Abstract
BACKGROUND & AIMS
Krüppel-like factor 4 (Klf4) is a putative gastric tumor suppressor gene. Rare, villin-positive progenitor cells in the gastric antrum have multi-lineage potential. We investigated the function of Klf4 in these cells and in gastric carcinogenesis.
METHODS
We created mice with disruption of Klf4 in villin-positive antral mucosa cells (Villin-Cre+;Klf4fl/fl mice). Villin-Cre+;Klf4fl/fl and control mice were given drinking water with or without 240 ppm N-methyl-N-nitrosourea (MNU) at 5 weeks of age and thereafter on alternating weeks for a total of 10 weeks. Gastric mucosa samples were collected at 35, 50, or 80 weeks of age from mice that were and were not given MNU, and analyzed by histopathologic and molecular analyses. Findings were compared with those from human gastric tumor specimens.
RESULTS
Preneoplasia formed progressively in the antrum in 35- to 80-week-old Villin-Cre+;Klf4fl/fl mice. Gastric tumors developed in 29% of 80-week-old Villin-Cre+;Klf4fl/fl mice, which were located exclusively in the lesser curvature of the antrum. MNU accelerated tumor formation, and tumors developed significantly more frequently in Villin-Cre+;Klf4fl/fl mice than in control mice, at 35 and 50 weeks of age. Mouse and human gastric tumors had reduced expression of KLF4 and increased expression of FoxM1, compared with healthy gastric tissue. Expression of KLF4 suppressed transcription of FoxM1.
CONCLUSIONS
Inactivation of Klf4 in villin-positive gastric progenitor cells induces transformation of the gastric mucosa and tumorigenesis in the antrum in mice. Villin-Cre+;Klf4fl/fl have greater susceptibility than control mice to chemical-induced gastric carcinogenesis and increased rates of gastric tumor progression.
Keywords: stomach cancer, mouse model, carcinogen, genetic
Gastric cancer is one of the most common cancers worldwide.1 Its aggressive nature is related to a variety of intracellular events, including activation of oncogenes and inactivation of tumor suppressor genes.2,3 However, the specific sequence of molecular changes leading to gastric cancer remains unclear.1–3 Clinically relevant animal models would be particularly useful for further exploration of the molecular pathogenesis of gastric cancer and to serve as preclinical models for evaluation of therapeutic and chemoprevention strategies.1–3
The gastrointestinal tract epithelium is a continuously renewing tissue, which also is under continuous exposure to various kinds of carcinogens and injurious agents that can cause cellular stress and trigger epithelial transformation and tumorigenesis.4 The gastric epithelium in particular contains functionally distinct pyloric and fundic mucosal lineages.5 The geographically heterogeneous population of multiple cell types in each pyloric gland is generated by controlled division of gastric epithelial stem cells located in the glands.5 Recent studies revealed that the majority of the pyloric glands are functionally monoclonal in the gastric antrum of adult mice and humans; specifically, all of the cellular progeny in the glandular gastric epithelium arise from a single stem cell.6–9 Gastric progenitor cell (GPC) studies have identified a rare subpopulation of murine GPCs with robust Villin expression predominantly in the antrum.6,10 These rare Villin-expressing GPCs are quiescent in the unstimulated stomach; however, they undergo both symmetric and asymmetric division and replace multiple entire pyloric glands during proinflammatory insults.6
Villin is an actin-bundling protein found in the apical brush border of absorptive tissue.11 Villin is also one of the first structural genes to be transcriptionally activated in the embryonic intestinal endoderm.10 The Villin gene is initially expressed in the intestinal hindgut endoderm 9 days post coitum during gut tube closure. Villin expression then rapidly extends throughout the small and large intestines and distal stomach.12–14 At 16 days post coitum, intestinal cells have their highest levels of Villin expression, whereas neighboring stomach cells have low levels of Villin expression.14 The promoter specificity in adult tissues has led to the use of Cre recombinase-expressing transgenic mouse models.10 Researchers have identified several cis-regulatory sequences that drive the Villin promoter.10,15,16
