Abstract
Sox4 is a transcription factor that regulates various developmental processes. Here we show that Sox4 was induced by TGF-β and negatively regulated the transcription factor GATA-3, the master regulator of function of T helper type 2 (TH2) cells, by two distinct mechanisms. First, Sox4 bound directly to GATA-3, preventing its binding to GATA-3 consensus DNA sequences. Second, Sox4 bound to the promoter region of the gene encoding interleukin 5 (IL-5), a TH2 cytokine, and prevented binding of GATA-3 to this promoter. TH2 cell–driven airway inflammation was modulated by alterations in Sox4 expression. Thus, Sox4 acted as a downstream target of TGF-β to inhibit GATA-3 function, TH2 differentiation and TH2 cell–mediated inflammation.
Elucidating the mechanisms by which naive CD4+ T cells differentiate into effector helper T cells is crucial for understanding T cell–dependent immune responses. Functionally distinct helper T cell subsets have been reported, including the TH1, TH2, TH9, TH17, TH22 and iTreg subsets1–8, and several transcription factors that regulate differentiation into these subsets have been identified, including T-bet1–8, GATA-3 (refs. 9,10) and RORγt and RORα11,12 for differentiation into TH1 cells, TH2 cells and TH17 cells, respectively. T lymphocytes have abundant expression of GATA-3, and its expression is required for the development of T cells in the thymus. In peripheral CD4+ T cells, interleukin 4 (IL-4)-mediated activation of the transcription factor STAT6 induces the expression of mRNA encoding GATA-3, which drives TH2 differentiation 13,14. GATA-3 binds to various regulatory regions in loci encoding TH2 cytokines and induces chromatin remodeling15–17. In addition, GATA-3 binds to the Il5 promoter and acts as a transcription factor for this gene18,19.
TGF-β is a pleiotropic cytokine that contributes to the maintenance of immune homeostasis through inhibition of the proliferation, differentiation, activation and effector function of cells of the immune system20. Mice with T cell–specific disruption of TGF-β signaling develop inflammation as a result of constitutive activation of T cells21,22. Depending on the cytokine environment, TGF-β induces the differentiation of peripheral CD4+ T cells into anti-inflammatory regulatory T cells (Treg cells) and also into proinflammatory TH17 and TH9 cells1–8. In contrast, TGF-β inhibits the differentiation of naive CD4+ T cells into effector TH1 and TH2 cells23,24. The molecular mechanisms of TGF-β-mediated inhibition of TH1 and TH2 differentiation remain unclear.
TGF-β mediates its biological functions by binding to type 1 and type 2 receptors for TGF-β, both of which are serine-threonine kinases20,25. Binding to these receptors induces the phosphorylation of proteins of the Smad family of signal transducers and the localization of Smad proteins to the nucleus. Eight Smad proteins have been identified in vertebrates; these are grouped into the following three categories: five receptor-associated Smad proteins (Smad1, Smad2, Smad3, Smad5 and Smad8), one common Smad protein (Smad4) and two inhibitory Smad proteins (Smad6 and Smad7). After TGF-β receptor–induced phosphorylation, Smad2 and Smad3 associate with Smad4, translocate to the nucleus and induce the transcription of target genes by binding to Smad-binding motifs.
Transcription factors of the Sox family (‘Sry-related high-mobility-group (HMG) box’) have key roles in the regulation of transcription during developmental processes, including early embryogenesis, sex determination, neural development, chondrogenesis, cardiac development and hematopoiesis26,27. The HMG box is critical for the function of Sox4 through its role in binding to DNA, bending DNA and protein interactions. Sox proteins can pair with various transcription factors28,29. Lymphocytes have high expression of Sox4, and Sox4 regulates T cell differentiation in the thymus and the population expansion of pro-B cells30,31. However, its expression profile in peripheral T cells and roles in immune responses have yet to be fully elucidated.
Here we investigated the role of Sox4 in peripheral CD4+ T cells and found that Sox4 was a downstream target of the TGF-β signaling pathway that negatively regulated TH2 differentiation and TH2 cell–dependent allergic airway inflammation. Sox4 inhibited the function of GATA-3 via two distinct mechanisms. Therefore, Sox4 is a unique negative regulator of GATA-3 function and a critical regulator of TH2 differentiation and TH2 cell–dependent immune responses.
RESULTS
Regulation of Sox4 expression by TGF-β
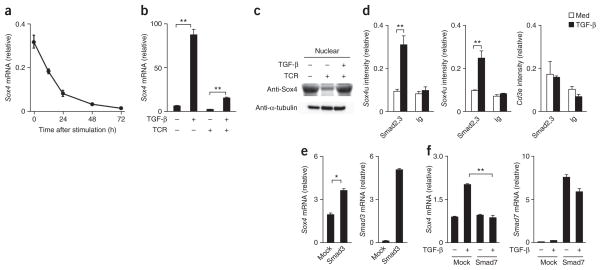
We detected substantial Sox4 mRNA in naive CD4+ T cells, but its expression decreased rapidly after stimulation mediated by the T cell antigen receptor (TCR; Fig. 1a). The treatment of naive CD4+ T cells with TGF-β for 24 h induced significant upregulation of the expression of Sox4 mRNA, which we also observed in CD4+ T cells stimulated with monoclonal antibody (mAb) to the TCR (Fig. 1b). We confirmed TGF-β-mediated induction of Sox4 at the protein level by immunoblot analysis (Fig. 1c). Sox4 belongs to the group C subfamily of Sox proteins, which includes the following three members in mice and humans: Sox4, Sox11 and Sox12. Although we detected low expression of Sox11 and moderate expression of Sox12 in activated CD4+ T cells, we noted no obvious effect of treatment with TGF-β (Supplementary Fig. 1a). Among the other helper T cell subsets, the expression of Sox4 mRNA was highest in iTreg cells and lowest in TH2 cells (Supplementary Fig. 1b). We confirmed high expression of Sox4 protein in iTreg cells by immunoblot analysis (Supplementary Fig. 1c).
Figure 1.
TGF-β-induced Sox4 expression in CD4+ T cells.
(a) Quantitative RT-PCR analysis of Sox4 mRNA in naive CD4+ T cells stimulated for 0–72 h with immobilized mAb to TCRβ in the presence of IL-2; results are presented relative to the expression of Hprt mRNA (encoding hypoxanthine guanine phosphoribosyl transferase).
(b) Quantitative RT-PCR analysis of Sox4 mRNA in naive CD4+ T cells left inactivated (TCR−) or activated for 24 h with mAb to TCR (TCR+) in the presence (+) or absence (−) of TGF-β. (c) Immunoblot analysis of Sox4 in the nuclear fraction of naive CD4+ T cells cultured as in b; analysis with antibody to α-tubulin (Anti-α-tubulin) serves as a loading control for the cytosolic fraction (bottom). (d) ChIP assay of the binding of Smad2-Smad3 (Smad2,3) or immunoglobulin (Ig; control) to a 5′ region upstream of the Sox4 locus (Sox4u) or the promoter of the gene encoding the invariant signaling protein CD3ε (Cd3e) in naive CD4+ T cells cultured for 12 h in the presence of medium (Med) or TGF-β; results are presented relative to those of input DNA. (e) Quantitative RT-PCR analysis of Sox4 and Smad3 mRNA in naive CD4+ T cells mock transduced (Mock) or transduced to express Smad3, then stimulated with mAb to TCR, followed by stimulation for 24 h with TGF-β (presented as in a). (f) Quantitative RT-PCR analysis of Sox4 and Smad7 mRNA in naive CD4+ T cells mock transduced or transduced to express Smad7, then stimulated with mAb to TCR, followed by no stimulation (−) or stimulation with TGF-β (+; presented as in a). *P < 0.05 and **P < 0.01 (Student’s t-test). Data are representative of four (a; mean and s.d. of three samples), three (b,d) or two (c,e,f) independent experiments with similar results (error bars (b,d–f, s.d.).
