulatory role that DNA methylation plays, one might expect to see DMRs near known or suspected regulatory sites such as near transcription start sites, particularly coinciding with CpG islands, and transcription factor binding sites. Indeed, these regions tend to be enriched with DMRs in both rats and humans, although some of the details differ. Approximately 8% of DMRs in both rat and human intersect promoter regions (−2,000…+200 bp of the transcription start site; Table S1); however, whereas this intersection is statistically significant in humans (P < 0.001), it does not reach significance in rats (P > 0.14). When analogous regions at the 3′ ends of genes are included, the overlap of both human and rat DMRs is significant (P < 0.001 and P < 0.0032, respectively). In humans, this enrichment extends to 1,000 bp past transcription start sites (P < 0.001).
Interestingly, much of this enrichment in humans is explained by aDMR enrichment (P < 2 × 10−4, promoters; P < 0.034, 3′ ends of genes; P < 2 × 10−4, 1,000 bp after transcription start sites; P < 0.019, first exons) because cDMRs are depleted in nearly all of these regions (P < 0.039, promoters; P < 0.031, 1,000 bp after transcription start sites; P < 0.02, first exons). Supporting the regulatory nature of the sites targeted by aDMRs is the observation that they are enriched for methylation-sensitive transcription factor binding sinks (P < 0.08; Methods) and highly enriched with CpG sites (P < 4.4 × 10−14). Not surprisingly, cDMRS are depleted in these regions (P < 0.02) and depleted of CpG sites compared with the rest of the locus (P < 1.4 × 10−8).
In rat, such a simple characterization of lDMRs and hDMRs is not possible. lDMRs are enriched in some of these regions (P < 0.02, promoters; P < 0.003, 3′ ends of genes) but depleted in others (P < 0.019, 1,000 bp after transcription start sites; P < 0.0011, first exons). In contrast, hDMRs are enriched primarily in first exons (P < 0.0008) and, interestingly, also in last exons (P < 0.0038). On the other hand, depletion of lDMRs (rat) and cDMRs (human) are observed in last exons (P < 0.0063 and P < 0.021, respectively).
Differential Methylation Across NR3C1.
We have previously shown that NR3C1 gene expression is lower in the abuse group and that this decrease in expression associates with increased methylation levels in the promoter of a splice variant (1F) of the NR3C1 gene (4). The comprehensive mapping of the NR3C1 locus presented here identified a total of seven DMRs in and around NR3C1: two upstream cDMRs, four aDMRs within the first and second introns, and one aDMR downstream of the gene (Fig. S2). The increased number of aDMRs compared with cDMRs is consistent with the repression of NR3C1 in the abuse group. In rats there are similar DMRs throughout the gene, with the majority being lDMRs (figure 4a in ref. 12), also consistent with the repression of NR3C1 in the low-LG group.
Conserved Methylation Sensitivity in the Protocadherin Families of Genes.
Notable methylation differences in the NR3C1 locus of both rats and humans are located downstream of NR3C1 within the α-, β-, and γ-protocadherin (PCDH) gene clusters. All three clusters together are highly enriched for aDMRs (P < 2 × 10−4) and depleted of cDMRs (P < 0.0014). Of the three clusters, α-PCDH is most enriched for DMRs (P < 0.054) and, particularly, for aDMRs (P < 2 × 10−4). Fig. S3 depicts the methylation differences within the PCDH gene clusters. Similarly to aDMRs, lDMRs are highly enriched in the PCDH gene clusters (P < 0.01).
These methylation differences observed in human hippocampus are of interest because the protocadherin families of genes are known to be regulated by promoter methylation (16–18) and have been implicated in synaptic function and neuronal connectivity (19–22).
Regions That Lack Differential Methylation.
The existence of DMR clusters implies the existence of regions lacking methylation differences. For example, despite the fact that gene promoters are enriched with DMRs compared with other genomic regions (P < 0.001), only 28 of the 171 gene promoters (−2000…+200 bp around the transcription start site of a gene) contain a DMR. That leaves a lot of gene promoters unaffected by differential methylation, despite the fact that DMRs are widely distributed across the locus. For some reason, these promoters were “avoided.” In fact, there are eight regions of more than 100 Kb within the NR3C1 locus that contain no differentially methylated sites (Table S2). Permutation tests show that the expected number of such regions is only 2.2, with a maximum of six found in 1,000 such tests (DMR positions were randomly permuted within the locus). Three of the eight regions contained no genes, and one of the eight regions contained at least 10 genes, more than three times the number genes expected. Hence, these methylation profiles identify large gene-rich and gene-poor regions without any DMRs, evidence for a widespread but selective effect on DNA methylation levels within the NR3C1 locus.
