Abstract
The enoylreductase (ER) is the final common enzyme from modular polyketide synthases (PKSs) to be structurally characterized. The 3.0 Å resolution structure of the didomain comprised of the ketoreductase (KR) and ER from the second module of the spinosyn PKS reveals that ER shares an ~600 Å2 interface with KR distinct from that of the related mammalian fatty acid synthase (FAS). In contrast to the ER domains of the mammalian FAS, the ER domains of the second module of the spinosyn PKS do not make contact across the twofold axis of the synthase. This monomeric organization may have been necessary in the evolution of multimodular PKSs to enable acyl carrier proteins (ACPs) to access each of their cognate enzymes. The isolated ER domain showed activity towards a substrate analog, enabling the contributions of its active site residues to be determined.
Modular polyketide synthases (PKSs) are megasynthases that synthesize biologically-active polyketides such as the antibacterial erythromycin, the antifungal amphotericin, and the insecticide spinosyn1,2. Each module along the PKS assembly line harbors a set of enzymes responsible for extending the polyketide chain as well as setting the chemistries and configurations of the newly added α- and β-carbons. While studying enzymes that naturally operate within a megasynthase is inherently challenging, these component enzymes must be more thoroughly characterized to decipher the mechanisms by which polyketides are synthesized. Our efforts have focused on the processing enzymes – a ketoreductase (KR) that stereoselectively reduces the β-keto group, a dehydratase (DH) that performs an elimination reaction to generate a double bond, and an enoylreductase (ER) that stereoselectively reduces that double bond3–7.
Relatively little structural and functional information exists for ER, even though the atomic-resolution structures of each of the other common PKS domains have been determined and in vitro studies have been conducted with those isolated domains4,7–10. The crystal structure of the nucleotide-binding subdomain of the ER from PpsC of the phthiocerol biosynthetic pathway reveals little about the active site or the overall architecture of ER11. Experiments to identify the key ER residues through mutagenesis of engineered triketide lactone synthases have been performed, and though residues mediating stereoselectivity were found, a point mutation that abolished activity was not identified12,13.
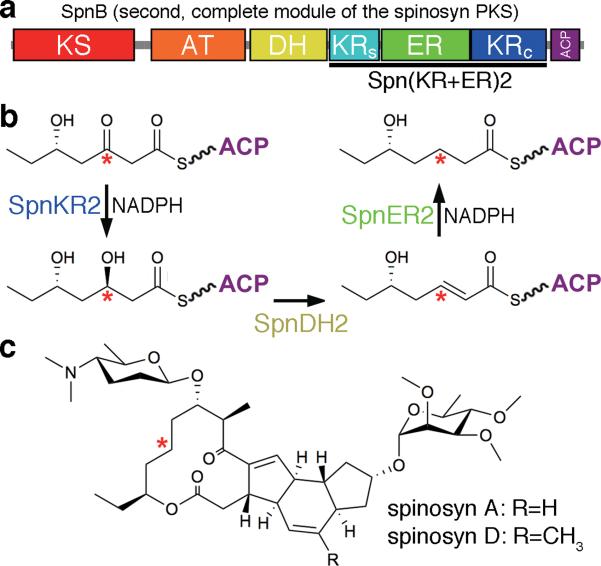
The processing enzymes from the second module of the spinosyn PKS - SpnKR2, SpnDH2, and SpnER2 (named by their PKS and module of origin) - sequentially process a triketide intermediate during the synthesis of the insecticide spinosad (Fig. 1)14. While the SpnER2 domain was not amenable to crystallization, we hypothesized that the Spn(KR+ER)2 didomain might be since the ER domain inserts into a loop connecting the KR subdomains and shares an interface with KR within the mammalian fatty acid synthase (FAS)15. Indeed, crystals were obtained that diffracted to 3.0 Å. The structure shows that SpnKR2 and SpnER2 both fold similar to their mammalian FAS counterparts15–17. Surprisingly, however, the domains form a completely different interface. The conformation adopted by helix αF from SpnER2 precludes the dimerization mode observed for the ER of the mammalian FAS. The three-dimensional organization of the KR and ER domains, confirmed by small-angle x-ray scattering (SAXS), reveals that the complete modules of multimodular PKSs possess a different architecture than the mammalian FAS. The first in vitro biochemical studies of an ER domain isolated from its PKS show that SpnER2 retains activity on the substrate analog crotonyl-pantetheine and that a lysine contributes most to catalysis.
Figure 1. A complete module from the spinosyn PKS.
(a) Domain composition of the second module of the spinosyn PKS (SpnMod2) and location of the Spn(KR+ER)2 didomain (approximate sequence scale). KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KRs, ketoreductase structural subdomain; ER, enoylreductase; KRc, ketoreductase catalytic subdomain; ACP, acyl carrier protein. (b) The indicated methylene group in the spinosyn backbone is generated by the processing enzymes in SpnMod2. (c) The complex polyketides spinosyn A and spinosyn D comprise the insecticide spinosad.
