Abstract
We recently presented clear evidence that the major low-phosphate-inducible phosphatase of the duckweed Spirodela oligorrhiza is a glycosylphosphatidylinositol (GPI)-anchored protein, and, to our knowledge, is the first described from higher plants (N. Morita, H. Nakazato, H. Okuyama, Y. Kim, G.A. Thompson, Jr. [1996] Biochim Biophys Acta 1290: 53–62). In this report the purified 57-kD phosphatase is shown to be a purple metalloenzyme containing Fe and Mn atoms and having an absorption maximum at 556 nm. The phosphatase activity was only slightly inhibited by tartrate, as expected for a purple acid phosphatase (PAP). Furthermore, the protein cross-reacted with an anti-Arabidopsis PAP antibody on immunoblots. The N-terminal amino acid sequence of the phosphatase was very similar to those of Arabidopsis, red kidney bean (Phaseolus vulgaris), and soybean (Glycine max) PAP. Extracts of S. oligorrhiza plants incubated with the GPI-specific precursor [3H]ethanolamine were treated with antibodies raised against the purified S. oligorrhiza phosphatase. Radioactivity from the resulting immunoprecipitates was specifically associated with a 57-kD band on sodium dodecyl sulfate-polyacrylamide gels. These results, together with previous findings, strongly indicate that the GPI-anchored phosphatase of S. oligorrhiza is a PAP.
Animal and fungal cells contain a diverse assortment of membrane proteins, which are anchored to the outside surface of the plasma membrane solely by a covalently linked GPI moiety (Englund, 1993). Among the proteins anchored in this way by a GPI chain are protozoan coat proteins, lymphoid antigens, hydrolytic enzymes, cell adhesion molecules, receptors for small molecules, the scrapie prion protein, and a wide variety of other functionally distinct proteins (Low, 1989).
In contrast to the more than 150 examples of GPI-anchored proteins now known in animals and yeast, until recently, there have been no indications that this type of protein anchorage occurs in algae or higher plants. Reports of a GPI-anchored nitrate reductase in Chlorella saccharophila (Stöhr et al., 1995) and in sugar beet (Kunze et al., 1997), a low-phosphate-inducible, GPI-anchored alkaline phosphatase in the duckweed Spirodela oligorrhiza (Morita et al., 1996), and unidentified GPI-anchored proteins in tobacco (Takos et al., 1997) have appeared. In none of the above instances has the reported GPI-anchored protein and its anchoring structure been fully characterized.
The lipid moiety of the S. oligorrhiza phosphatase anchor has been tentatively identified as a ceramide (Morita et al., 1996). Following polypeptide synthesis, glycosylation, GPI anchor attachment, and transport to the cell surface, the terminal lipid is cleaved off in vivo, leaving behind a cell wall-localized phosphatase still linked to the ethanolamine-containing fragment of the GPI chain. This GPI-anchored S. oligorrhiza phosphatase retaining part of its GPI anchor is a 100-kD homodimer consisting of 57-kD subunits (Nakazato et al., 1997a). It is interesting that, whereas the enzyme's pH optimum for catalysis was about 8.0 in crude extracts, it decreased to about 7.0 during purification procedures (Nakazato et al., 1997a), bringing its original designation as an alkaline phosphatase into question. In this paper we present convincing evidence that the major low-phosphate-inducible phosphatase of S. oligorrhiza is a GPI-anchored PAP. Nakazato et al. (1997b) presented a preliminary report expressing this conclusion, which was presented at the XIII International Plant Nutrition Colloquium, September 13 to 19, 1997, in Tokyo, Japan.
MATERIALS AND METHODS
Materials
Spirodela oligorrhiza plants were grown in modified Hoagland medium (Posner, 1967) containing either 1.5 (+P plants) or 0 (−P plants) mm KH2PO4 for 2 to 3 weeks at 25°C under a 16-h daylength with illumination from fluorescent lamps (80 μE m−2 s−1). Harvested plants were stored at −30°C until use. Wheat germ and bovine alkaline phosphatases were purchased from Sigma. [1,2-3H]Ethanolamine hydrochloride (15 Ci/mmol) was obtained from Moravek Biochemicals (Brea, CA).