To determine whether TGF-β-mediated activation of the Smad signaling pathway was involved in the induction of Sox4 expression, we did chromatin-immunoprecipitation (ChIP) analysis with an antibody to Smad2 and Smad3. We detected significant binding of Smad2-Smad3 to the Sox4 promoter (−480 base pairs from the transcription start site) and an upstream region of the Sox4 locus (−1,000 base pairs from the transcription start site; Fig. 1d). Retrovirus vector–mediated expression of Smad3 in developing TH2 cells significantly enhanced the TGF-β-induced expression of Sox4 mRNA (Fig. 1e). Furthermore, the retroviral expression of an inhibitory Smad protein, Smad7, resulted in the inhibition of TGF-β-induced Sox4 expression in activated CD4+ T cells (Fig. 1f). These results indicated that TGF-β induced Sox4 expression through activation of the Smad signaling pathway.
Inhibition of TH2 differentiation by TGF-β-induced Sox4
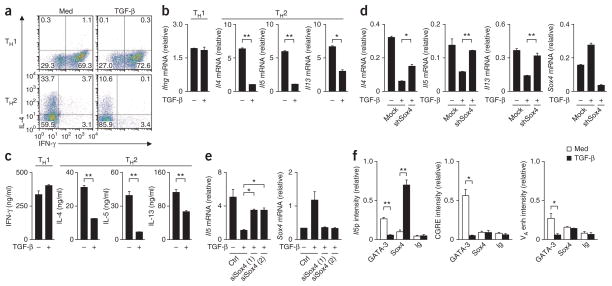
TGF-β inhibits the proliferation and activation of CD4+ T cells and the differentiation of CD4+ T cells into helper T cells32. Although both TH1 differentiation and TH2 differentiation were inhibited by TGF-β, TH2 differentiation was ‘preferentially’ inhibited (data not shown). In particular, when we added TGF-β to cells cultured under TH1- or TH2-polarizing conditions 48 h after the initial stimulation of the TCR, TH2 differentiation was selectively inhibited, whereas TH1 differentiation was left intact (Fig. 2a). We confirmed lower expression of Il4, Il5 and Il13 and normal induction of Ifng (which encodes interferon-γ (IFN-γ)) by quantitative RT-PCR (Fig. 2b) as well as lower expression of the corresponding proteins by enzyme-linked immunosorbent assay (ELISA; Fig. 2c). Although TGF-β had potent inhibitory effects on TH2 differentiation, inhibition of the expression of Gata3 mRNA and GATA-3 protein was moderate under these conditions (Supplementary Fig. 2a, b). To assess the involvement of Sox4 in the TGF-β-mediated inhibition of TH2 differentiation, we did a series of knockdown experiments. Inhibition of the expression of Il4, Il5 and Il13 by TGF-β was restored significantly by the introduction of short hairpin RNA (shRNA) specific for Sox4 (Fig. 2d). The inhibition of Il5 expression by TGF-β was also reproducibly diminished by knockdown of Sox4 expression mediated by small interfering RNA (siRNA; Fig. 2e). Knockdown of Sox4 in TH1 cells did not result in the induction of IL-4-producing TH2 cells (data not shown). By ChIP assay, we assessed the effect of treatment with TGF-β on the binding of GATA-3 to loci encoding TH2 cytokines. Binding of GATA-3 to the Il5 promoter, the conserved GATA-3-response element (CGRE) of genes encoding TH2 cytokines, and the DNase-hypersensitive site VA in the Il4 enhancer was abrogated by treatment with TGF-β (Fig. 2f). We observed TGF-β-dependent binding of Sox4 to the Il5 promoter but not to the CGRE or the VA site (Fig. 2f). These results indicated that Sox4 mediated the TGF-β-mediated inhibition of TH2 differentiation and expression of TH2 cytokines.
Figure 2.
Involvement of Sox4 in the TGF-β-mediated inhibition of TH2 differentiation. (a) Intracellular staining of IL-4 and IFN-γ in cells cultured under TH1- or TH2-polarizing conditions with medium alone or TGF-β. Numbers in quadrants indicate percent cells in each throughout. (b) Quantitative RT-PCR analysis of cytokine-encoding mRNA in TH1 and TH2 cells left untreated (−) or treated with TGF-β (+), then restimulated for 24 h with immobilized anti-TCR (presented as in Fig. 1a). (c) ELISA of cytokines in supernatants of the cells in b. (d) Quantitative RT-PCR analysis of cytokine-encoding mRNA in cells treated with control shRNA (Mock) or Sox4-specific shRNA (shSox4) and left untreated or treated with TGF-β (presented as in Fig. 1a). (e) Quantitative RT-PCR analysis of Il5 and Sox4 mRNA in cells treated with control siRNA (Ctrl) or Sox4-specific siRNA (siSox4 (1) and siSox4 (2)) and left untreated or treated with TGF-β (presented as in Fig. 1a). (f) ChIP assay of the binding of GATA-3, Sox4 and immunoglobulin to the Il5 promoter (Il5p), the CGRE and the VA site in the Il4 enhancer (VA enh) in developing TH2 cells cultured with medium alone or TGF-β (presented as in Fig. 1d). *P < 0.05 and **P < 0.01 (Student’s t-test). Data are representative of three (a,b,d,e) or two (c,f) independent experiments with similar results (error bars (b–f), s.d.).
Suppression of TH2 differentiation by enforced expression of Sox4
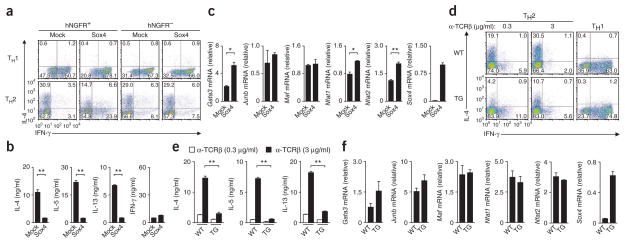
To further investigate the role of Sox4 in differentiation into various helper T cell subsets, we ectopically expressed Sox4 in TH1 cells and TH2 cells via a retroviral vector. Ectopic expression of Sox4 resulted in loss of IL-4-producing cells under TH2-differentiating conditions, whereas the generation of IFN-γ-producing cells under either TH1- or TH2-polarizing conditions was augmented (Fig. 3a). The differentiation of cells infected with empty vector (mock-infected cells) was not altered (Fig. 3a). Sox4-transduced TH2 cells had lower expression of Il4, Il5 and Il13 mRNA (Supplementary Fig. 3a) and IL-4, IL-5 and IL-13 protein (Fig. 3b) but slightly higher expression of Ifng mRNA than did mock-infected TH2 cells (Supplementary Fig. 3a). The introduction of Sox4 also inhibited the TH2 differentiation of T-bet-deficient CD4+ T cells (Supplementary Fig. 3b), which indicated that the inhibition of TH2 differentiation by Sox4 was not dependent on T-bet expression. There was less histone H3 acetylated at Lys9 (H3K9ac) and H3K14ac, which are associated with transcriptionally active chromatin, at the Il4, Il5 and Il13 promoters, but more at the Ifng promoter, in Sox4-transduced TH2 cells than in mock-infected TH2 cells (Supplementary Fig. 3c). Sox4-transduced TH2 cells had slightly higher, not lower, expression of Gata3 mRNA and Nfat1 and Nfat2 mRNA (which encode transcription factors of the NFAT family) than did mock-infected TH2 cells, whereas their expression of Junb mRNA (which encodes the transcription factor JunB) and Maf mRNA (which encodes the transcription factor c-Maf) was not affected (Fig. 3c). These results indicated that Sox4 negatively regulated TH2 differentiation and the expression of mRNA encoding TH2 cytokines without inhibiting Gata3 expression.