RESULTS
The overall architecture of Spn(KR+ER)2
The structure of the Spn(KR+ER)2 didomain (SpnB residues 1216–1989, Fig. 1a) was solved to 3.0 Å resolution through selenomethionine incorporation and multi-wavelength anomalous dispersion (MAD) phasing (Fig. 2, Supplementary Results, Supplementary Table 1). The two Spn(KR+ER)2 didomains in the asymmetric unit adopt the same conformation with 0.6 Å Cα r.m.s.d. (Supplementary Fig. 1a). Although the SpnKR2 domains have similar average B-factors (49 Å2 vs. 56 Å2), the average B-factor of one SpnER2 domain is highly elevated compared to the other (96 Å2 vs. 62 Å2, Supplementary Fig. 1b). The 60 C-terminal residues of each Spn(KR+ER)2 molecule in the asymmetric unit are domain-swapped, resulting in the close association of SpnKR2 catalytic subdomains and the occlusion of the SpnKR2 NADPH-binding sites (Supplementary Fig. 2). Nonphysiological domain-swapping is sometimes observed in crystals of truncated proteins18,19. Aside from the four residues spanning the domains, the swapped C-terminal residues are folded as expected from related KR structures and possess B-factors equivalent to the surrounding protein3,5,6,7.
Figure 2. KR+ER architecture.
(a) The crystal structure of Spn(KR+ER)2 shows the interface between the structural and catalytic subdomains of its KR (KRs and KRc) and ER, which is mostly formed by hydrophilic residues. The N- and C-terminal linkers help mediate the interface, Trp506 inserting into a hydrophobic pocket of KRc. (b) The structure of the porcine FAS shows a very different interface between its KR and ER and how ER dimerizes across the twofold axis of the FAS. The C-terminal linker is longer than that of Spn(KR+ER)2 (29 vs. 8 residues). (c) SAXS data were obtained for Spn(KR+ER)2 and compared to the theoretical curves predicted by the program CRYSOL for a model generated from the Spn(KR+ER)2 crystal structure (red curve), and for a model generated by superposing SpnKR2 and SpnER2 on the KR and ER from the porcine FAS (blue curve). (d) An ab initio molecular envelope was generated from the SAXS curve (by the program DAMMIF), which was much better fit by the model generated from the Spn(KR+ER)2 crystal structure (using the program SUPCOMB).
The interface between SpnER2 and SpnKR2 is very different from the KR/ER interface of the mammalian FAS and ~50% larger (~600 Å2) (Figs. 2a and 2b, Supplementary Figs. 3 and 4)14. Contacts between the catalytic subdomain of SpnKR2 and the substrate-binding subdomain of SpnER2 form the majority of the interface, which is primarily comprised of unconserved, hydrophilic residues (Supplementary Figs. 3–5). SpnER2 inserts into a loop between the catalytic and structural subdomains of SpnKR2; the N- and C-terminal ends of the insertion loop, referred to here as the N- and C-terminal linkers, help mediate the interface between SpnKR2 and SpnER2. The N-terminal linker (residues 176–189, numbering based on first residue visible in the electron density maps) spans from the C-terminal end of the SpnKR2 structural subdomain to β1 of the SpnER2 substrate-binding domain via unconserved residues; the C-terminal linker (residues 503–510) connects β9 of the SpnER2 substrate-binding domain to the N-terminal end of the SpnKR2 catalytic subdomain via semi-conserved residues. While the N-terminal linkers are comparable in length to the mammalian FAS (~13 residues), the C-terminal linker is much shorter than its mammalian FAS counterpart (8 vs. ~29 residues) and is constrained by a conserved hydrophobic interaction with the KR catalytic subdomain (mediated by Trp506 in Spn(KR+ER)2) (Fig. 2a and 2b, Supplementary Figs. 4–6)15. Within the mammalian FAS, the separation of the C-terminal end of β9 of ER (residue 1855) and the N-terminal end of the KR catalytic subdomain (residue 1886) is 36 Å. The short C-terminal linker of multimodular PKSs cannot span this distance, preventing KR and ER from forming the orientation observed in the mammalian FAS.
To investigate whether the conformation of the Spn(KR+ER)2 didomain in solution is similar to that observed in the crystal structure (corrected for domain-swapping), small-angle X-ray scattering (SAXS) was performed (Fig. 2c and Supplementary Fig. 7)20. The experimental scattering curve closely matched the scattering curve predicted by the program CRYSOL for the Spn(KR+ER)2 monomer from the crystal structure21. The scattering curve predicted from a model generated through the superposition of SpnKR2 and SpnER2 on the KR and ER domains of the porcine FAS did not match the experimentally-obtained scattering curve. The radius of gyration (Rg) determined from the SAXS data through the Guinier approximation (29.5 Å) matches that calculated from the Spn(KR+ER)2 crystal structure (30.2 Å) but not that calculated from the model generated through the superposition of SpnKR2 and SpnER2 on the KR and ER domains of the porcine FAS (36.7 Å). The ab initio molecular envelope generated from the SAXS data by the program DAMMIF indicates a compact two-domain structure consistent with the Spn(KR+ER)2 crystal structure but not with the model generated using the orientations of KR and ER in the porcine FAS (Fig. 2d and Supplementary Fig. 7)22,23.