Purification of the Phosphatase from S. oligorrhiza
Purification of the S. oligorrhiza phosphatase was carried out as described previously (Nakazato et al., 1997a). Phosphatase enzymatic activity was assayed as described by Nakazato et al. (1997a). The electrophoretically purified phosphatase was used as the experimental material.
Electrophoresis
Proteins were analyzed by SDS-PAGE according to the method of Laemmli (1970), using gels with either 5% polyacrylamide (type NPU-5L, Atto, Tokyo, Japan) or a linear gradient of 5% to 20% polyacrylamide (type NPG-520L, Atto). Samples were applied to the gels in 10 mm Tris-HCl, pH 6.8, containing 20% glycerol, 1% SDS, and 0.02% bromphenol blue. For denaturing conditions, 5% 2-mercaptoethanol was added and the samples were boiled for 5 min. The proteins were detected by silver staining. The images shown in Figures 2 and 4 were scanned and uniformly enhanced to provide better definition.
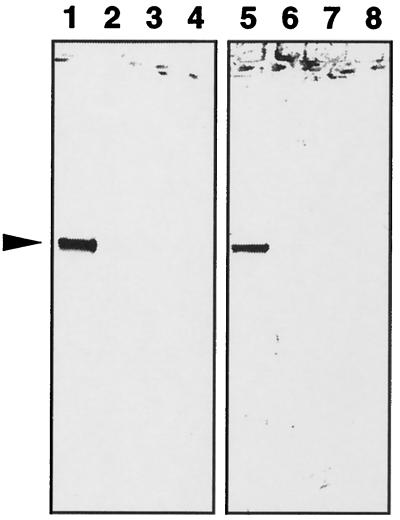
Figure 2.
Immunoblot analysis of the S. oligorrhiza phosphatase and other phosphatases using an anti-S. oligorrhiza phosphatase antibody (lanes 1–4) and an anti-Arabidopsis PAP antibody (lanes 5–8). Lanes 1 and 5, 2 μg of purified S. oligorrhiza phosphatase; lanes 2 and 6, 2 μg of bovine alkaline phosphatase; lanes 3 and 7, 2 μg of wheat germ acid phosphatase; and lanes 4 and 8, 2 μg of potato acid phosphatase. The arrow indicates the position of the S. oligorrhiza PAP band.
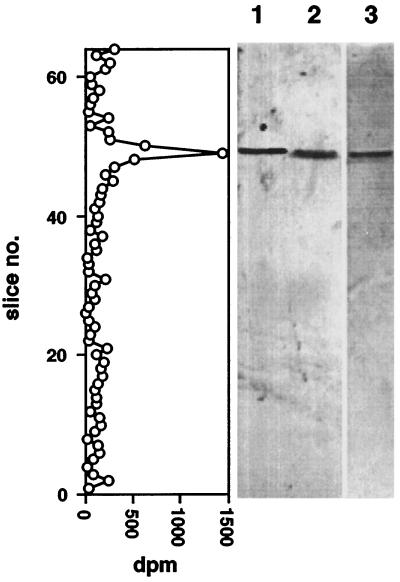
Figure 4.
Association of [3H]ethanolamine with S. oligorrhiza proteins immunoprecipitated by anti-N-terminal peptide antibodies. Right, SDS-PAGE gel. Immunoblots of 2 μg of purified S. oligorrhiza PAP (lane 1) and 2 μg of solubilized immunoprecipitate from protein extract of [3H]ethanolamine-labeled S. oligorrhiza (lane 2). Lane 3 shows the banding pattern of a Coomassie blue-stained gel lane containing 2 μg of the same solubilized immunoprecipitate used for lane 2. Left, The tracing indicates the radioactivity recovered in 1-mm slices of lane 3.
Amino Acid Sequencing
Five milligrams of the purified phosphatase was electrophoresed as described above and transferred onto a PVDF membrane. Bands corresponding to the 57-kD protein were excised from the membrane. The protein was sequenced by the Hokkaido University Analytical Center (Sapporo, Japan) on an automatic gas-phase sequencer (model 477A/120A, Applied Biosystems).