Discussion
There is growing evidence for association between variation in early life experience and differential methylation in several genes. Past studies focused on documented or highly predicted regulatory regions around the transcription start sites of these genes. Such an approach might miss important DMRs and ignore the larger scope of the DNA methylation response to environmental cues. Our recent study of the epigenetic response to maternal care in rats (12) determined that the epigenetic response is in fact not limited to a few sites but affects broader genomic regions. We asked here to what extent this broad response might be conserved across species. Hence, we performed a human study similar to our rat study covering the 6.5-Mb region centered at the NR3C1 gene, wherein we examined differences in DNA methylation in the hippocampi of subjects who committed suicide and experienced severe abuse during childhood vs. control individuals with negative histories of abuse. Similarly to our rat study, we showed that differential methylation is not restricted solely to NR3C1 promoters but instead appears at many sites throughout the NR3C1 locus, both within as well distant from promoters (Fig. 1).
Although there are many methylation differences, they are not uniformly distributed across the locus, and our analysis describes several levels of structural organization of this association with early life experience showing surprising agreement with our parallel rat analysis. The differences in DNA methylation tend to concentrate in specific regions relative to transcription start sites and in specific regions containing dozens of genes within the NR3C1 locus (Fig. 1), suggesting high-level organization. Especially remarkable was the discovery of a division of the entire 6.5-Mb locus into two major domains characterized by genomic sites with reduced DNA methylation in the abuse group upstream to the NR3C1 locus and genomic sites with increased in DNA methylation downstream to the NR3C1 locus (Fig. 1).
A strikingly significant number of DMRs can be found in the promoters of the protocadherin families of genes (Fig. 1) in both humans and rats, supporting the hypothesis that protocadherins play a key role in the response to early life experience. This hypothesis is consistent with previous findings that the complex expression patterns of the protocadherins are regulated by DNA methylation leading to differential promoter activation and alternative pre-mRNA splicing (16–18). That our methylation differences were observed in the hippocampus and that protocadherins have been implicated in synaptic function and neuronal connectivity (19–22) suggests that regional DNA methylation may play a role in concert with NR3C1 changes in neuronal rewiring in response to early life experience.
The potential regulatory roles of the methylation differences that do not map to regulatory sites, such as transcription start sites and methylation-sensitive transcription factor binding sites, are more difficult to characterize. However, the conserved enrichment of methylation differences around these regulatory sites in humans and rats supports the existence of a regulatory role for other methylation differences yet to be elucidated.
Not surprisingly, given the important differences between rats and humans and the nature of their early life environments, comparison between the rat and human methylation changes associated with early life experience was neither simple nor straightforward. For example, the methylation profiles do not support a direct analogy at the individual base level between low maternal care in rats and childhood abuse in humans. However, such an analogy was not the purpose of our study. Instead, we reasoned that a cross-species comparison of DNA methylation associated with variation in early life environment would identify genomic regions beyond the promoters regions of NR3C1 that are epigenetically labile in response to a range of early life experiences. Our results support this hypothesis. In both rats and humans, we identified a broad but selective response to early life experience that is enriched in suspected regulatory regions, exhibits evidence of a long-range coordination between distant sites, and seems to particularly target the regulation of the protocadherin families of genes, suggesting that these genes may also be involved in the response to early life experience. Such a conserved response motivates the development of novel experimental approaches to understand how these DNA methylation modulations affect genome function.
Methods
Methods related to the human samples only are provided here because methods and analyses related to the rat samples have already been published (12).
Ethics Statement.
Studies with human subjects were approved by the McGill University institutional review board, and signed informed consent was obtained from next of kin. All procedures involving rodents were performed according to guidelines developed by the Canadian Council on Animal Care, and the protocol was approved by the McGill University Animal Care Committee.
Subjects and Tissue Preparation.