Oligomerization state
The architecture of PKS modules as determined by limited-proteolysis/size-exclusion experiments and structural biology has generally correlated well with the architecture of the mammalian FAS: KS and DH are dimeric, and AT, KR, and ACP are monomeric4,7,9,10,15,24. However, studies of the oligomerization state of the ERs of modular PKSs have been seemingly contradictory: while the crystals structures of the mammalian FAS and the nucleotide-binding subdomain of the ER from PpsC show these ERs making dimeric contacts via the interface anticipated for medium-chain dehydrogenase/reductase (MDR) enzymes, limited-proteolysis of DEBS2 (the second polypeptide of the erythromycin PKS) produced a monomeric EryER424. A size-exclusion study of Ery(DH+KR+ER)4 suggests this tridomain is dimeric; however, this result may not address the oligomerization state of EryER4 since it is possible that EryDH4 alone mediates dimerization4,7.
Spn(KR+ER)2, SpnER2, and SpnKR2 were analyzed by size-exclusion chromatography, and each migrates as a monomer (~90, ~40, ~60 kDa, respectively; 83, 36, 50 kDa predicted) (Supplementary Fig. 8). The oligomerization states of these proteins were also examined through sedimentation velocity experiments (Supplementary Fig. 9)25. For each Spn(KR+ER)2, SpnER2, and SpnKR2, the van Holde-Weischet integral distribution plots show essentially identical sedimentation coefficient distributions for three different concentrations examined, indicating the absence of any mass-action-based oligomerization effects. A global genetic algorithm/Monte Carlo (GA/MC) analysis was performed to confirm the composition of and identify the molecular weights and anisotropies for each protein. The molecular weights determined from the GA/MC analysis are in excellent agreement with the known monomeric molecular weights of Spn(KR+ER)2, SpnER2, and SpnKR2. The molecular weight of Spn(KR+ER)2 was also estimated to be 81 kDa from the SAXS data26, providing further confirmation that Spn(KR+ER)2 is a monomer in solution.
SpnKR2 structure
SpnKR2 consists of a structural and a catalytic subdomain (residues 1–175 and 511–753, respectively; both subdomains possess a Rossmann-like fold, and the KR domain belongs to the “complex” subfamily of the short-chain dehydrogenase/reductase (SDR) superfamily), aligning well with TylKR1, AmpKR2, and EryKR1 (1.8, 1.8, and 1.9 Å Cα r.m.s.d., respectively) (Supplementary Fig. 10a)3,5,7,16. Compared to these isolated KRs, the structural subdomain of SpnKR2 is missing the first helix and is ~30 residues shorter7. Within the catalytic subdomain, the active site residues Lys621, Ser643, Tyr656, and Asn660 are in the same orientations observed in other reductase-competent KRs (Supplementary Fig. 10b)3,5,6,7. NADP+ is bound in the dinucleotide-binding site of the catalytic subdomain as observed in the ternary complex of AmpKR26. Adjacent to the active site, SpnKR2 possesses a Leu-Asp-Asp motif, employed by B-type KRs to help stereoselectively generate D-hydroxyl groups, as well as a lid helix that is one turn longer than the lid helices of KRs from modules that do not contain other β-carbon processing enzymes (Supplementary Fig. 10c)3. In contrast to their equivalents in most other KR structures, both of these elements are well-ordered and make contacts to active site residues as well as one another, as illustrated by Arg701 of the lid helix binding the second aspartate of the Leu-Asp-Asp motif through a salt-bridge (the interactions between these elements may have been promoted in the crystal structure by the lid helix being the first element of the C-terminal domain-swap) (Supplementary Fig. 2c).
SpnER2 structure
The ERs of modular PKSs and FASs belong to the acyl-CoA reductase family within the MDR superfamily; thus, SpnER2 possesses two characteristic MDR subdomains (Fig. 3a)17. Residues 190–307 and 448–501 comprise the substrate-binding subdomain, while residues 308–447 constitute the nucleotide-binding subdomain that binds NADPH through a Rossmann fold15. The ERs of modular PKSs are structurally distinct from their functional analogs in bacterial fatty acid biosynthesis (FabI, FabL, and FabV), which are SDRs27,28. SpnER2 is most structurally similar to quinone oxidoreductases (DALI server; PDB Codes: 2J8Z, 3JYN, 1QOR; 2.1 Å, 2.3 Å, 2.4 Å Cα r.m.s.d., respectively)29 SpnER2 is also highly structurally related to the ER of the mammalian FAS (PDB Code: 2VZ8; 2.1 Å Cα r.m.s.d.), a trans- acting enoylreductase in the lovastatin biosynthetic pathway (PDB Code: 3B70; 2.6 Å Cα r.m.s.d.), and 2-enoyl thioester reductases from mitochondrial fatty acid synthesis pathways (PDB Codes: 2VCY, 1N9G, 1GUF; 3.1 Å, 3.7 Å, 3.7 Å Cα r.m.s.d., respectively)15,30,31,32 The boundaries of the SpnER2 domain are defined by two β-strands (β1 and β9) in the substrate-binding subdomain. The substrate-binding and nucleotide-binding subdomains are oriented relative to one another largely by αA, which spans both subdomains and is bent through the insertion of a side-chain carboxylate (from Glu250) at its third turn. While the nucleotide-binding subdomain of SpnER2 adopts a classical Rossmann fold, the nine β-strands of the substrate-binding subdomain form two β-sheets (β1 and β2 create a small, antiparallel β-sheet and β3–β9 build a highly-twisted, mixed β-sheet) surrounded by four α-helices (α1, αA, α2, α3).