Metal Analysis
To determine the content of metals, the purified phosphatase (0.37–1.37 mg in 10 mm Tris-HCl buffer, pH 6.8) was dialyzed overnight against distilled water. The concentration of the enzyme was adjusted to 0.15 mg/mL with 1 n HNO3. The contents of Fe, Mn, Zn, Cu, Tl, Pb, Ni, and Co were determined on triplicate samples by microwave-induced plasma/quadrupole MS (Douglas and French, 1981). As a control, a solution of 10 mm Tris-HCl containing no phosphatase protein was analyzed.
Immunological and Blotting Methods
Immunoblotting was performed using anti-S. oligorrhiza phosphatase and anti-Arabidopsis PAP antibodies (a generous gift from Thomas D. McKnight, Texas A & M University, College Station). Two micrograms of the purified phosphatase was electrophoresed under denaturing conditions and transferred onto a PVDF membrane. Immunological detection by immunoblotting was carried out with the purified rabbit anti-PAP IgG, followed by goat anti-rabbit IgG conjugated to horseradish peroxidase (Bio-Rad). Biotinylated Mr standards (Bio-Rad) were visualized after they were coupled to avidin-linked horseradish peroxidase.
Affinity Purification of the Anti-S. oligorrhiza Phosphatase Antibody
Antiserum to the S. oligorrhiza-purified phosphatase was obtained as previously described (Nakazato et al., 1997a). Purification of the anti-S. oligorrhiza antibody was carried out with a HiTrap-affinity column (Pharmacia). Seven milligrams of the 19-amino acid N-terminal peptide from the S. oligorrhiza phosphatase was coupled to beads for the column. Then, the anti-S. oligorrhiza phosphatase serum was loaded onto the column, and the purified antibodies were eluted with 50 mm Gly, pH 2.5. The eluted fraction (6 mL) was concentrated to 1 mL with a Molcut LGC ultrafiltration device (Millipore). This affinity-purified antibody was designated “anti-N-terminal peptide antibody.”
Radioisotope Labeling and Immunoprecipitation
About 500 mg of −P plants was floated on two 5-mL aliquots of −P medium containing 0.75 mCi of [3H]ethanolamine. After the samples were exposed to 18 h of continuous light, proteins were extracted from labeled plants. Proteins were precipitated from the crude extract with cold acetone for 3 h at −30°C and then redissolved and reprecipitated with acetone to remove traces of [3H]ethanolamine. The proteins were spun down and resuspended in 1 mL of buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 5 mm EDTA, 0.2% SDS, and 1 mm PMSF). Proteins were precipitated with cold acetone, redissolved in buffer, and again precipitated with cold acetone. Five microliters of the anti-N-terminal peptide antibody was added, and the mixture was incubated at 4°C for 16 h with agitation. Immune complexes were precipitated by the addition of 50 μL of 10% protein A-Sepharose (Sigma) and incubated at 4°C for 3 h. The supernatant after centrifugation was subjected to a second round of immunoprecipitation. The combined precipitates were resuspended in 50 μL of SDS-sample buffer and boiled for 4 min to disrupt the complexes.
Measurement of Radioactivity
The immunoprecipitated proteins were separated by SDS-PAGE on 5% polyacrylamide gels. For measurement of radioactivity, the gel was dried, and then the lane containing radioactive proteins was sliced into 1-mm sections with a cutting device. The gel slices were each put into a vial and dissolved with 2 mL of 30% H2O2. Radioactivity was measured in a scintillation counter.
RESULTS
Amino Acid Sequence
Following purification of the phosphatase (Nakazato et al., 1997a), the first 19 amino acids of its N terminus were sequenced. The sequence A-V-D-M-P-L-N-A-D-V-F-R-V-P-P-G-Y-N-A is very similar to that of the PAPs of red kidney bean (Phaseolus vulgaris; Klabunde et al., 1994), Arabidopsis (accession no. U48448), and soybean (Glycine max; LeBansky et al., 1991; Fig. 1). The identities to the red kidney bean, Arabidopsis, and soybean phosphatase domains were 79%, 63%, and 69%, respectively. This finding tentatively identified the S. oligorrhiza phosphatase as a PAP.