Hippocampal samples obtained from the Quebec Suicide Brain Bank included 12 suicide subjects with histories of severe childhood abuse and 12 controls with validated negative histories of childhood abuse who did not differ in postmortem interval, sex, age at death, and brain pH (all P > 0.05). Psychiatric diagnoses were obtained by means of the Structured Clinical Interview for DSM-III-R (23) interview adapted for psychological autopsies, which is a validated method to reconstruct psychiatric and developmental history by means of extensive proxy-based interviews, as outlined elsewhere (24). To be considered in this study, all suicide subjects had to have a positive history of severe childhood sexual and/or physical abuse or severe neglect, as determined by most severe scores in the respective scales of the structured Childhood Experience of Care and Abuse (25) questionnaire adapted for psychological autopsies (26). Conversely, controls had to have validated evidence of negative lifetime histories of abuse and/or neglect.
All samples were from male suicide and control subjects of French-Canadian origin. Samples were dissected at 4 °C and stored in plastic vials at −80 °C until analysis. All samples were processed and analyzed blind to demographic and diagnostic variables. To be included in this study, all subjects had to die suddenly, with no medical or paramedic intervention and no prolonged agonal period. Suicide as the cause of death was determined by the Quebec Coroner’s Office.
DNA Immunoprecipitation and Microarray Hybridization.
The procedure for methylated DNA immunoprecipitation was adapted from previously published work (27–29). The amplification (Whole Genome Amplification kit; Sigma) and labeling reaction (CGH labeling kit; Invitrogen), and all of the steps of hybridization including washing and scanning were performed according to the Agilent protocol for chip-on-chip analysis. Microarrays were hybridized in triplicate for each sample.
Quantitative Real-Time PCR of Immunoprecipitated Samples.
Gene-specific real-time PCR validation of microarray was performed for DNA methylation enrichment (30) for the same samples used for microarray experiments. Triplicate reactions were performed, and relative concentration was determined as a ratio of the crossing point threshold (Ct). The average concentration for each set of replicates was plotted along with its SEM. Primers for each amplicon are given in Table S3.
Microarray Design and Analysis.
Custom 44K tiling arrays were designed using eArray (Agilent). Probes of ∼55 bp were selected to tile all unique regions within ∼3.25 MB upstream and downstream of the NR3C1 gene described in Ensembl (version 44) at 100-bp spacing. Probe intensities were extracted from microarray scan images using Agilent’s Feature Extraction 9.5.3 Image Analysis Software and analyzed using the R software environment for statistical computing (31). Background corrected log-ratios of the bound (Cy5) and input (Cy3) microarray channel intensities were computed for each microarray. Microarrays were normalized to one another using quantile normalization (32).
All genomic coordinates are given with respect to the hg18 human genome assembly.
In some cases, DNA methylation levels at genomic locations were estimated from microarray probe intensities. In these cases, a Bayesian convolution algorithm was used to incorporate probe values from nearby probes (13).
Differential methylation between groups was determined in two stages to ensure both statistical significance and biological relevance. In the first stage, linear models implemented in the “limma” package (33) of Bioconductor (34) were used to compute a modified t statistic at the individual probe level. An individual probe was called differentially methylated if the significance of its t statistic was at most 0.05 (uncorrected for multiple testing) and the associated difference of log-normalized means between the groups was at least 0.5. Given that the DNA samples were sonicated into 200- to 700-bp fragments before hybridization, we assumed that probes within 500 bp should have approximately similar probe scores. Therefore, in the second stage, we computed differential statistics for 1,000-bp intervals from the differential statistics of the probes that they contained. The intervals tiled the entire 6.5-Mb region under investigation at 500-bp spacing. Differential significance of these intervals was determined using the Wilcoxon rank-sum test comparing t statistics of the probes within the interval against those of all of the probes on the microarray. Significance levels were then adjusted to obtain false discovery rates. An interval was called differentially methylated if it satisfied each of the following: (i) its false discovery rate was at most 0.2, and (ii) the 1,000-bp interval contained at least one probe called differentially methylated. The first requirement ensured that several probes in the interval had similar group differences, and the second requirement ensured that the difference was not simply weakly distributed across the entire interval and consequently difficult to validate. Intervals satisfying these tests were called differentially methylated regions (DMRs). Consecutive DMRs for which the difference of means showed greater methylation in the abused group were called aDMRs and the converse cDMRs. Consecutive a/cDMRs were coalesced into single a/cDMRs.