Figure 3. The structure of SpnER2.
(a) SpnER2 is an MDR enzyme comprised of a substrate-binding domain (residues 190–307 and 448–501) and a nucleotide-binding subdomain (residues 308–447). (b) A superposition of SpnER2 (green) and ER from the porcine FAS (cyan) shows the different organization of the region surrounding βF, which usually forms the dimerization interface in MDR enzymes. (c) Conserved SpnER2 active site residues form specific interactions with a bound NADP+ molecule. Next to where the 4-pro-R hydride of NADPH would be positioned during the reduction reaction (indicated by *) are the lysine and aspartate implicated in catalysis as well as a tyrosine known to mediate stereoselectivity.
MDRs usually dimerize via βF and secondary structural elements that surround it17. The surrounding elements in the ER from the mammalian FAS are αF, βG, and αG14. SpnER2 possesses αF but not βG or αG. Quinone oxidoreductases share a similar one-helix architecture to SpnER2; however, interactions between Asp405 and Arg426 as well as Leu408 and His435 position helix αF of SpnER2 in an orientation that is seemingly incompatible with dimerization (Figs. 3b, Supplementary Figs. 5, 6, and 11a)33. The highly-related trans- acting lovastatin enoylreductase (PDB Code: 3B70) is the only other MDR enzyme to have crystallized as a monomer.
The NADPH binding site is located in a deep cleft between the subdomains of SpnER2 (Figs. 3c, Supplementary Fig. 11b and 11c). The pyrophosphate moiety of the nicotinamide coenzyme is bound to the highly-conserved pyrophosphate-binding motif, GlyGlyValGlyMetAla (residues 335–340) (Supplementary Fig. 5)15. The conserved residues Lys360 and Arg375 form salt bridges with the adenine ribose phosphate. Elements from the substrate-binding subdomain, such as Phe231 from helix α1 and His494 from the loop following helix α3, also interact with the bound nicotinamide coenzyme. Thus, the 4-pro-R hydride of NADPH is positioned to add to the β-carbon of a trans- α,β-unsaturated polyketide substrate, in contrast to KRs, which catalyze the addition of the 4-pro-S hydride6.
SpnER2 function
The structures of both the ER from the porcine FAS and SpnER2 reveal an invariant lysine/aspartate pair (Lys422 and Asp444 in SpnER2) ~6 Å from the nicotinamide hydride15. They may directly participate in substrate protonation, stabilize reaction intermediates, or aid in binding the substrate (Fig. 3c). Structural and functional analysis of many MDRs suggest that a tyrosine following α1 is the proton source during the reduction reaction; however, only half of the ERs from modular PKSs possess the corresponding tyrosine (Supplementary Fig. 5). When present, it is known to play the major role in setting an S configuration at the α-carbons of α-substituted polyketide intermediates12.
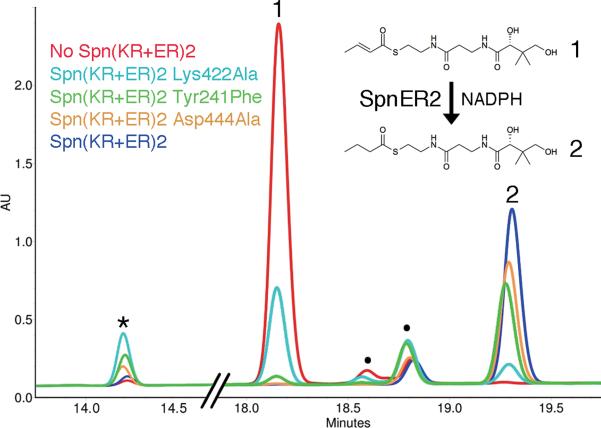
The activities and stereocontrol of isolated KRs have been extensively analyzed in vitro34–37, while all functional assays of ER domains have been limited to in vivo assays within the context of complete modules12,13. Thus, Spn(KR+ER)2 was assayed for in vitro activity towards the trans- α,β-unsaturated crotonyl-pantetheine, which mimics its natural substrate, trans-(5S)-hydroxyhept-2-enoyl-SpnACP2 thioester (Figs. 1b and 4). Indeed, Spn(KR+ER)2 generates the expected product, butyryl-pantetheine (verified by mass spectrometry; Supplementary Fig. 12). A recent study attempted to determine the catalytic residues of the ERs of modular PKSs employing an in vivo assay; however, significant activity was preserved in each of the generated point mutants13. We sought to identify the catalytic residues of ER through an in vitro assay with crotonyl-pantetheine. The structure of the mammalian FAS, the structure of Spn(KR+ER)2, and previous studies guided the selection of residues to be examined12,13,15. Tyr241Phe, Lys422Ala, and Asp444Ala point mutants were expressed and purified using the same protocol as described for wild-type Spn(KR+ER)2. Along with the anticipated butyryl-pantetheine product, a side-product possessing the molecular weight of β-hydroxybutyryl-pantetheine was generated (Fig. 4). That the Lys422Ala mutant produced the least butyryl-pantetheine suggests that the lysine is most critical for the reductase activity of SpnER2.