Figure 3.
Enforced expression of Sox4 inhibits TH2 differentiation. (a) Staining of IL-4 and IFN-γ in CD4+ T cells left uninfected (hNGFR−) or infected (hNGFR+), and transduced with empty vector (mock transduced; Mock) or with a Sox4-expressing retroviral vector (Sox4) and then cultured for 3 d under TH1- or TH2-polarizing conditions. (b) ELISA of cytokines in TH2 cells mock transduced (Mock) or transduced with a Sox4-expressing retroviral vector (Sox4) and restimulated with immobilized mAb to TCRβ. (c) Quantitative RT-PCR analysis of mRNA encoding various transcription factors (vertical axes) in TH2 cells transduced as in b (presented as in Fig. 1a). (d) Staining of IL-4 and IFN-γ in wild-type (WT) and Sox4-transgenic (TG) naive CD4+ T cells cultured under TH2-polarizing conditions with various concentrations (above plots) of anti-TCRβ (α-TCRβ) or under TH1-polarizing conditions (far right). (e) ELISA of cytokines in supernatants of wild-type and Sox4-transgenic TH2 cells generated in vitro and stimulated for 16 h with immobilized mAb to TCRβ at a concentration of 0.3 or 3 μg/ml (key). (f) Quantitative RT-PCR analysis of mRNA encoding various transcription factors (vertical axes) in wild-type and Sox4-transgenic CD4+ T cells (presented as in Fig. 1a). *P < 0.05 and **P < 0.01 (Student’s t-test). Data are representative of three (a,b,d–f) or four (c) independent experiments with similar results (error bars (b,c,e,f), s.d.).
To further investigate the role of Sox4 in peripheral T cell function, we generated mice with transgenic expression of Sox4 under the control of the T cell–specific Lck distal promoter (Sox4-transgenic mice; Supplementary Fig. 4a). Sox4 mRNA expression was approximately tenfold higher in both CD4+ splenic T cells and CD8+ splenic T cells from Sox4-transgenic mice than in those from wild-type mice (Supplementary Fig. 4b). Although Sox4-transgenic mice had slightly fewer CD4+ T cells in the spleen and thymus than did wild-type mice, cell-surface expression of TCRs and receptors for cytokines, as well as antigen-induced proliferative responses, were within the normal range in these mice (Supplementary Fig. 4c–e). There was less generation of IL-4-producing Sox4-transgenic CD4+ T cells than of IL-4-producing wild-type CD4+ T cells under TH2 conditions, whereas the generation of IFN-γ-producing Sox4- transgenic CD4+ T cells was modestly enhanced (Fig. 3d). The generation of IFN-γ-producing Sox4-transgenic CD4+ T cells cells under TH1 conditions was enhanced (Fig. 3d), whereas the generation of IL-17-producing TH17 cells, transcription factor Foxp3–expressing iTreg cells and TH9 cells was not affected by the transgene encoding Sox4 (Supplementary Fig. 4f). We confirmed by ELISA the lower production of IL-4, IL-5 and IL-13 by Sox4-transgenic CD4+ T cells than by wild-type cells (Fig. 3e). There was less H3K9ac and H3K14ac at the Il4, Il5 and Il13 promoters in Sox4-transgenic TH2 cells than in wild-type cells (Supplementary Fig. 4g). Although the expression of Gata3 mRNA was higher in Sox4-transgenic cells than in wild-type cells (not statistically significant; Fig. 3f), wild-type and Sox4-transgenic TH2 cells had a similar amount of GATA-3 protein (Supplementary Fig. 4h). There was no significant difference between wild-type and Sox4-transgenic cells in their expression of other TH2 cell–associated transcription factors (Fig. 3f). These results indicated that TH2 differentiation was impaired without affecting the expression of TH2-associated transcription factors, including GATA-3.
Sox4 inhibits GATA-3 function by distinct mechanisms
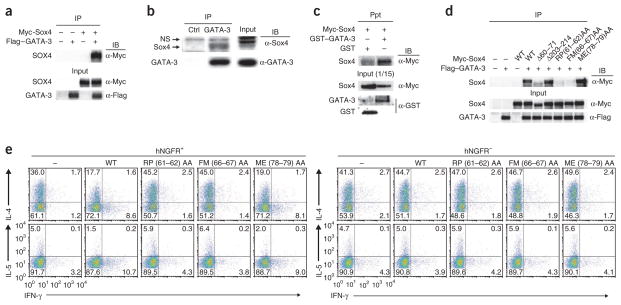
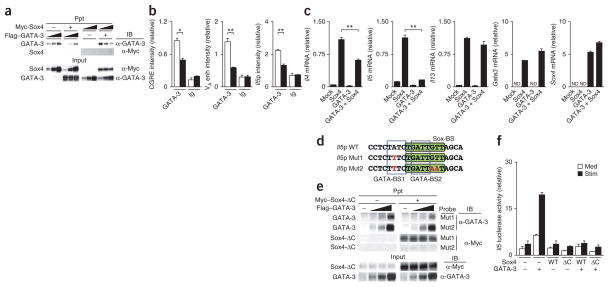
To identify the molecular mechanisms by which Sox4 inhibits TH2 differentiation, we assessed the possible physical association of Sox4 with GATA-3 in three different experimental systems. Sox4 immunoprecipitated with GATA-3 in mixtures of lysates of 293T cells (human embryonic kidney T cells) transfected to express Myc-tagged Sox4 or Flag-tagged GATA-3 (Fig. 4a). We also observed the association of Sox4 with GATA-3 in TG40 cells, a mouse T cell line with substantial endogenous expression of both Sox4 and GATA-3 protein (Fig. 4b). In addition, glutathione S-transferase–tagged recombinant GATA-3 directly interacted with Myc-tagged recombinant Sox4, as detected in a precipitation assay with glutathione 4B Sepharose beads (Fig. 4c). Next we generated several Myc-tagged Sox4 mutants to determine which domains of Sox4 were important for its association with GATA-3 (Supplementary Fig. 5a). The association between GATA-3 and a Sox4 mutant lacking the amino-terminal region was much weaker than that of GATA-3 and wild-type Sox4; also, a mutant in which both the amino-terminal region and HMG box were deleted and a mutant in which the carboxy-terminal region, including the transactivation domain, was deleted each completely failed to associate with GATA-3 in 293T cells (Supplementary Fig. 5a). This indicated that both the HMG box and carboxy-terminal portion of Sox4 were important for the association with GATA-3. The arginine at position 61 (Arg61), proline at position 62 (Pro62), phenylalanine at position 66 (Phe66) and methionine at position 67 (Met67) are perfectly conserved among proteins of the Sox family. We generated several Sox4 mutants (Supplementary Fig. 5b) and examined their association with GATA-3 in 293T cells. A Sox4 mutant with deletion of amino acids 60–71 showed less association with GATA-3 than did wild-type Sox4, but the association with GATA-3 was unaffected by the deletion of amino acids 203–214 (Fig. 4d). Substitution of Arg61 and Pro62 or of Phe66 and Met67 with alanine abolished the association of Sox4 with GATA-3 (Fig. 4d), but substitution of the methionine at position 78 (Met78) and glutamic acid at position 79 (Glu79) with alanine had no effect on the binding of Sox4 to GATA-3. These results indicated that Arg61, Pro62, Phe66 and Met67 in the amino-terminal region of the HMG box in Sox4 were important for the association of Sox4 with GATA-3.