Figure 1.
Comparison of the 19-amino acid N-terminal sequence of S. oligorrhiza PAP with equivalent domains of other plant PAPs. Asterisks indicate amino acids identical to those in S. oligorrhiza.
Immunoblotting
Immunoblotting of the purified phosphatase was performed using an affinity-purified anti-S. oligorrhiza phosphatase antiserum and an anti-Arabidopsis PAP antiserum. As shown in Figure 2, the Arabidopsis PAP antiserum cross-reacted strongly with the S. oligorrhiza phosphatase. As controls, wheat germ and bovine alkaline phosphatases did not cross-react with either of the antisera.
Metal Content
When calculating the metal content, a phosphatase molecular mass of 57 kD was used. However, a significant but unquantified proportion of this mass is known to be carbohydrate. The metal content was determined on triplicate samples. As shown in Table I, Fe and Mn were detected at 0.3 and 0.25 mol mol−1 phosphatase subunit, respectively, whereas Zn was detected at 0.10 mol mol−1 phosphatase subunit. Levels of Ni, Co, Cu, Tl, and Pb were negligible (data not shown).
Table I.
Metal content of phosphatase from S. oligorrhiza
| Metal | Content |
|---|---|
| mol mol−1 subunit | |
| Mn | 0.250 ± 0.06 |
| Fe | 0.298 ± 0.04 |
| Zn | 0.100 ± 0.03 |
Absorption Spectra
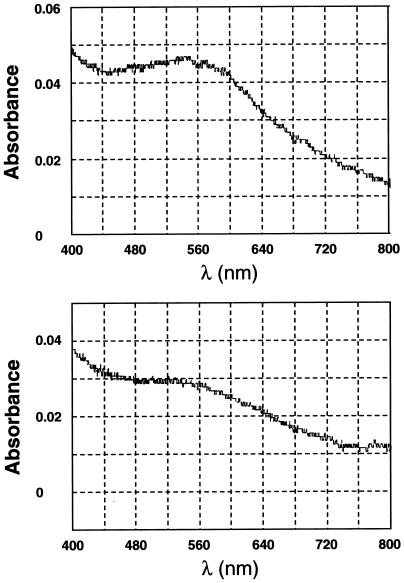
As analyzed by Beck et al. (1986), the red kidney bean PAP contained one atom of Fe and one atom of Zn per subunit, and its absorption spectrum showed a maximum at 560 nm. Solutions of the purified S. oligorrhiza phosphatase were distinctly purple. The visible absorption spectrum of the protein dissolved in 50 mm Tris-maleic acid buffer, pH 8.5, at 0.08 mg/mL is shown in Figure 3. The absorption maximum was approximately 556 nm. Although A556 persisted in the presence of sodium dithionite, the intensity of absorbance was significantly decreased, and the absorption maximum shifted to lower wavelengths as expected for reduced metal ions.
Figure 3.
Top, Absorption spectrum of purified S. oligorrhiza phosphatase (0.08 mg/mL) in 50 mm Tris-maleic acid buffer, pH 8.5. Bottom, Absorption spectrum of above preparation after 10 mm sodium dithionite was added.
Insensitivity to Tartrate Inhibition
Of all of the animal acid phosphatases, only the PAPs are resistant to inhibition by tartrate (Vincent and Averill, 1990). Plant PAPs, although sharing little structural homology with the animal enzymes (Vincent and Averill, 1990), are also only slightly inhibited by tartrate (Sugiura et al., 1981). The S. oligorrhiza acid phosphatase was inhibited by 15% in the presence of 50 mm tartrate, and inhibition increased only slowly with increasing tartrate concentrations (Table II). If a phosphatase is tartrate sensitive, 10 mm of the inhibitor is generally enough to cause a much larger loss of activity (Cashikar et al., 1997).
Table II.
Effect of different concentrations (mm) of sodium-potassium tartrate on phosphatase activity
| 0 mm | 10 mm | 50 mm | 100 mm | 200 mm | 400 mm | 600 mm |
|---|---|---|---|---|---|---|
| μmol min−1 mg−1 protein | ||||||
| 640 (100%) | 530 (82%) | 550 (86%) | 520 (81%) | 490 (77%) | 480 (75%) | 430 (67%) |
The activity was assayed using p-nitrophenylphosphate as a substrate at pH 8.0.