Figure 4. Functional analysis of SpnER2.
SpnER2 reduces the α,β-unsaturated substrate analog crotonyl-pantetheine. To determine the relative contributions of SpnER2 active site residues to catalysis, three Spn(KR+ER)2 mutants were generated. Unmutated Spn(KR+ER)2 and each of the point mutants were incubated with crotonyl-pantetheine for 2 days prior to analysis by reversed-phase HPLC. The negative control (red) does not contain Spn(KR+ER)2. Unmutated Spn(KR+ER)2 (dark blue) completely converts crotonyl-pantetheine to butyryl-pantetheine. Asp444Ala (yellow) and Tyr241Phe (green) mutants are still active, while the Lys422Ala mutant (cyan) was much less active, revealing the importance of the lysine in catalysis. Each reaction contains trace impurities (●). Reactions with ER mutants generate larger quantities of a byproduct (*) with a mass equivalent to that of β-hydroxybutryl-pantetheine. See also Supplementary Fig. 12 and Supplementary Table 3.
We sought to determine the steady-state kinetic parameters of the reduction reaction catalyzed by SpnER2 and its point mutants on the substrate crotonyl-pantetheine (Supplementary Table 2). Since we observed that standalone SpnKR2 could catalyze the hydration of crotonyl-pantetheine, the Tyr241Phe, Lys422Ala, and Asp444Ala mutations were introduced into standalone SpnER2. The catalytic efficiency of reduction reaction of wild-type SpnER2 (kcat(R)/Km = 0.11 ± 0.02 M−1s−1; kcat(R) = 0.0085 ± 0.0007 s−1 and Km = 74 ± 14 mM) is similar to the catalytic efficiencies of isolated KR domains for diketide-N-acetylcysteamine thioesters (e.g. EryKR1: kcat/Km = 7 M−1s−1; kcat = 0.26 ± 0.01 s−1 and Km=35 ± 4 mM)35 but much lower than the ER domain of the intact Rattus norvegicus FAS for crotonyl-CoA (kcat/Km=1.6 × 106 M−1s−1; kcat = 9.8 ± 0.5 s−1 and Km = 6.5 ± 0.6 μM)38. The Asp444Ala mutant was slightly more catalytically efficient than wild-type SpnER2 (kcat(R)/Km = 0.30 ± 0.04 M−1s−1 The Tyr241Phe mutant was less efficient (kcat(R)/Km = 0.08 ± 0.01 M−1s−1), while the Lys422Ala mutant was the least catalytically efficient of the mutants (kcat(R)/Km = 0.018 ± 0.004 M−1s−1). Each of the SpnER2 point mutants was more active at catalyzing the hydration side-reaction than wild-type SpnER2 (Supplementary Table 3).
DISCUSSION
The order of PKS enzymes within a module and the reactions they catalyze closely parallel those of the mammalian FAS1,2. Nonetheless, the combinations of β-carbon processing enzymes present within PKS modules enable a much wider range of products to be generated. Determining the architectures and mechanisms of these enzymes are necessary steps towards elucidating how these complicated synthetic machines generate chemically diverse polyketides. This structural and functional report of a KR+ER didomain from a complete PKS module represents a large step towards this goal.
While no mechanism for the ERs from mammalian FASs or modular PKSs has been generally accepted, it is thought that NADPH transfers a hydride to the β-carbon of the substrate and that a proton is donated by either a general acid or a solvent molecule to the α-carbon (Figs. 3c and 4)2,15. That Lys422 was found to be the most crucial for catalysis makes sense as it is structurally conserved in the ERs of both mammalian FASs and modular PKSs, while the tyrosine is absent in the ERs from mammalian FASs (and half of the ERs from modular PKSs) and the aspartate resides in a different location within the ER of the mammalian FAS (Supplementary Figs. 5 and 6)15.
The mechanisms by which some ERs from modular PKSs set stereochemistry are made less mysterious through the structure of SpnER2. The tyrosine that plays the principal role in setting an S configuration at a substituted α-carbon (equivalent to Tyr241 in SpnER2) lies on the opposite side of the active site cleft from the lysine15. The lysine and tyrosine are within 5 Å of one another, and both residues are in appropriate positions to protonate the α-carbon of a bound polyketide substrate during the reduction reaction (Fig. 3c). Mutation of this tyrosine has been observed to result in an R configuration being set at the α-carbon of an α-substituted substrate, suggesting that in the absence of the tyrosine, the lysine is able to donate a proton from the opposite side of the polyketide substrate12. From the mutagenesis results presented here, the lysine serves a key role even when the tyrosine is present, possibly lowering the pKa of the tyrosine hydroxyl group so that it may readily donate a proton to the substrate. We have also observed the reduction of 2-methylbut-2-enoyl-pantetheine by SpnER2; how stereocontrol is enforced by the ERs of modular PKSs on this substrate is under investigation.