Increasing evidence has established the relationship between chronic inflammation and gastric cancer.2,3 Studies have suggested that Krüppel-like factor 4 (KLF4) plays an important role in mediating proinflammatory responses and that KLF4 expression is markedly induced by interferon-γ in macrophages and human colon cancer cell lines.17 KLF4 is a zinc-finger transcription factor, and KLF4 mRNA expression is found primarily in postmitotic, terminally differentiated epithelial cells in organs such as the skin, lungs, and gastrointestinal tract.18 Accumulating clinical, experimental, and mechanistic evidence shows that KLF4 is a potential tumor suppressor in patients with various cancers, including gastric cancer.19–21
Our recent study has shown that loss of KLF4 and overexpression of FoxM1 are evident in human gastric cancer and altered expression and function of FoxM1 contribute to gastric carcinogenesis.22 FoxM1 is a member of the Forkhead box transcription factor family.23 At the mRNA level, FoxM1 expression is ubiquitously expressed in mouse embryonic tissues and in proliferating mouse adult tissues but is extinguished in differentiated cells.23 Recent studies have suggested that FoxM1 is required to couple the S and M phases of the cell cycle in part through regulating transcription of genes essential for cell cycle progression, including cyclin D1 and p27.24,25 FoxM1 overexpression has been linked to oncogenesis.25,26 However, the mechanistic role of FoxM1 in gastric carcinogenesis and its causal link to altered KLF4 function are unknown.
In the present study, we sought to determine the functional significance of Klf4 inactivation in Villin-positive progenitor cells (GPCs) in gastric carcinogenesis. Our results clearly showed a protective role for Klf4 in these cells from transformation and carcinogenesis.
Materials and Methods
Detailed materials and methods are described in the Supplementary Methods.
Mouse Strains, Derivations, and Maintenance
The derivation and use of Klf4-LoxP, Villin-Cre, and Foxa3-Cre mice were described previously.10,21,27 All strains are on C57BL/6 genetic background. The animals were maintained in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care International in accordance with the current regulations and standards of the US Department of Agriculture, US Department of Health and Human Services, and National Institutes of Health.
Genotyping and Identification of the Rearranged Alleles
Genomic DNA isolated from tail clippings from the study mice was assayed for the presence of Klf4-flox using polymerase chain reaction (PCR) with three primers in the klf4 gene:21 exon 1, 5′-ctgggcccccacattaatgag-3′; exon 2, 5′-tcgctgacagccatgtcagac; and intron 3, 5′-ccagcagagccgttctggctg-3′. PCRs were carried out using the GoTaq PCR system (Promega, Madison, WI). In this report, the “floxed deletion” was referred as “deletion”, unless specifically stated or indicated otherwise.
Identification of the Villin-Cre Transgene
A mouse model with a 12.4-kb fragment of Villin promoter that drives Cre gene expression was used in this study.10 The presence of the Villin-Cre transgene in specific tissues was identified using PCR amplification with specific primers for the 12.4-kb VilCre transgene (The Jackson Laboratory) (forward, 5′-gtgtgggacagagaacaaacc-3′; reverse, 5′-acatcttcaggttctgcggg-3′) and Cre gene (forward, 5′-gcggcatggtgcaagttgaat-3′; reverse, 5′-cgttcaccggcatcaacgttt-3′). Also, a 205-bp fragment of Klf4 was amplified with the following primers and used as an internal control: forward, 5′-caaatgttgcttgtctggtg-3′; reverse, 5′-tcagtcgagtgcacagttt-3′.