Statistically significant enrichment or depletion of DMRs in specific regions such as CpG islands or gene promoters was computed using permutation tests on the locations of DMRs. More specifically, the statistic used the number of base pairs overlapping between DMRs and the regions in question, for example CpG islands. A distribution for this statistic was computed by repeatedly (1,000 times) randomly assigning theoretically possible coordinates (based on the locations of probes) to the DMRs and then calculating the overlap between the regions and the newly located DMRs.
Methylation-sensitive transcription factor sinks were computed by position weight matrices for specific transcription factors from the Transfac (35) and Jaspar (36) databases, including AP2, CBF, CREB, ETS, FOXP3, GABP, GATA1, NF-kappaB, NGFIA/EGR1, NR3C1, P53, RUNX, SP1, SP3, TCF, and USF1. There is evidence that the activity of each of these transcription factors is affected by the presence or absence of DNA methylation (3, 37–48). For some of these transcription factors, the databases contained multiple identical position weight matrices. To avoid having these matrices bias the identification of transcription factor sinks in favor of a single transcription factor, we removed one position weight matrix for any pair whose targets overlapped 75% of the time. Transcription factor targets were identified by scanning the sequence with a second-order position weight matrix adjusting log-likelihoods with the 500-bp sequence background context (49). Sites with a log-likelihood greater than 14 were called binding sites. To make the remaining computation to identify transcription factor sinks more efficient, the genome was then partitioned into 100-bp segments, and the binding score for each transcription factor in each segment was set to the maximum log-likelihood in that segment. Each segment was called a transcription factor sink if its transcription factor scores were significantly higher than average as determine by the Wilcoxon rank sum test (P ≤ 1 × 10−9 or 6 × 10−5 after Bonferroni correction).
The variability of a probe was quantified as the number of sample pairs for which all normalized replicate log-ratios for one sample were at least 0.5 greater than all normalized replicate log-ratios for the other sample.
Overall homology between human and rat were computed using the lastz program (50). Specifically, the lastz program was used with default settings to align the human and rat NR3C1 locus sequences. The percentage of homology was given as the percentage of human sequence that was successfully mapped to the rat sequence.
Rat DMR sequences, all 1,000 bp long, were mapped onto the human NR3C1 using BLAT (51) with the following settings: tile_size = 10, step_size = 10, min_match = 2, min_score = 400, min_identity = 75, and max_intron = 250.
Fig. 4 illustrates the correlation of methylation differences across various genomic distances as Pearson correlations of modified t statistics computed by limma for all pairs of probes at specified distances (with a 10% tolerance). Error bars denote 95% confidence intervals obtained from 1,000 bootstraps composed of randomly selected probe pairs with replacement. The gray rectangle denotes the 95% confidence interval for correlations of probe pairs independent of their distance. Independence was simulated by with 500 random permutations of the probe coordinates.
All microarray data are MIAME compliant, and the raw data have been deposited in the Gene Expression Omnibus.
Supplementary Material
Acknowledgments
This study was supported by grants from the Canadian Institutes of Mental Health and the Sackler Foundation (to M.J.M. and M.Szyf).
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Biological Embedding of Early Social Adversity: From Fruit Flies to Kindergartners,” held December 9–10, 2011, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at www.nasonline.org/biological-embedding.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA microarray data reported in this paper have been deposited into the Gene Expression Omnibus, www.ncbi.nlm.nih.gov/geo (accession no. GSE38352).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121260109/-/DCSupplemental.