Figure 4.
Physical association of Sox4 with GATA-3. (a) Immunoprecipitation (IP) and immunoblot analysis (IB) of the association of Sox4 and GATA-3 in 293T cells left untransfected (−) or transfected (+) to express Myc-tagged Sox4 (Myc-Sox4) and/or Flag-tagged GATA-3 (Flag–GATA-3); below (Input), parallel analysis of total cell lysates (without immunoprecipitation). (b) Immunoprecipitation and immunoblot analysis of the association of endogenous GATA-3 with endogenous Sox4 in TG40 cells (Input (middle right), as in a). NS, nonspecific band. (c) Precipitation assay (Ppt) of the association of Sox4 and GATA-3 in cells transfected to express Myc-tagged recombinant Sox4 (Myc-Sox4), glutathione S-transferase–tagged recombinant GATA-3 (GST–GATA-3) and/or glutathione S-transferase alone (GST), probed with anti-Myc; below (Input), immunoblot analysis of an aliquot of input samples (1/15 volume) with anti-Myc or anti-GST. (d) Immunoprecipitation and immunoblot analysis of the association of Sox4 and GATA-3 in 293T cells mock transfected (–) or transfected to express Flag-tagged GATA-3 and Myc-tagged wild-type Sox4 (WT) or mutant Sox4 lacking amino acids 60–71 (Δ60–71) or amino acids 203–214 (Δ203–214) or with substitution of alanine residues for arginine and proline (RP(61–62)AA), phenylalanine and methionine (FM(66–67)AA) or methionine and glutamic acid (ME(78–79)AA); Input (below), as in a. (e) Staining of IL-4, IL-5 and IFN-γ in TH2 cells left untransduced (hNGFR−) or transduced (hNGFR+) and mock transfected or transfected to express wild-type or mutant Sox4 (as in d), assessed 3 d after retroviral infection. Data are representative of at least three independent experiments with similar results.
To further examine the effect of the Sox4 mutants described above on TH2 differentiation, we expressed those mutants in primary developing TH2 cells through the use of retrovirus vectors. The Sox4 mutants with substitution of Arg61 and Pro62 or of Phe66 and Met67 with alanine did not inhibit the differentiation of IL-4 and IL-5 producing TH2 cells, but the Sox4 mutant with substitution of Met78 and Glu79 with alanine did have this effect (Fig. 4e). Thus, the association of Sox4 with GATA-3 was required for the Sox4-mediated inhibition of TH2 differentiation. To identify the domains of GATA-3 critical for its association with Sox4, we generated various GATA-3 mutants (Supplementary Fig. 5c). A GATA-3 mutant with deletion of the amino-terminal finger and another with deletion of the carboxy-terminal finger showed much weaker association with Sox4 than did wild-type GATA-3 (Supplementary Fig. 5c), whereas the association of GATA-3 with Sox4 in 293T cells was eliminated by deletion of the zinc-finger domain and carboxy-terminal region of GATA-3 (Supplementary Fig. 5c). These results indicated that the zinc-finger domains of GATA-3 were important for its association with Sox4.
Next we assessed the effect of Sox4 on the DNA-binding activity of GATA-3 by a precipitation assay. The binding of GATA-3 to an oligonucleotide containing a consensus GATA-binding site was decreased in a dose-dependent manner in the presence of Sox4 (Fig. 5a), which indicated that Sox4 interfered with the binding of GATA-3 to the GATA-binding consensus motif. To assess whether Sox4 inhibited the binding of GATA-3 to DNA in primary TH2 cells, we did a ChIP assay with Sox4-transgenic TH2 cells. We detected less binding of GATA-3 to its target regions in Sox4-transgenic TH2 cells than in wild-type primary TH2 cells (Fig. 5b). In addition, we examined the effect of retroviral expression of Sox4 and GATA-3 in developing TH1 cells on TH2 differentiation and the expression of TH2 cytokines. Transduction of GATA-3 alone induced the expression of TH2 cytokines33, whereas the coexpression of GATA-3 and Sox4 resulted in lower expression of TH2 cytokines (Fig. 5c). Ectopic expression of Sox4 lead to more inhibition of Il5 transcription than of Il4 transcription or Il13 transcription (Fig. 5c).
Figure 5.
Sox4 interferes with the binding of GATA-3 to DNA.
(a) Precipitation assay of the binding of GATA-3 to a GATA-binding consensus motif in 293T cells transfected to express Myc-tagged Sox4 and/or Flag-tagged GATA-3 (Input (below), immunoblot analysis of whole-cell lysates without precipitation (control). Wedges indicate threefold ‘titration’ of input lysates. (b) ChIP analysis of the binding of GATA-3 and immunoglobulin at the CGRE, Il4 VA site and Il5 promoter region in Sox4-transgenic TH2 cells (presented as in Fig. 1d). (c) Quantitative RT-PCR analysis of mRNA encoding TH2 cytokines in TH2 cells mock infected (Mock; far left) or infected with retroviral vector encoding Sox4 or GATA-3 alone (middle) or together (GATA-3 + Sox4; far right) and stimulated with immobilized mAb to TCRβ (presented as in Fig. 1a). ND, not detected. (d) Probes used in e, containing the wild-type Il5 promoter (Il5p WT) or Il5 promoter with mutation (red) of GATA-binding site 1 (GATA-BS1 (blue outline)) alone (Il5p Mut1) or in combination with mutation of the Sox-binding site (Sox-BS (green); Il5p Mut2), which overlaps GATA-binding site 2 (GATA-BS2 (blue outline)). (e) Precipitation assay of mixtures of lysates of 293T cells mock transfected (−) or transfected (+) to express a Myc-tagged Sox4 mutant lacking the carboxy-terminal region (Myc–Sox4-ΔC) or various concentrations (wedges) of Flag-tagged GATA-3, assessed with the mutated probes in d (Input (below), as in a). (f) Luciferase activity in M12 B cells left untransfected (−) or transfected (+) to express wild-type Sox4 or the Sox4 mutant in e, plus GATA-3, as well as a firefly luciferase reporter for the Il5 promoter, then left unstimulated (Med) or stimulated (Stim) with the phorbol ester PMA (30 ng/ml) and dibuteryl cAMP (100 μM); results are presented relative to renilla luciferase activity. *P < 0.05 and **P < 0.01 (Student’s t-test). Data are representative of at least three independent experiments with similar results (mean and s.d. of three samples in c; mean and s.d. in b and f).
Analysis of the Il5 promoter sequence showed a conserved Sox-binding site that overlapped a GATA-binding site (Fig. 5d). To assess the DNA sequence required for the binding of Sox4 to the Il5 promoter region, we generated oligonucleotides of the Il5 promoter containing single-nucleotide mutation of GATA-binding site 1a only or of GATA-binding site 1a and the Sox-binding site. To exclude possible confounding effects due to the interaction of Sox4 with GATA-3, we used the Sox4 mutant lacking the carboxy-terminal region that failed to associate with GATA-3 (Supplementary Fig. 5a). That Sox4 mutant inhibited the binding of GATA-3 to the Il5 promoter with mutation of GATA-binding site 1a only but did not inhibit the binding of GATA-3 to the Il5 promoter with mutation of both GATA-binding site 1a and the Sox-binding site (Fig. 5e). Both wild-type Sox4 and the mutant Sox4 efficiently suppressed Il5 promoter activity in M12 mouse B cells (Fig. 5f). These results indicated that Sox4 was able to inhibit Il5 expression independently of its association with GATA-3.