Treatment Protocol for N-Methyl-N-Nitrosourea
The alkylating agent N-methyl-N-nitrosourea (MNU; Sigma Chemical Co., St. Louis, MO), which is widely used to study gastric carcinogenesis,28 was dissolved in distilled water at 240 ppm and freshly prepared three times a week for administration to the mice in drinking water in light-shielded bottles.28 Five-week-old mice were given the MNU-containing drinking water on alternating weeks for a total of 10 weeks of exposure. The mice were killed using CO2 asphyxiation and underwent a thorough postmortem examination at the age of 35, 50, or 80 weeks.
Statistical Analysis
The two-tailed Fisher exact test was used to determine the significance of the tumor incidence in each group. The two-tailed χ2 or Fisher exact test was used to determine the significance of the difference between the covariates of KLF4 and FoxM1 expression. In all of the tests, P values less than .05 were considered statistically significant. The SPSS software program (version 12.0; SPSS Inc., Chicago, IL) was used for the statistical analyses.
Results
Villin-Cre–Mediated Deletion of the Klf4 Gene in Mice
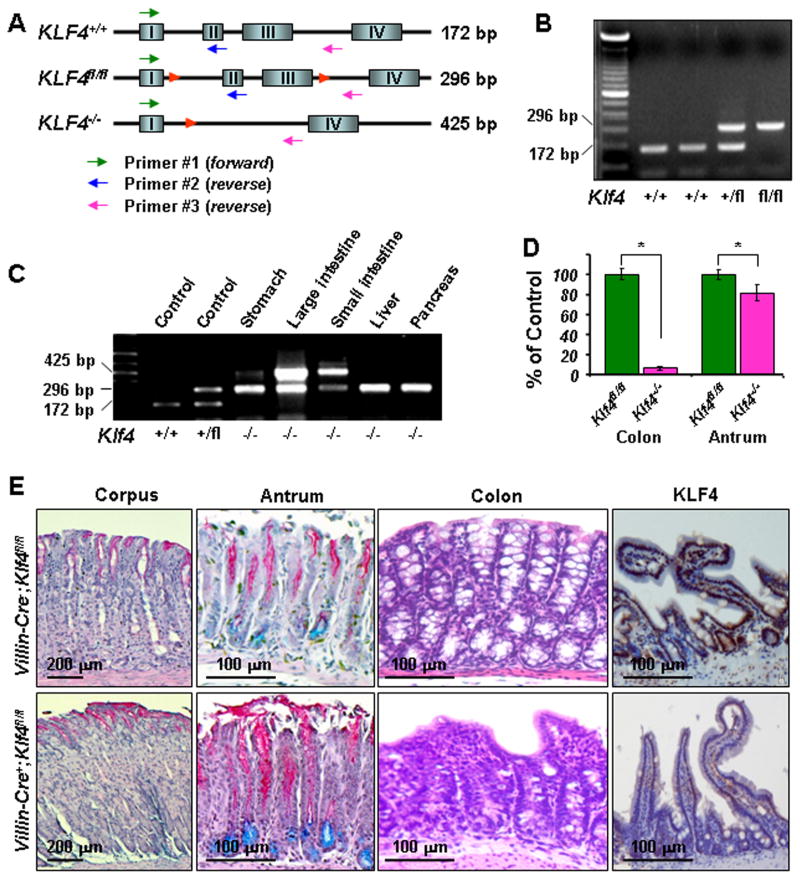
The lesser curvature of the antrum is the most common site of gastric tumor formation in humans3 and certain mouse models.28 Strikingly, recently identified rare Villin-positive progenitor cells are localized in the antrum and exhibit multilineage potential as GPCs.6 Although Klf4 is regarded as a putative gastric cancer suppressor gene, the functional significance of Klf4 inactivation in these Villin-positive cells is unknown. To determine the effects of Klf4 ablation in Villin-positive cells on gastric mucosal transformation and tumorigenesis, we genetically generated and verified Villin-Cre+;Klf4fl/fl mice (Figures 1A and 1B) using previously published procedures.6,10,21,27 In Villin-Cre transgenic mice, we detected both Villin and Cre protein predominantly in the intestines (sFigure 1A), including the duodenum but rarely in the adjacent antrum (sFigure 2A). However, we observed an increase in Villin-positive cells in the antrum in Villin-Cre+;Klf4fl/fl mice (sFigure 2A) and a further rapid and patchy increase in Villin-Cre+;Klf4fl/fl mice upon treatment with MNU (sFigure 2B). This differing pattern of Villin expression in these tissues was consistent with the differing efficacy of Villin-Cre–mediated Klf4 deletion in them (Figure 1C). Villin-Cre+;Klf4fl/fl mice survived to at least 80 weeks of age. Our quantitative PCR analysis revealed that Klf4 was deleted in more than 95% of the cells in the intestines and less than 20% cells in the antrum of a Villin-Cre+;Klf4fl/fl mouse (Figure 1D). Morphologically, we observed no discernible changes in the corpus mucosa, whereas loss of both Klf4 expression in small intestinal mucosa and goblet cells in the colonic mucosa further confirmed the expected function of the Villin-Cre transgene in Klf4fl/fl mice (Figure 1E).