[3H]Ethanolamine Labeling
About 500 mg of −P plants was labeled with [3H]ethanolamine, a universal precursor of the GPI anchor, following the general procedure used previously (Morita et al., 1996). After 18 h of labeling, proteins were extracted from labeled plants and immunoprecipitated with anti-N-terminal peptide antibody.
After the samples were boiled to separate the immune complexes, the resulting radiolabeled samples were separated by SDS-PAGE on a 5% gel. The presence of PAP was detected by immunoblot analysis. As shown in Figure 4, a 57-kD band was detected by the antibodies. Radioactivity recovered in 1-mm slices from a separate, Coomassie blue-stained gel lane was associated only with the same 57-kD band, indicating that the phosphatase contained covalently bound [3H]ethanolamine. A second independent experiment gave similar results.
DISCUSSION
One of the most widespread and extensively studied GPI-anchored proteins of animals is alkaline phosphatase (Low, 1989). When the major inducible phosphatase of S. oligorrhiza was first found to contain a GPI anchor and to have a pH optimum of 7.0 to 8.0 (Morita et al., 1996), it was assumed to be structurally similar to animal alkaline phosphatase. However, upon purification certain properties of the S. oligorrhiza phosphatase, such as a shift downward in its pH optimum to 7.0 (Nakazato et al., 1997a), suggested that it may be a different enzyme. Additional data confirm that the S. oligorrhiza enzyme is a PAP closely resembling the PAPs of kidney bean (Klabunde et al., 1994), Arabidopsis (accession no. U48448), and soybean (LeBansky et al., 1991). Clear evidence for this comes from the strong similarity of the 19 N-terminal amino acids of the purified S. oligorrhiza phosphatase to the N-terminal domains of the red kidney bean and Arabidopsis PAP genes. The S. oligorrhiza N terminus is also very similar to a shorter soybean PAP domain. Preliminary findings indicate that the deduced amino acid sequence of the S. oligorrhiza clone is strikingly similar to the Arabidopsis PAP gene throughout (Nakazato et al., 1997b).
In contrast, the S. oligorrhiza N-terminal peptide showed no homology with PAP from animal sources. This is not surprising, since the animal and plant PAPs, although functionally and spectroscopically similar and known to have similar secondary structure (Klabunde et al., 1994), have distinctly different primary structures (Vincent and Averill, 1990).
An anti-Arabidopsis PAP antibody and the anti-S. oligorrhiza antiserum both cross-reacted with the S. oligorrhiza phosphatase but not with wheat germ phosphatase (an Mn-containing acid phosphatase), potato acid phosphatase, or bovine alkaline phosphatase (Fig. 2). These results, together with the other findings described above, leave no doubt that the S. oligorrhiza phosphatase is a PAP.
Metal analysis of the S. oligorrhiza phosphatase gave values of 0.3 mol of Fe, 0.25 mol of Mn, and 0.1 mol of Zn per mol of subunit (Table I). These absolute values may be misleading because the S. oligorrhiza phosphatase contains an undetermined quantity of covalently bound carbohydrate, but the relative proportions of metals are interesting. According to Beck et al. (1986), the red kidney bean PAP contains one atom of Fe and one atom of Zn per subunit, giving it an absorption maximum of 560 nm. The slightly lower 556 nm absorption maximum of the S. oligorrhiza enzyme may reflect its mixed metal content. As in the red kidney bean PAP, sodium dithionite caused a partial bleaching of the S. oligorrhiza PAP absorbance and shifted its absorption maximum to lower wavelengths (Fig. 3).
Metals contained in PAPs vary from species to species. Sugiura et al. (1980) found a PAP from sweet potato tubers that contained Mn, giving that enzyme maximal absorbance at 515 nm. Subsequent analyses of the sweet potato phosphatase showed that the native enzyme contains traces of Fe as well as Mn (Kawabe et al., 1984). More detailed studies will be necessary to determine the precise metal stoichiometry in the S. oligorrhiza phosphatase.