Sox4 controls TH2 cell–dependent airway inflammation
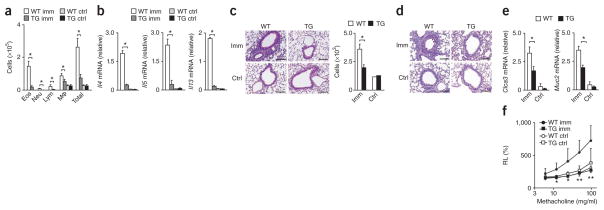
Next we investigated the role of Sox4 in TH2 differentiation in an in vivo model of airway inflammation. We immunized wild-type and Sox4-transgenic mice with ovalbumin (OVA) and aluminum hydroxide (as an adjuvant) and then challenged the mice with OVA by inhalation. We observed less infiltration of inflammatory cells, including eosinophils, in the bronchioalveolar lavage (BAL) fluid of OVA-immunized Sox4-transgenic mice than in that of OVA- immunized wild-type mice (Fig. 6a). The expression of Il4, Il5 and Il13 mRNA in cells of the BAL fluid was also very low in Sox4-transgenic mice (Fig. 6b). Sox4-transgenic mice had fewer mononuclear cells infiltrating the peribronchiolar regions of the lung than did wild-type mice (Fig. 6c). Sox4-transgenic bronchioles had less mucus hyperproduction and goblet-cell metaplasia than did wild-type bronchioles, as assessed by staining with periodic acid–Schiff reagent (Fig. 6d), and Sox4- transgenic mice had lower expression of Clca3 mRNA (encoding the calcium-activated chloride channel CLCA3) and Muc2 mRNA (encoding the mucin protein Mucin2) in the lungs than did wild-type mice (Fig. 6e). No obvious methacholine-induced airway hyper-responsiveness was induced in the Sox4-transgenic mice (Fig. 6f).
Figure 6.
Attenuated OVA-induced allergic airway inflammation in Sox4-transgenic mice. (a) Quantification of eosinophils (Eos), neutrophils (Neu), lymphocytes (Lym), macrophages (Mφ) and total cells in BAL fluid from wild-type and Sox4-transgenic mice (n = 5 per group) left unimmunized (ctrl) or immunized with OVA (imm). *P < 0.001 (analysis of variance (ANOVA) and Bonferroni test). (b) Quantitative RT-PCR analysis of Il4, Il5 and Il13 mRNA in cells from the BAL fluid of mice as in a. *P < 0.01 (Student’s t-test). (c,d) Microscopy (c (left) and d) of lungs from mice as in a, fixed and stained with hematoxylin and eosin (c) or periodic acid–Schiff reagent (d). Original magnification, ×200 (scale bars, 10 μm). Right (c), quantification of leukocytes in the peribronchiolar region (cells per mm2). *P < 0.01 (Student’s t-test). (e) Quantitative RT-PCR analysis of Clca3 and Muc2 mRNA in lungs from mice as in a (n = 3 per group). (f) Airway resistance of mice as in a (n = 6 per group) treated with various concentrations of methacholine (horizontal axis), presented as lung resistance (RL) relative to the lung resistance without methacholine treatment. *P < 0.01 and **P < 0.001 (ANOVA and Bonferroni test). Data are representative of three independent experiments with similar results (mean and s.d. in a–c,e,f).
We also knocked down Sox4 with shRNA in DO11.10 TH2 cells (which have transgenic expression of an OVA-specific TCRαβ) and evaluated the ability of these cells to cause airway inflammation. After restimulation of the TCR, we found higher expression of Il4 and Il5 mRNA in DO11.10 TH2 cells transfected with Sox4-specific shRNA than in those transfected with control shRNA (Supplementary Fig. 6a). We intravenously injected DO11.10 TH2 cells transfected with control or Sox4-specific shRNA into wild-type BALB/c mice, followed by intranasal administration of OVA34. The infiltration of the BAL fluid with inflammatory cells, including eosinophils, as well as airway hyper-responsiveness, were much greater in the group given cells transfected with Sox4-specific shRNA than in the group given cells transfected with control shRNA (Supplementary Fig. 6b, c).
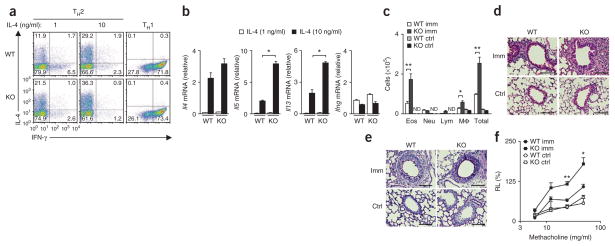
To generate mice with T cell–specific deficiency in Sox4, we crossed mice with loxP-flanked Sox4 alleles (Sox4fl/fl mice)35 to mice with transgenic expression of Cre recombinase driven by the promoter of the gene encoding CD4 (which results in T cell–specific deletion of loxP-flanked genes) to produce Sox4fl/flCD4-Cre offspring. The number of CD4+ T cells, cell-surface expression of TCR and cytokine receptors and the proliferative responses of naive CD4+ T cells induced by stimulation with mAb to TCR plus mAb to CD28 were within the normal range in Sox4fl/flCD4-Cre mice (Supplementary Fig. 7a–c). Sox4fl/flCD4-Cre naive CD4+ T cells cultured under TH2 conditions generated more IL-4-producing cells than did wild-type cells, whereas the generation of IFN-γ-producing cells under TH1 conditions was similar in cells of each genotype (Fig. 7a). The generation of IL-17-producing TH17 cells, Foxp3-expressing iTreg cells and IL-9-producing TH9 cells was not affected by the genotype of the cells (Supplementary Fig. 7d). We detected significantly higher expression of Il5 and Il13 mRNA in Sox4fl/flCD4-Cre TH2 cells than in wild-type TH2 cells (Fig. 7b). The expression of GATA-3 protein was similar in TH2 cells of each genotype (Supplementary Fig. 7e).
Figure 7.
Enhanced OVA-induced allergic airway inflammation in mice with CD4+ T cell–specific Sox4 deficiency. (a) Staining of IL-4 and IFN-γ in wild-type naive CD4+ T cells (WT) and Sox4fl/flCD4-Cre naive CD4+ T cells (KO) cultured under TH2-polarizing conditions with various concentrations (above plots) of IL-4 or under TH1-polarizing conditions (far right). (b) Quantitative RT-PCR analysis of Il4, Il5, Il13 and Ifng mRNA in wild-type and Sox4fl/flCD4-Cre TH2 cells generated in vitro and treated with IL-4 at a concentration of 1 or 10 ng/ml (key) and stimulated with immobilized mAb to TCRβ (presented as in Fig. 1a). *P < 0.01 (Student’s t-test). (c) Quantification of eosinophils, neutrophils, lymphocytes, macrophages and total cells of the BAL fluid from wild-type or Sox4fl/flCD4-Cre mice (n = 5 per group) left unimmunized or immunized with OVA. *P < 0.05 and **P < 0.01 (ANOVA and Bonferroni test). (d) Microscopy of lungs from the mice in c, fixed and stained with hematoxylin and eosin. Quantification of mononuclear cells (per mm2): wild-type, 552 ± 37.6 (unimmunized) or 2,654 ± 308.6 (immunized); Sox4fl/flCD4-Cre, 474.7 ± 80.5 (unimmunized) or 5,694 ± 574.7 (immunized). P < 0.01, wild-type immunized versus Sox4fl/flCD4-Cre immunized (Student’s t-test). (e) Microscopy of lungs from the mice in c, fixed and stained with periodic acid–Schiff reagent. Original magnification (d,e), ×200 (scale bars, 10 μm). (f) Airway resistance of lungs from the mice in c (n = 6 per group; presented as in Fig. 6f). *P < 0.01 and **P < 0.001 (ANOVA and Bonferroni test). Data are representative of three (a,b) or two (c–f) independent experiments with similar results (mean and s.d. in b,c,f).