Figure 1. Villin-Cre–mediated Klf4 deletion in the gastric antrum in mice.
(A) Schematic diagram of the genomic structure of the Klf4 gene with wild-type (Klf4+/+), floxed (Klf4fl/fl), and disrupted (Klf4−/−) alleles. Shown are LoxP sites (red triangles), PCR primer positions (arrows), and expected PCR products using three primers: 172bp for the wild-type allele, 296bp for the floxed allele, and 425bp for the deleted allele. (B) Genotyping using mouse tail DNA at the age of 4 weeks. PCR screening for Klf4 revealed a band of 172bp for the wild-type allele and 296bp for the floxed allele. (C) Detection of Villin-Cre–mediated rearrangement of the Klf4 allele in different tissues in mice at the age of 20 weeks. DNA was extracted from different tissues from a previously genotyped Klf4−/− mouse. PCR analysis indicated various levels of rearrangement of the Klf4 gene (425bp for the deleted allele). (D) The efficacy of Klf4 deletion in the colon and gastric antrum in Klf4fl/fl and Klf4−/− mice was measured using quantitative PCR analysis of colon and antral DNA from those mice at the age of 35 weeks. (E) PAS/Alcian blue staining of gastric corpus and antral mucosa specimens obtained from Villin-Cre−;Klf4fl/fl and Villin-Cre+;Klf4fl/fl mice at the age of 50 weeks. Villin-Cre+;Klf4fl/fl antral mucosa exhibited increases in the number of both PAS- and Alcian blue-positive cells. Hematoxylin and eosin staining of colons obtained from Villin-Cre−;Klf4fl/fl and Villin-Cre+;Klf4fl/fl mice. Goblet cells were nearly absent from Villin-Cre+;Klf4fl/fl colonic mucosa, whereas a normal contour and numerous goblet cells were observed along the crypts and surface epithelium in Villin-Cre−;Klf4fl/fl mice. Positive Klf4 staining in the small intestinal mucosa cells of Villin-Cre−;Klf4fl/fl mice but negative in that of Villin-Cre+;Klf4fl/fl mice.