The structures of the monomeric TE from the mammalian FAS and the dimeric TE from multimodular PKSs revealed that structural differences exist between these related megasynthases8,39. The monomeric state of SpnER2 also speaks to the divergent evolution of multimodular PKSs and mammalian FASs. A dimeric ER2 was likely possessed by their common ancestor; however, when some PKSs evolved so that a single polypeptide contained multiple modules, disruption of the ER dimer interface may have been necessary to enable the ACPs, restricted by a dimeric element immediately downstream, to access each of their cognate enzymes. The DH dimer interface within a multimodular PKS is quite different from the DH dimer interface of the mammalian FAS; as with an evolutionary transition to a monomeric ER, a more flattened DH dimer in multimodular PKSs may have facilitated the access of ACPs to their cognate enzymes (especially KS from the same module)4,40. No structural information is available for how a DH domain in a multimodular PKS interfaces with KR and ER; the presence of DH could result in KR and ER adopting an interface different from that observed in the Spn(KR+ER)2 crystal structure. The homodimeric oligomerization state of the nucleotide-binding subdomain of the ER from phthiocerol synthase PpsC seems to argue against an evolutionary transition to monomeric ERs within multimodular PKSs (PDB Code: 1PQW; the interface is equivalent to that of the ER in the mammalian FAS)11. However, PpsC is more related to mammalian FASs on a sequence level than modules from multimodular PKSs -the C-terminal linker as well as the sequence surrounding βF are very similar to that of the mammalian FAS (Fig. 3b, Supplementary Figs. 4–6)15. Perhaps the architecture of some monomodular PKSs such as PpsC resembles the mammalian FASs more than the modules of multimodular PKSs.
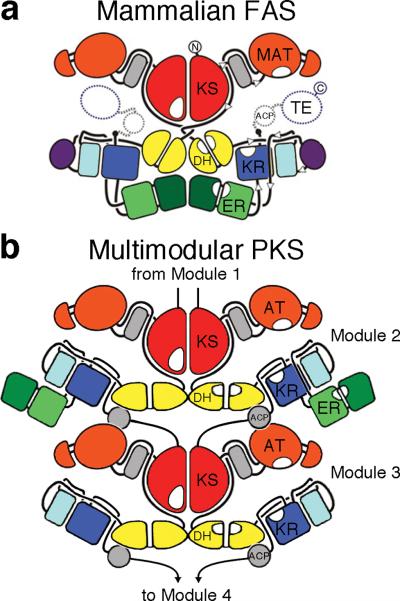
Models of complete PKS modules have been constructed based on the structure of the mammalian FAS1,15. However, the KR/ER interface observed in Spn(KR+ER)2, the short C-terminal linker between ER and KR, the altered tertiary structure of the MDR dimerization region, and the flattened architecture of the DH dimer of multimodular PKSs indicate that the three-dimensional organization of the processing enzymes of multimodular PKSs is quite different from that of the mammalian FAS (Fig. 2, Supplementary Figs. 4–8)4,15. Indeed, the twofold axis of a multimodular PKS does not contain enough space for ER domains to form dimeric contact since the ACPs, geometrically restricted by ~15-residue linkers to KR and a downstream dimeric element, require this space to access KS from the same module (the mammalian FAS and some monomodular PKSs do not encounter this geometric problem since their ACPs are not C-terminally linked to a dimeric domain). Thus, we propose that within multimodular PKSs the “reductive loop” is comprised of a dimeric DH, two monomeric KR, and two monomeric ER, enabling the ACPs to utilize the space occupied in the mammalian FAS by the dimeric ER and preserving the twofold symmetry of the synthase (Fig. 5). While the crystal structure of a complete PKS module remains elusive, the structural and functional dissection of its component enzymes provides insights into the architectures and mechanisms of these complex megasynthases.
Figure 5. Synthase schematic.
The structure of the mammalian FAS revealed that KS, DH, and ER comprise the dimer interface. The data presented here indicate that the ERs of multimodular PKSs are monomeric and loop out from the twofold axis. ACPs from a complete module of a multimodular PKS access all of the enzymes within that module as well as a downstream dimeric KS or dimeric TE. Already restrained by the short peptide that joins them to the downstream KS or TE, ACPs would be prevented from accessing their cognate enzymes by a dimeric ER. In the illustrated mycolactone multimodular PKS, the ACPs of the complete second module can access each of their cognate enzymes since ER is looped out. Artwork was reproduced with permission from N. Ban15.
METHODS
Cloning, expression, and purification
The DNA encoding Spn(KR+ER)2 and SpnER2 was amplified from Saccharopolyspora spinosa genomic DNA and inserted into pET28b. Reverse PCR was used to remove the ER domain from the Spn(KR+ER)2 to generate DNA fragment encoding SpnKR2. All proteins were expressed in Escherichia coli BL21(DE3) and purified by affinity chromatography and gel filtration chromatography. Selenomethionine-labeled protein was obtained by the pathway inhibition method. Further experimental details are in Supplementary Methods.