In the airway inflammation model, we observed significantly more infiltration of the BAL fluid by inflammatory cells, including eosinophils, in OVA-immunized Sox4fl/flCD4-Cre mice than in OVA-immunized wild-type mice (Fig. 7c). The expression of Il5 and Il13 mRNA in cells of the BAL fluid was slightly but notably higher Sox4fl/flCD4-Cre mice than in wild-type mice (Supplementary Fig. 7f); Sox4fl/flCD4-Cre mice had more mononuclear cells infiltrating the peribronchiolar regions of the lungs than did wild-type mice (Fig. 7d), and Sox4fl/flCD4-Cre mice had more mucus hyper-production and goblet-cell metaplasia in the bronchioles than did wild-type mice (Fig. 7e). There was also significantly more airway hyper-responsiveness in Sox4fl/flCD4-Cre mice than in wild-type mice (Fig. 7f). Together these results indicated that Sox4 regulated TH2 cell–mediated allergic airway inflammation and airway hyper-responsiveness.
DISCUSSION
Here we have demonstrated that Sox4 was induced by TGF-β and downregulated TH2 differentiation through its role in the negative regulation of GATA-3 function via two distinct mechanisms. First, Sox4 associated with GATA-3 and inhibited the binding of GATA-3 to its DNA-binding sequence, which resulted in impaired GATA-3-induced TH2 differentiation. Second, Sox4 directly bound to the Il5 promoter and interfered with the binding of GATA-3 to DNA, which led to the repression of Il5 transcription. Therefore, Il5 expression was negatively regulated by Sox4 through the inhibition of both TH2 differentiation and transcription.
During the initiation of TH2 differentiation, stimulation of the TCR seemed to downregulate Sox4 expression and facilitated the GATA-3 function of inducing TH2 differentiation and Il5 transcription. In the presence of TGF-β, however, Sox4 was induced and suppressed GATA-3 function, which resulted in the inhibition of TH2 differentiation and Il5 transcription. The expression of Sox4 seemed to depend on the balance between the strength of TCR stimulation and the amount of TGF-β.
Overexpression of Sox4 in developing TH1 cells induced more IFN-γ-producing cells. However, we observed no apparent change in TH1 differentiation in cultures of Sox4-deficient TH1 cells. Therefore, although it is likely that Sox proteins, including Sox4, control TH1 differentiation under some conditions, we obtained no solid evidence indicating that Sox4 directly controlled TH1 differentiation.
TGF-β-dependent induction of SOX4 mRNA has been demonstrated in a human pituitary tumor cell line36. TGF-β induces Sox4 expression in glioma-initiation cells, and Sox4 is suggested to maintain the stem-cell qualities of the glioma-initiation cells via the induction of Sox2 expression37. Although the expression of Sox2 was undetectable in naive CD4+ T cells, Sox4 mRNA expression was high in these cells. Therefore, Sox4 may have an important role in maintaining the quiescent state and/or the multilineage differentiation potential of naive CD4+ T cells.
Several transcription factors, including ROG (‘repressor of GATA’), FOG (‘friend of GATA’) and LEF1, have been reported to associate with GATA-3 and modulate GATA-3 function38–40. ROG binds to the carboxy-terminal finger of GATA-3 and FOG binds to the amino-terminal finger of GATA-3, whereas both zinc fingers of GATA-3 are required for its association with LEF1. Similar to that40, both zinc fingers of GATA-3 seemed to be involved in its association with Sox4. Both Sox4 and LEF1 belong to the HMG box–containing family of transcription factors, and the HMG box of these molecules is required for the association with GATA-3 (ref. 40). This is consistent with the proposal that heterodimerization occurs via the HMG box of Sox molecules and the DNA-binding domain of partner transcription factors27,41,42. Thus, although detailed structural analysis of the interaction between the Sox HMG box and the GATA-3 zinc fingers is required, Sox proteins may be able to associate with GATA proteins and thus may be important in the regulation of transcription factors of the GATA family.
We found that a Sox-binding motif overlapped one of the GATA-binding motifs in the Il5 promoter. Sox4 itself did not activate the Il5 promoter in M12 mouse B cells, but Sox4 interfered with the binding of GATA-3 to DNA via directly binding to the Sox-binding motif. A similar example of overlapping binding sites for Sox and GATA in the promoter of the gene encoding fibroblast growth factor 3 (Fgf3) has been reported43. In this case, both Sox7 and GATA-4 independently bind to and activate the Fgf3 promoter, which indicates that Sox7 and GATA-4 are competitive activators of Fgf3 transcription. In contrast, Sox4 seemed to compete with GATA-3 to inhibit Il5 transcription.
TCR-induced proliferative responses of naive CD4+ cells are required for TH1 and TH2 differentiation. In addition, it takes 2–3 d to induce GATA-3 expression similar to that in differentiated TH2 cells44. Therefore, we added TGF-β to the TH2-differentiation cultures 48 h after the initial TCR stimulation to address the role of TGF-β in TH2-specific processes more selectively. In our experimental system, the inhibition of GATA-3 expression by TGF-β was marginal, whereas we observed selective inhibition of TH2 differentiation. TGF-β is known to control several processes during the development of TH2 cells, such as the proliferation of naive CD4+ T cells20. TGF-β is also known to strongly inhibit the development of TH2 cells even in the presence of exogenous IL-4 (refs. 23,24). However, the effect of TGF-β on TH1 development is less clear. TGF-β is known to inhibit IL-12-dependent TH1 differentiation 45, whereas the IFN-γ-induced development and/or enhancement of TH1 cells is not perturbed but is instead enhanced in the presence of TGF-β46.
CD4+ T cells cultured under TH2 conditions in the presence of TGF-β ‘preferentially’ differentiate into TH9 cells47,48. We found that the efficiency of the generation of TH9 cells by either Sox4-transgenic or Sox4fl/flCD4-Cre T cells was not altered and the induction of Foxp3 was not impaired in Sox4-deficient CD4+ T cells. Thus, Sox4 may have only a limited role in TH9 and iTreg differentiation. In summary, Sox4 is a downstream target of TGF-β and, through its role as a negative regulator of GATA-3, functions a critical regulator of TH2 differentiation and TH2 cell–dependent immune responses.
ONLINE METHODS
Mice
Mice with transgenic expression of Sox4 under the control of the distal promoter of the mouse Lck gene were generated on a C57BL/6 background in the Department of Immunology of Chiba University. The distal Lck promoter was used to minimize the effect of Sox4 overexpression on T cell development in the thymus, as this promoter is reported to be active from the late CD4+CD8+ double-positive stage to the CD4+ or CD8+ single-positive stage of thymocyte development. Sox4-deficient mice35 established by V.L. were backcrossed to C57BL/6 mice eight times. Mice with transgenic expression of Cre under the control of the Cd4 promoter, on a C57BL/6 background, were from Jackson Laboratory. DO11.10 mice (with transgenic expression of an OVA-specific TCR-αβ) were provided by D. Loh. C57BL/6 and BALB/c mice were from Clea. All mice were maintained under specific pathogen–free conditions and were used at 6–10 weeks of age. All experiments with mice received approval from the Chiba University Administrative Panel for Animal Care. All animal care was in accordance with the guidelines of Chiba University.