Spontaneous Tumors in the Gastric Antrum in Villin-Cre+;Klf4fl/fl Mice
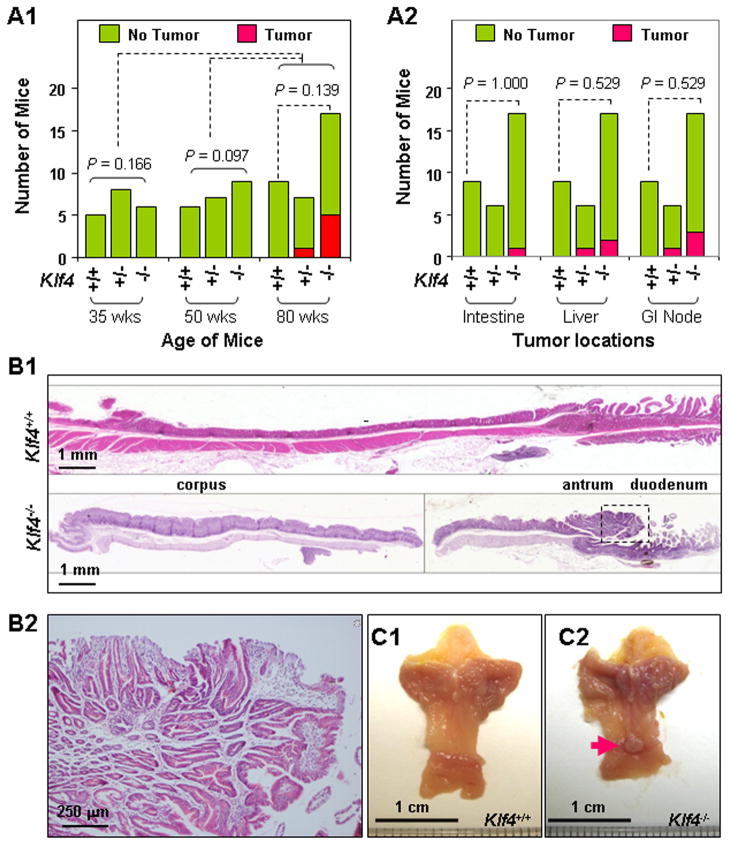
We then systematically examined gastric tumor formation in six different mouse strains: Villin-Cre+;Klf4fl/fl, Villin-Cre+;Klf4+/fl, Villin-Cre+;Klf4+/+, Villin-Cre−;Klf4fl/fl, Villin-Cre−;Klf4+/fl, and Villin-Cre−;Klf4+/+. No visible tumors formed in the stomachs of any of the mice by the age of 35 or 50 weeks (Figure 2A1). At the age of 80 weeks, 29% (5/17) of the Villin-Cre+;Klf4fl/fl mice and 14% (1/7) of the Villin-Cre+;Klf4+/fl mice had gastric tumors (Figure 2A1), and small percentages of these mouse strains also had tumors in other organs (Figure 2A2) and their significance and underlying mechanisms are unclear. In contrast, no visible tumors formed in the stomachs or other organs in the control mouse strains (Figure 2A2), including Villin-Cre+;Klf4+/+, Villin-Cre−;Klf4+/fl, and Villin-Cre−;Klf4+/+ mice without MNU treatment (data not shown). More importantly, we found that all of the gastric tumors were located in the lesser curvature of the antrum (Figures 2B and 2C). Histological examination of gastric antral tumors showed that they were adenomas. The polypoid adenomatous lesions demonstrated elongated pits and enlarged glandular structures which led to additional branching and interglandular bridging in the thickened lamina propria and the intraepithelial compartments (Figure 2B2). Given the unique location (antrum) and time frame (advanced age) of gastric tumor formation, our model is highly relevant to human gastric cancer in the antrum.3
Figure 2.
Spontaneous gastric tumor development in the antrum in Villin-Cre+;Klf4fl/fl mice. (A) Tumor incidence in the stomach (A1) and other locations (A2) in Villin-Cre−;Klf4fl/fl (Klf4+/+), Villin-Cre+;Klf4+/fl (Klf4+/−), and Villin-Cre+;Klf4fl/fl (Klf4−/−) mice at the ages of 35, 50, and 80 weeks. (B) Representative photographs of macroscopic views of the entire gastric mucosa in 80-week-old Klf4+/+ and Klf4−/− mice (B1) and microscopic views of an antral tumor in the Klf4−/− mouse (B2). (C) Gross morphology of stomachs obtained from 80-week-old Klf4+/+ (C1) and Klf4−/− (C2) mice. A visible antral tumor in a Klf4−/− mouse stomach is indicated by a red arrow.