Radiolabeling with specific precursors of the GPI anchor has been a common approach to the identification of GPI-anchored proteins of animals (Englund, 1993) and, more recently, of two of the few plants examined (Stöhr et al., 1995; Morita et al., 1996). For the present study we incubated S. oligorrhiza with [3H]ethanolamine as described previously in characterization studies (Morita et al., 1996), but we used the resulting extracts for radioimmunoprecipitation of the phosphatase. As previously observed (Morita et al., 1996), most of the cellular radioactivity was present in phosphatidylethanolamine, a major membrane phospholipid, or as unincorporated ethanolamine. The immunopurified protein accounted for 6% of the total phosphatase activity and 1.6% of protein radioactivity. SDS-PAGE and immunoblot analysis (Fig. 4) showed clearly that the immunoprecipitated protein was indeed the 57-kD phosphatase and that it was radiolabeled. Our previous work with S. oligorrhiza (Morita et al., 1996) demonstrated that radioactivity incorporated into proteins from added [3H]ethanolamine is not randomized into other metabolites. This result supports our previous conclusion that ethanolamine is covalently bound to the phosphatase.
With only a limited number of partially complete characterizations of plant GPI-anchored proteins currently available, the field is in its infancy. PAPs of the type described here are widely distributed plant proteins that are localized on the cell surface and in the cell wall (Cashikar et al., 1997), as would be expected of a GPI-anchored protein. Although the synthesis of plant PAP is markedly induced in phosphate-deficient environments, these conditions also favor the turnover of PAP (Cashikar and Rao, 1996) and possibly the rapid cleavage of its GPI anchor (Morita et al., 1996). PAP stability is greatly enhanced by its enzymatic reaction product, Pi (Cashikar and Rao, 1996). Because of the newly recognized role of the GPI anchor in signal transduction (Robinson, 1997) and receptor cycling (Maxfield and Mayor, 1997), it is important to continue exploring how this protein modification may contribute to the regulation of metabolism in plant tissues.
ACKNOWLEDGMENTS
We are grateful to Dr. T. D. McKnight (Texas A & M University, College Station) for providing the antiserum against Arabidopsis PAP and the cDNA clone of Arabidopsis PAP. We also appreciate the synthesis and provision of the N-terminal oligopeptide of S. oligorrhiza PAP by Drs. C. Mazur and L. Wolfe (U.S. Environmental Protection Agency, Athens, GA).
Abbreviations:
- GPI
glycosylphosphatidylinositol
- PAP
purple acid phosphatase
Footnotes
This work was supported in part by grants from the U.S. Department of Agriculture (nos. 93-37304-9228 and 96-35304-3629 to G.A.T.).
LITERATURE CITED
- Beck J, McConachie LA, Summors AC, Arnold WN, De Jersey J, Zerner B. Properties of a purple phosphatase from red kidney bean: a zinc-iron metalloenzyme. Biochim Biophys Acta. 1986;869:61–68. [Google Scholar]
- Cashikar AG, Kumaresan R, Rao NM. Biochemical characterization and subcellular localization of the red kidney bean purple acid phosphatase. Plant Physiol. 1997;114:907–915. doi: 10.1104/pp.114.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar AG, Rao NM. Unfolding pathway in red kidney bean acid phosphatase is dependent on ligand binding. J Biol Chem. 1996;271:4741–4746. doi: 10.1074/jbc.271.9.4741. [DOI] [PubMed] [Google Scholar]
- Douglas DJ, French JB. Elemental analysis with a microwave-induced plasma/quadrupole mass spectrometer system. Anal Chem. 1981;53:37–41. [Google Scholar]