CD4+ T cell–differentiation cultures
CD4+ T cells with a naive phenotype (CD44loCD62Lhi) were purified with a FACSAria cell sorter (Becton Dickinson), which yielded a purity of >98%, and were used as naive CD4+ T cells. Naive CD4+ T cells (1.5 × 106) were stimulated for 2 d with immobilized mAb to TCRβ (10 μg/ml or 1 μg/ml; H57-597; BioLegend) plus soluble mAb to CD28 (1 μg/ml; 37.5; BioLegend) in the presence of IL-2 (2.5 ng/ml; Peprotech), IL-12 (3 ng/ml; PeproTech) and mAb to IL-4 (5 μg/ml; 11B11; BioLegend), for TH1 conditions, or in the presence of IL-2 (2.5 ng/ml), IL-4 (10 ng/ml; PeproTech) and mAb to IFN-γ (5 μg/ml; R4-6A2; BioLegend), for TH2 conditions. Cells were cultured for an additional 3 d without stimulation of the TCR in the presence of the original cytokines. Where needed, TGF-β (10 ng/ml; PeproTech) was added to the second culture. Cultured cells were restimulated for 6 h with mAb to TCRβ (10 μg/ml), and intracellular staining was done as described34.
Expression plasmids and retrovirus-mediated gene transfer
Flag-tagged GATA-3 mutants (pFlag-CMX2-GATA-3) and Myc-tagged Sox4 mutants (pCMV Myc-Sox4) were generated by PCR-based mutation. Expression plasmids were transfected into 293T cells with Fugene reagentaccording to the manufacturer’s protocol (Roche). The method for the generation of retrovirus supernatant and infection of developing TH2 cells has been described49. Infected cells were detected by staining with mAb to human nerve growth factor receptor (C40-1457; BD Bioscience).
Knockdown analysis
A Sox4-containing microRNA-adapted retroviral vector (MSCV/LTRmiR30-PIG; vector pLMP; Open Biosystems) was used for Sox4 shRNA. Naive CD4+ T cells were cultured for 2 d under TH2 conditions and then infected with retrovirus vector containing control shRNA (pLMP-hNGFR) or Sox4 shRNA (pLMP-shSox4-hNGFR) and were cultured in the presence of TGF-β (10 ng/ml). Then, 3 d after infection, infected cells (positive for human nerve growth factor receptor (hNGFR)) were purified and subjected to further analysis. The siRNA was introduced into primary CD4+ T cells by electroporation with a Mouse T cell Nucleofector Kit and Nucleofector I (Amaxa). Naive CD4+ T cells were transfected with 675 pmol control (random) siRNA or siRNA specific for Sox4 (Applied Biosystems) and were cultured under the appropriate conditions. At 4 d after transfection, cultured cells were analyzed by quantitative RT-PCR.
Quantitative RT-PCR
Quantitative RT-PCR was done as described34. Primers and TaqMan probes for the detection of mouse Gob5, Mac5a-Mac5c, Sox4, Gata3, Nfat1, Nfat2, Maf, Junb, Il4, Il5, Il13, Ifng and Hprt were from Applied Biosystems.
ChIP assay
ChIP assays were done as described49. Antibody to H3K9ac-H3K14ac (06-595; Upstate Biotechnology), antibody to H3 trimethylated at Lys4 (ab8580; Abcam), mAb to GATA-3 (HG3-31; Santa Cruz Biotechnology), anti-Smad2-Smad3 (ab28379; Abcam), mAb to Smad2-Smad3 (D7G7; Cell Signaling) and anti-Sox4 antiserum (AB5803; Millipore) were used. For the detection of specific genome regions, the Roche Universal Probe Library System was used. The specific primers and TaqMan probes for the detection of the Sox4 and Cd3e were as follows: Sox4 U forward, 5′-CGGGAGACAATGGGTAAG AA-3′, and reverse, 5′-CCAAAGGATAGATGGGTTCG-3′; universal probe 12 (4-685-113; Roche); Sox4 P forward, 5′-TGCACCAAAGGCTGATTC TT-3′, and reverse: 5′-TTCTGCTTAAAAGCCGAGTGA-3′; universal probe 26 (4-687-574; Roche); CD3ε P forward, 5′-ACACTTCCTGTGTGG GGTTC-3′, and reverse, 5′-CTGAAGAAGGCACCAGACG-3′; and universal probe 16 (4-686-896; Roche). Other specific primers and TaqMan probes used have been described34.
Luciferase reporter assay
The luciferase assay for Il5 promoter activity was done in an M12 B cell line as described18 with a firefly luciferase reporter (pGL3; Promega) and a renilla luciferase plasmid (pRL; Promega). Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega).
Precipitation assay
Lysates of transfected 293T cells were incubated with biotinylated oligonucleotides and bound proteins were eluted and separated by SDS-PAGE, then analyzed by immunoblot with specific antibodies. Oligonucleotide probes for the precipitation assay were as follows: GATA-3 consensus, 5′-CACTTGATAACAGAAAGTGATAACT CT-3′; Il5p Mut1, 5′-CCCTCTATCTGATTAATAGCA-3′; and Il5p Mut2. 5′-CCTCTTTCTGATTGTTAGCA-3′.
Immunoprecipitation and immunoblot analysis
Immunoprecipitation and immunoblot were done as described40. The TG40 T cell line50 was used for coimmunoprecipitation. A mAb to GATA-3 (HG3-31; Santa Cruz Biotechnology), anti-Sox4 antiserum (C-20; Santa Cruz Biotechnology), mAb to Flag (M2; Sigma-Aldrich) and mAb to Myc (PL14; MBL, Japan) were used for immunoblot analysis. After immunoprecipitation with mAb to Flag, the immunoprecipitates were eluted with 3X FLAG peptide (F4799; Sigma-Aldrich) and were then separated by electrophoresis.
Glutathione S-transferase precipitation assay
Glutathione S-transferase–tagged recombinant GATA-3 (1.5 μg; Abnova) and Myc-tagged recombinant Sox4 (1.5 μg; Origene) were mixed with glutathione Sepharose 4B (GE Healthcare) in binding buffer (20 mM Tris-HCl (pH 7.4), 250 mM NaCl, 1% Triton X-100, 0.25% sodium deoxycholate) and incubated for 1 h at 4 °C. Coprecipitated Sox4 was separated by electrophoresis and visualized by immunoblot analysis with mAb to Myc. Glutathione S-transferase protein (ab70456; Abcam) served as a control.
OVA-induced allergic airway inflammation and hyper-responsiveness
Mice were immunized intraperitoneally with 250 μg OVA in 2 mg aluminum hydroxide gel (alum) on days 0 and 7 and then challenged with aerosolized OVA in saline (10 mg/ml) on days 14, 16, 21 and 23. At 2 d after the final inhalation of OVA, cells from BAL fluid and lung samples for histological examination were prepared as described49. Airway hyper-responsiveness was assessed by measurement of the change in lung resistance and dynamic compliance in response to increasing doses of inhaled methacholine as described49. For transfer experiments, naive DO11.10 CD4+ T cells were stimulated for 2 d under neutral conditions and then infected with empty retroviral vector or retroviral vector containing Sox4-specific shRNA. Then, 3 d after infection, the infected cells were purified and stimulated for another 5 d with mAb to TCRβ plus mAb to CD28 under TH2 conditions. The infected TH2 cells (1 × 106) were transferred intravenously into BALB/c mice and the recipient mice were challenged by inhalation of OVA twice on days 1 and 3. The infiltration of inflammatory cells into BAL fluid and airway hyper-responsiveness were assessed on day 4.