Susceptibility of Villin-Cre+;Klf4fl/fl Mice to Chemical Gastric Carcinogenesis
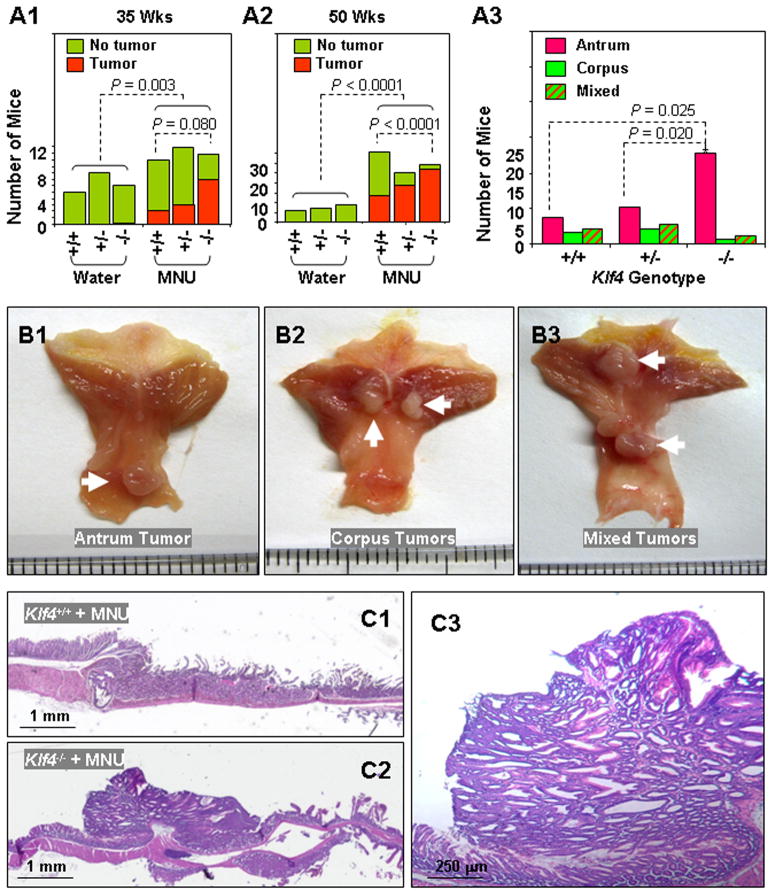
To further determine the functional significance of Klf4 inactivation in gastric carcinogenesis, we treated the Villin-Cre+;Klf4fl/fl, Villin-Cre+;Klf4+/fl, and Villin-Cre−;Klf4fl/fl mice with MNU, which is widely used to induce gastric adenomas and adenocarcinomas in mice. The incidences of gastric tumors in the MNU-treated mice were significantly higher than those in the matched control mice that did not receive MNU treatment (Figures 3A1 & 3A2). However, the incidences were higher in Villin-Cre+;Klf4fl/fl mice than in Villin-Cre−;Klf4fl/fl mice, which strongly suggested that Villin-Cre+;Klf4fl/fl mice had increased susceptibility to chemical carcinogenesis in the stomach. Although we observed various locations of gastric tumors, the predominant location was the antrum (Figures 3A3, 3B & 3C).
Figure 3.
Induction of gastric tumor development in the antrum in Villin-Cre+;Klf4fl/fl mice. (A) Incidence (A1 & A2) and locations (A3) of gastric tumor formation in Villin-Cre−;Klf4fl/fl (Klf4+/+), Villin-Cre+;Klf4+/fl (Klf4+/−), and Villin-Cre+;Klf4fl/fl (Klf4−/−) mice at the ages of 35 and 50 weeks with or without MNU-based treatment. (B) Gross morphology of visible tumors (white arrows) in the gastric antrum (B1), corpus (B2), or both (“mixed,” B3) in 50-week-old Klf4−/− mice. (C) Representative photographs of macroscopic views of the entire gastric mucosa in 50-week-old Klf4+/+ (C1) and Klf4−/− (C2) mice and a microscopic view of an antral tumor in the Klf4−/− mouse (C3).