Site-directed mutagenesis
The GeneTailor Site-Directed Mutagenesis System (Invitrogen) was used to generate all the mutants (verified by sequencing) as described in Supplementary Methods. The mutants were purified by the same protocol used for the native protein.
Protein size determination
The molecular weight of each protein was first estimated by size exclusion chromatography and then confirmed by analytical ultracentrifugation as described in Supplementary Methods.
Crystallization and structure determination
Crystals of Spn(KR+ER)2 were grown by sitting drop vapor diffusion. Three data sets (peak, inflection, and remote) from a crystal of selenomethionine-labeled Spn(KR+ER)2 were collected at Advanced Light Source (ALS) Beamline 5.0.2 and processed by HKL200041. The structure was solved to 3.40 Å resolution by multiwavelength anomalous dispersion phasing using the program Phenix42. A 3.0 Å-resolution dataset collected at ALS Beamline 8.2.1 from a crystal of native Spn(KR+ER)2 was used to iteratively build and refine the model using the programs Coot and Refmac543,44. Further details are provided in Supplementary Methods.
SAXS data collection and analysis
SAXS data were collected on a Rigaku BioSAXS-1000 (Kratky camera, with a 2.5kW FRE+ source). The radius of gyration (Rg) and I(0) were evaluated using the Guinier approximation (0.0223 Å−1 < q < 0.0437 Å−1). The program GNOM was used to obtain the distance distribution function, P(r), and to determine the value for the Rg from the entire scattering profile22. A molecular weight was estimated from this data26. The program CRYSOL predicted the scattering curves from two models of Spn(KR+ER)2 and compared them to the experimentally-obtained SAXS curve21. Ab initio envelopes were generated by the program DAMMIF23, filtered with the program DAMAVER45, and aligned to each model using the program SUPCOMB46. Further data collection and processing details are in Supplementary Methods.
Functional assays
Spn(KR+ER)2 and its point mutants were incubated with substrate mimic crotonyl-pantetheine and the resulting reactions were analyzed by reversed-phase HPLC and LC/MS. The details of substrate synthesis, reaction conditions, and product analysis can be found in Supplementary Methods.
Kinetic analysis
A kinetic model derived from standard Michaelis–Menten kinetics was employed to analyze the reduction and hydration reactions catalyzed by standalone SpnER2 and its point mutants. NADPH-linked assays were used to determine the kinetic parameters of enoylreduction. The consumption of NADPH was monitored, and initial velocities were plotted against substrate concentration and fit to the kinetic model to obtain Km and kcat(R). To compare the hydration/reduction activities of SpnER and its point mutants, the ratio of hydrated to reduced product generated by each enzyme was estimated by LC/MS. Further details are reported in Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Arthur F. Monzingo for helping with in-house diffraction experiments and Christopher D. Fage for helping optimize crystals. Synchrotron data were obtained at the Advanced Light Source Beamlines 8.2.1 and 5.0.2. Financial support was provided by Welch Foundation Grant F-1712 (A.T.K.) as well as the Sealy and Smith Foundation (to M.A.W. and to the Sealy Center for Structural and Molecular Biology at the University of Texas Medical Branch). The development of the UltraScan software is supported by the National Institutes of Health through grant RR022200 (B.D.). Supercomputer time allocations were provided through National Science Foundation grant TG-MCB070038 (B.D.). We acknowledge the support of the San Antonio Cancer Institute grant P30 CA054174 for the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio.
Footnotes
J.Z. performed all of the studies except for the functional assays (conducted by D.G.), analytical ultracentrifugation (conducted by B.D.), and SAXS (conducted by M.A.W.). All authors contributed to writing the manuscript and creating the figures.
Competing financial interests The authors declare no competing financial interests.
Accession codes Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 3SLK.