References
- 1.Francis D, Diorio J, Liu D, Meaney MJ. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286:1155–1158. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert R, et al. Burden and consequences of child maltreatment in high-income countries. Lancet. 2009;373:68–81. doi: 10.1016/S0140-6736(08)61706-7. [DOI] [PubMed] [Google Scholar]
- 3.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 4.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murgatroyd C, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 7.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGowan PO, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS ONE. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 10.Coolen MW, et al. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat Cell Biol. 2010;12:235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 12.McGowan PO, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS ONE. 2011;6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Down TA, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasic B, et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi M, et al. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J Biol Chem. 2008;283:12064–12075. doi: 10.1074/jbc.M709648200. [DOI] [PubMed] [Google Scholar]
- 18.Yagi T. Clustered protocadherin family. Dev Growth Differ. 2008;50(Suppl 1):S131–S140. doi: 10.1111/j.1440-169X.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- 19.Bass T, Ebert M, Hammerschmidt M, Frank M. Differential expression of four protocadherin alpha and gamma clusters in the developing and adult zebrafish: DrPcdh2gamma but not DrPcdh1gamma is expressed in neuronal precursor cells, ependymal cells and non-neural epithelia. Dev Genes Evol. 2007;217:337–351. doi: 10.1007/s00427-007-0145-4. [DOI] [PubMed] [Google Scholar]
- 20.Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: About cell death, synapses and processing. Curr Opin Cell Biol. 2005;17:446–452. doi: 10.1016/j.ceb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki ST. Recent progress in protocadherin research. Exp Cell Res. 2000;261:13–18. doi: 10.1006/excr.2000.5039. [DOI] [PubMed] [Google Scholar]
- 22.Weiner JA, Wang X, Tapia JC, Sanes JR. Gamma protocadherins are required for synaptic development in the spinal cord. Proc Natl Acad Sci USA. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spitzer RL, Williams JB, Gibbon M, First MB. The Structured Clinical Interview for DSM-III-R (SCID). I: History, rationale, and description. Arch Gen Psychiatry. 1992;49:624–629. doi: 10.1001/archpsyc.1992.01820080032005. [DOI] [PubMed] [Google Scholar]
- 24.Dumais A, et al. Risk factors for suicide completion in major depression: A case-control study of impulsive and aggressive behaviors in men. Am J Psychiatry. 2005;162:2116–2124. doi: 10.1176/appi.ajp.162.11.2116. [DOI] [PubMed] [Google Scholar]
- 25.Bifulco A, Brown GW, Harris TO. Childhood Experience of Care and Abuse (CECA): A retrospective interview measure. J Child Psychol Psychiatry. 1994;35:1419–1435. doi: 10.1111/j.1469-7610.1994.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 26.Zouk H, Tousignant M, Seguin M, Lesage A, Turecki G. Characterization of impulsivity in suicide completers: Clinical, behavioral and psychosocial dimensions. J Affect Disord. 2006;92:195–204. doi: 10.1016/j.jad.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 28.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 29.Brown SE, Szyf M. Dynamic epigenetic states of ribosomal RNA promoters during the cell cycle. Cell Cycle. 2008;7:382–390. doi: 10.4161/cc.7.3.5283. [DOI] [PubMed] [Google Scholar]
- 30.Sadikovic B, et al. In vitro analysis of integrated global high-resolution DNA methylation profiling with genomic imbalance and gene expression in osteosarcoma. PLoS ONE. 2008;3:e2834. doi: 10.1371/journal.pone.0002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 32.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 33.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Vol 1. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 34.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matys V, et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res. 2008;36(Database issue):D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McPherson LA, Baichwal VR, Weigel RJ. Identification of ERF-1 as a member of the AP2 transcription factor family. Proc Natl Acad Sci USA. 1997;94:4342–4347. doi: 10.1073/pnas.94.9.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McPherson LA, Weigel RJ. AP2alpha and AP2gamma: A comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040–4049. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polansky JK, et al. Methylation matters: Binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl) 2010;88:1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas ME, Crider KS, Powell DR, Kapoor-Vazirani P, Vertino PM. Methylation-sensitive regulation of TMS1/ASC by the Ets factor, GA-binding protein-alpha. J Biol Chem. 2009;284:14698–14709. doi: 10.1074/jbc.M901104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutchins AS, et al. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 44.Siegfried Z, Cedar H. DNA methylation: A molecular lock. Curr Biol. 1997;7:R305–R307. doi: 10.1016/s0960-9822(06)00144-8. [DOI] [PubMed] [Google Scholar]
- 45.Nabilsi NH, Broaddus RR, Loose DS. DNA methylation inhibits p53-mediated survivin repression. Oncogene. 2009;28:2046–2050. doi: 10.1038/onc.2009.62. [DOI] [PubMed] [Google Scholar]
- 46.Zhu WG, et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol Cell Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aoki M, et al. Kidney-specific expression of human organic cation transporter 2 (OCT2/SLC22A2) is regulated by DNA methylation. Am J Physiol Renal Physiol. 2008;295:F165–F170. doi: 10.1152/ajprenal.90257.2008. [DOI] [PubMed] [Google Scholar]
- 48.Choy MK, et al. Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated. BMC Genomics. 2010;11:519. doi: 10.1186/1471-2164-11-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stormo GD. DNA binding sites: Representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 50.Harris RS. University Park, PA: Pennsylvania State Univ; 2007. Improved pairwise aligment of genomic DNA. PhD dissertation. [Google Scholar]
- 51.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.