Statistical analysis
Student’s t-test was used. ANOVA and the Bonferroni test was used for multiple comparisons of different groups.
Supplementary Material
Acknowledgments
We thank D. Loh (Washington University School of Medicine) for DO11.10 mice; R. Kubo, A. Singer and D. Singer for comments and criticisms in the preparation of this manuscript; and K. Sugaya, H. Asou, S. Norikane, M. Kato and T. Ito for technical assistance. Supported by the Global Center for Education and Research in Immune System Regulation and Treatment, City Area Program (Kazusa/Chiba Area) of the Ministry of Education, Culture, Sports, Science and Technology of Japan, Japan Science and Technology Agency Core Research for Evolutional Science and Technology, Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology, the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants-in-Aid for Scientific Research on Priority Areas 22021008 and 22021011; Scientific Research (B) 21390147 and 23390075), the Ministry of Health, Labor and Welfare of Japan, the Uehara Memorial Foundation, the Mochida Foundation, the Naito Foundation and the Takeda Science Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
M.K. and M.Y. designed and did experiments, analyzed data and wrote the manuscript; K.S., S.T., A.O., R.S., S.M., D.T., H.H. and C.I. designed and did experiments and edited the manuscript; V.L. established Sox4 deficient mice; and T.N. conceptualized the research, directed the study and wrote and edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Note: Supplementary information is available in the online version of the paper.
References
- 1.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33–36. doi: 10.1016/j.cell.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Soroosh P, Doherty TA. Th9 and allergic disease. Immunology. 2009;127:450–458. doi: 10.1111/j.1365-2567.2009.03114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witte E, Witte K, Warszawska K, Sabat R, Wolk K. Interleukin-22: A cytokine produced by T, NK and NKT cell subsets, with importance in the innate immune defense and tissue protection. Cytokine Growth Factor Rev. 2010;21:365–379. doi: 10.1016/j.cytogfr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 8.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 9.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov II, et al. The orphan nuclear receptor RORαt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORαα and RORγ. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onodera A, et al. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J Exp Med. 2010;207:2493–2506. doi: 10.1084/jem.20100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei L, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 16.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama T, Yamashita M. Initiation and maintenance of Th2 cell identity. Curr Opin Immunol. 2008;20:265–271. doi: 10.1016/j.coi.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Schwenger GT, et al. GATA-3 has dual regulatory functions in human interleukin-5 transcription. J Biol Chem. 2001;276:48502–48509. doi: 10.1074/jbc.M107836200. [DOI] [PubMed] [Google Scholar]
- 19.Inami M, et al. CD28 costimulation controls histone hyperacetylation of the interleukin 5 gene locus in developing th2 cells. J Biol Chem. 2004;279:23123–23133. doi: 10.1074/jbc.M401248200. [DOI] [PubMed] [Google Scholar]
- 20.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-β regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 21.Gorelik L, Flavell RA. Abrogation of TGFβ signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 22.Nakao A, et al. Blockade of transforming growth factor β/Smad signaling in T cells by overexpression of Smad7 enhances antigen-induced airway inflammation and airway reactivity. J Exp Med. 2000;192:151–158. doi: 10.1084/jem.192.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorelik L, Fields PE, Flavell RA. Cutting edge: TGF-β inhibits Th type 2 development through inhibition of GATA-3 expression. J Immunol. 2000;165:4773–4777. doi: 10.4049/jimmunol.165.9.4773. [DOI] [PubMed] [Google Scholar]
- 24.Heath VL, Murphy EE, Crain C, Tomlinson MG, O’Garra A. TGF-β1 down-regulates Th2 development and results in decreased IL-4-induced STAT6 activation and GATA-3 expression. Eur J Immunol. 2000;30:2639–2649. doi: 10.1002/1521-4141(200009)30:9<2639::AID-IMMU2639>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 25.Ikushima H, Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 26.Smith JM, Koopman PA. The ins and outs of transcriptional control: nucleocytoplasmic shuttling in development and disease. Trends Genet. 2004;20:4–8. doi: 10.1016/j.tig.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 29.Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- 30.Schilham MW, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 31.Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. Eur J Immunol. 1997;27:1292–1295. doi: 10.1002/eji.1830270534. [DOI] [PubMed] [Google Scholar]
- 32.Li MO, Flavell RA. TGF-β: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HJ, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita M, et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Penzo-Méndez A, Dy P, Pallavi B, Lefebvre V. Generation of mice harboring a Sox4 conditional null allele. Genesis. 2007;45:776–780. doi: 10.1002/dvg.20358. [DOI] [PubMed] [Google Scholar]
- 36.Ruebel KH, et al. Effects of TGFβ1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine. 2008;33:62–76. doi: 10.1007/s12020-008-9060-3. [DOI] [PubMed] [Google Scholar]
- 37.Ikushima H, et al. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 38.Miaw SC, Choi A, Yu E, Kishikawa H, Ho IC. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity. 2000;12:323–333. doi: 10.1016/s1074-7613(00)80185-5. [DOI] [PubMed] [Google Scholar]
- 39.Fox AH, Kowalski K, King GF, Mackay JP, Crossley M. Key residues characteristic of GATA N-fingers are recognized by FOG. J Biol Chem. 1998;273:33595–33603. doi: 10.1074/jbc.273.50.33595. [DOI] [PubMed] [Google Scholar]
- 40.Hossain MB, et al. Lymphoid enhancer factor interacts with GATA-3 and controls its function in T helper type 2 cells. Immunology. 2008;125:377–386. doi: 10.1111/j.1365-2567.2008.02854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reményi A, et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wissmüller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami A, Shen H, Ishida S, Dickson C. SOX7 and GATA-4 are competitive activators of Fgf-3 transcription. J Biol Chem. 2004;279:28564–28573. doi: 10.1074/jbc.M313814200. [DOI] [PubMed] [Google Scholar]
- 44.Kimura M, et al. Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity. 2001;15:275–287. doi: 10.1016/s1074-7613(01)00182-0. [DOI] [PubMed] [Google Scholar]
- 45.Gorelik L, Constant S, Flavell RA. Mechanism of transforming growth factor β-induced inhibition of T helper type 1 differentiation. J Exp Med. 2002;195:1499–1505. doi: 10.1084/jem.20012076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smeltz RB, Chen J, Shevach EM. Transforming growth factor-β1 enhances the interferon-γ-dependent, interleukin-12-independent pathway of T helper 1 cell differentiation. Immunology. 2005;114:484–492. doi: 10.1111/j.1365-2567.2005.02115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dardalhon V, et al. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+IL-10+Foxp3− effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veldhoen M, et al. Transforming growth factor-β ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9–producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita M, et al. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med. 2008;205:1109–1120. doi: 10.1084/jem.20072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sussman JJ, Saito T, Shevach EM, Germain RN, Ashwell JD. Thy-1- and Ly-6-mediated lymphokine production and growth inhibition of a T cell hybridoma require co-expression of the T cell antigen receptor complex. J Immunol. 1988;140:2520–2526. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.