- Englund PT. The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- Kawabe H, Sugiura Y, Terauchi M, Tanaka H. Mn(III)-containing acid phosphatase. Properties of Fe(III)-substituted enzyme and function of Mn(III) and Fe(III) in plant and mammalian acid phosphatases. Biochim Biophys Acta. 1984;784:81–89. doi: 10.1016/0167-4838(84)90176-6. [DOI] [PubMed] [Google Scholar]
- Klabunde T, Stahl B, Suerbaum H, Hahner S, Karas M, Hillenkamp F, Krebs B, Witzel H. The amino acid sequence of the red kidney bean Fe(III)-Zn(II) purple acid phosphatase. Eur J Biochem. 1994;226:369–375. doi: 10.1111/j.1432-1033.1994.tb20061.x. [DOI] [PubMed] [Google Scholar]
- Kunze M, Riedel J, Lange U, Hurwitz R, Tischner R. Evidence for the presence of GPI-anchored PM-NR in leaves of Beta vulgaris and for PM-NR in barley leaves. Plant Physiol Biochem. 1997;35:507–512. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeBansky BR, McKnight TD, Griffing LR. Purification and characterization of a secreted purple phosphatase from soybean suspension cultures. Plant Physiol. 1991;99:391–395. doi: 10.1104/pp.99.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low MG. The glycosyl-phosphatidylinositol anchor of membrane proteins. Biochim Biophys Acta. 1989;988:427–454. doi: 10.1016/0304-4157(89)90014-2. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Mayor S (1997) Cell surface dynamics of GPI-anchored proteins. In F Haag, F Koch-Nolte, eds, ADP-Ribosylation in Animal Tissues. Plenum Press, New York, pp 365–370
- Morita N, Nakazato H, Okuyama H, Kim Y, Thompson GA., Jr Evidence for a glycosylinositolphospholipid-anchored alkaline phosphatase in the aquatic plant Spirodela oligorrhiza. Biochim Biophys Acta. 1996;1290:53–62. doi: 10.1016/0304-4165(95)00185-9. [DOI] [PubMed] [Google Scholar]
- Nakazato H, Okamoto T, Ishikawa K, Okuyama H. Purification and characterization of phosphatase inducibly synthesized in Spirodela oligorrhiza grown under phosphate-deficient conditions. Plant Physiol Biochem. 1997a;35:437–446. [Google Scholar]
- Nakazato H, Okamoto T, Nishikoori M, Washio K, Morita N, Haraguchi K, Thompson GA, Jr, Okuyama H. Characterization and cDNA cloning of the GPI-anchored phosphatase from Spirodela oligorrhiza. In: Ando T, Fujita K, Mae T, Matsumoto H, Mori S, Sekiya J, editors. Plant Nutrition—For Sustainable Food Production and Environment. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997b. pp. 229–230. [Google Scholar]
- Posner HB (1967) Aquatic vascular plants. In FA Witt, NK Wessels, eds, Methods in Developmental Biology. Crowell, New York, pp 301–317
- Robinson PJ (1997) Signal transduction via GPI-anchored membrane proteins. In F Haag, F Koch-Nolte, eds, ADP-Ribosylation in Animal Tissues. Plenum Press, New York, pp 365–370
- Stöhr C, Schuler F, Tischner R. Glycosyl-phosphatidylinositol-anchored proteins exist in the plasma membrane of Chlorella saccharophila (Krüger) Nadson: plasma membrane-bound nitrate reductase as an example. Planta. 1995;196:284–287. [Google Scholar]
- Sugiura Y, Kawabe H, Tanaka H. New manganese(III)-containing acid phosphatase. Evidence for an intense charge-transfer band and tyrosine phenolate coordination J Am Chem Soc. 1980;102:6581–6582. [Google Scholar]
- Sugiura Y, Kawabe H, Tanaka H, Fujimoto S, Ohara A. Purification, enzymatic properties and active site environment of a novel manganese(III)-containing acid phosphatase. J Biol Chem. 1981;256:10664–10670. [PubMed] [Google Scholar]
- Takos AM, Dry IB, Soole K. Detection of glycosyl-phosphatidylinositol-anchored proteins on the surface of Nicotiana tabacum protoplasts. FEBS Lett. 1997;405:1–4. doi: 10.1016/s0014-5793(97)00064-1. [DOI] [PubMed] [Google Scholar]
- Vincent JB, Averill BA. An enzyme with a double identity: purple acid phosphatase and tartrate-resistant acid phosphatase. FASEB J. 1990;4:3009–3014. doi: 10.1096/fasebj.4.12.2394317. [DOI] [PubMed] [Google Scholar]