References
- 1.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 2.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat. Prod. Rep. 2007;24:1041–72. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Keatinge-Clay A. Crystal structure of the erythromycin polyketide synthase dehydratase. J. Mol. Biol. 2008;384:941–53. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J, Keatinge-Clay AT. Structural and functional analysis of C2-type ketoreductases from modular polyketide synthases. J. Mol. Biol. 2011;410:105–17. doi: 10.1016/j.jmb.2011.04.065. [DOI] [PubMed] [Google Scholar]
- 6.Zheng J, Taylor CA, Piasecki SK, Keatinge-Clay AT. Structural and functional analysis of A-type ketoreductases from the amphotericin modular polyketide synthase. Structure. 2010;18:913–922. doi: 10.1016/j.str.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–48. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Tsai SC, et al. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc. Natl. Acad. Sci. USA. 2001;98:14808–13. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Kim CY, Mathews, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. USA. 2006;103:11124–9. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alekseyev VY, Liu CW, Cane DE, Puglisi JD, Khosla C. Solution structure and proposed domain domain recognition interface of an acyl carrier protein domain from a modular polyketide synthase. Protein Sci. 2007;16:2093–107. doi: 10.1110/ps.073011407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedelacq JD, et al. Experimental mapping of soluble protein domains using a hierarchical approach. Nucleic Acids Res. 2011;39:e125. doi: 10.1093/nar/gkr548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwan DH, et al. Prediction and manipulation of the stereochemistry of enoylreduction in modular polyketide synthases. Chem. Biol. 2008;15:1231–40. doi: 10.1016/j.chembiol.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Kwan DH, Leadlay PF. Mutagenesis of a modular polyketide synthase enoylreductase domain reveals insights into catalysis and stereospecificity. ACS Chem Biol. 2010;5:829–38. doi: 10.1021/cb100175a. [DOI] [PubMed] [Google Scholar]
- 14.Kirst HA. The spinosyn family of insecticides: realizing the potential of natural products research. J. Antibiot. (Tokyo) 2010;63:101–11. doi: 10.1038/ja.2010.5. [DOI] [PubMed] [Google Scholar]
- 15.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–22. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 16.Oppermann U, et al. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact. 2003;143–144:247–53. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 17.Persson B, Hedlund J, Jornvall H. Medium- and short-chain dehydrogenase/reductase gene and protein families : the MDR superfamily. Cell. Mol. Life Sci. 2008;65:3879–94. doi: 10.1007/s00018-008-8587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–99. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishan KV, Scita G, Wong WT, Di Fiore PP, Newcomer ME. The SH3 domain of Eps8 exists as a novel intertwined dimer. Nat. Struct. Biol. 1997;4:739–43. doi: 10.1038/nsb0997-739. [DOI] [PubMed] [Google Scholar]
- 20.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 21.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 22.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 23.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–6. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staunton J, et al. Evidence for a double-helical structure for modular polyketide synthases. Nat. Struct. Biol. 1996;3:188–92. doi: 10.1038/nsb0296-188. [DOI] [PubMed] [Google Scholar]
- 25.Demeler B, van Holde KE. Sedimentation velocity analysis of highly heterogeneous systems. Anal. Biochem. 2004;335:279–88. doi: 10.1016/j.ab.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 26.Fischer H, de Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle x-ray scattering measurement on a relative scale. J. Appl. Cryst. 2010;43:101–9. [Google Scholar]
- 27.Kim KH, et al. Crystal structures of Enoyl-ACP reductases I (FabI) and III (FabL) from B. subtilis. J. Mol. Biol. 2010;406:403–15. doi: 10.1016/j.jmb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Lu H, Tonge PJ. Mechanism and inhibition of the FabV enoyl-ACP reductase from Burkholderia mallei. Biochemistry. 2010;49:1281–9. doi: 10.1021/bi902001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen ZJ, et al. Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new insights into its substrate recognition properties. J. Mol. Biol. 2008;379:830–44. doi: 10.1016/j.jmb.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 31.Torkko JM, et al. Candida tropicalis expresses two mitochondrial 2-enoyl thioester reductases that are able to form both homodimers and heterodimers. J. Biol. Chem. 2003;278:41213–20. doi: 10.1074/jbc.M307664200. [DOI] [PubMed] [Google Scholar]
- 32.Airenne TT, et al. Structure-function analysis of enoyl thioester reductase involved in mitochondrial maintenance. J. Mol. Biol. 2003;327:47–59. doi: 10.1016/s0022-2836(03)00038-x. [DOI] [PubMed] [Google Scholar]
- 33.Thorn JM, Barton JD, Dixon NE, Ollis DL, Edwards KJ. Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH. J. Mol. Biol. 1995;249:785–99. doi: 10.1006/jmbi.1995.0337. [DOI] [PubMed] [Google Scholar]
- 34.Piasecki SK, et al. Employing modular polyketide synthase ketoreductases as biocatalysts in the preparative chemoenzymatic syntheses of diketide chiral building blocks. Chem. Biol. 2011;18:1331–40. doi: 10.1016/j.chembiol.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Siskos AP, et al. Molecular basis of Celmer's rules: stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases. Chem. Biol. 2005;12:1145–53. doi: 10.1016/j.chembiol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Baerga-Ortiz A. Directed mutagenesis alters the stereochemistry of catalysis by isolated ketoreductase domains from the erythromycin polyketide synthase. Chem. Biol. 2006;13:277–85. doi: 10.1016/j.chembiol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. The Biochemical Basis for Stereochemical Control in Polyketide Biosynthesis. J. Am. Chem. Soc. 2009;131:18501–11. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witkowski A, Joshi AK, Smith S. Characterization of the beta-carbon processing reactions of the mammalian cytosolic fatty acid synthase: role of the central core. Biochemistry. 2004;43:10458–66. doi: 10.1021/bi048988n. [DOI] [PubMed] [Google Scholar]
- 39.Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain. Proc. Natl. Acad. Sci. USA. 2004;101:15567–72. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akey DL, et al. Crystal structures of dehydratase domains from the curacin polyketide biosynthetic pathway. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr., Sweet RM, editors. Methods in Enzymology, Volume 276: Macromolecular Crystallography, Part A. Academic Press; New York, NY: 1997. pp. 307–26. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–7. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Volkov VV, Svergun DI. Uniqueness of ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–4. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozin M, Svergun D. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2000;34:33–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.