Loss of Integrity and/or Expression of the Klf4 Gene in Gastric Tumors
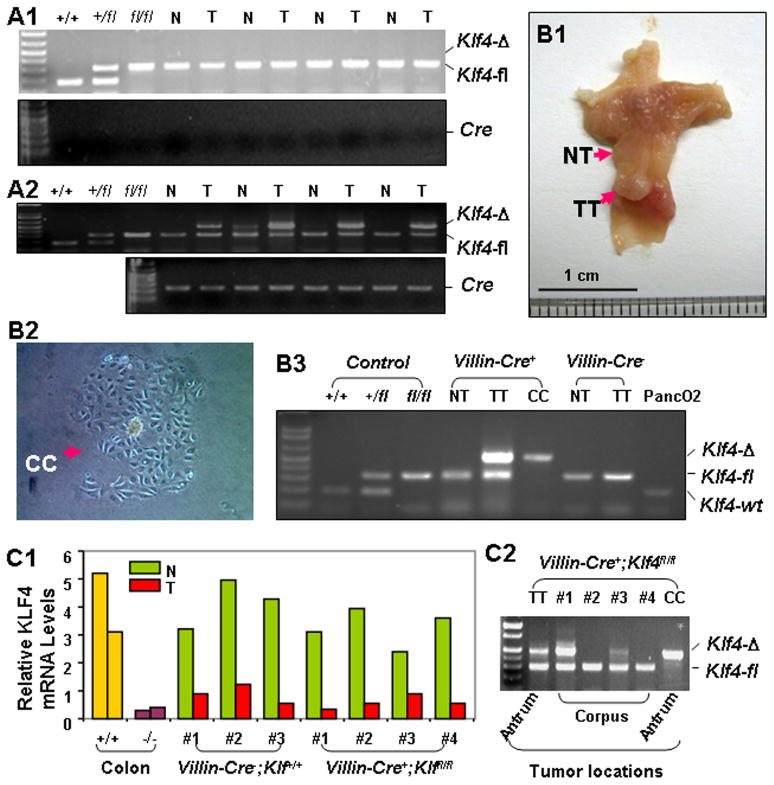
To determine whether Klf4 deletion was relevant to the development of gastric tumors in mice with MNU treatment, we performed PCR analysis using DNA from gastric tumors and matched corpus mucosal tissues obtained from Villin-Cre−;Klf4fl/fl and Villin-Cre+;Klf4fl/fl mice. Whereas gastric tumors from Villin-Cre−;Klf4fl/fl mice showed no Klf4 deletions (Figure 4A1), all of the gastric antral tumors (n = 4) from Villin-Cre+;Klf4fl/fl mice did exhibit Klf4 deletions (Figure 4A2). As expected, we observed no discernible Klf4 deletions in the matched corpus mucosal tissues in either mouse strain, i.e., Villin-Cre−;Klf4fl/fl mice (Figure 4A1) and Villin-Cre+;Klf4fl/fl mice (Figure 4A2).
Figure 4.
Analysis of the integrity and expression of the Klf4 gene in gastric tumors. (A) DNA was isolated from matched gastric tumor specimens (T) and nontumorous corpus mucosa specimens (N) obtained from Villin-Cre−;Klf4fl/fl (A1) and Villin-Cre+;Klf4fl/fl (A2) mice. PCR analysis was performed for genotyping of Klf4 alleles. (B) An individual antral tumor was obtained immediately after surgery (B1) and divided into two parts: one for primary culture (CC, B2) and one for DNA extraction (TT). DNA was also extracted from a nontumorous corpus mucosa specimen (NT). PCR analysis was performed for genotyping of Klf4 alleles (B3). (C) Klf4 mRNA expression and genetic integrity in corpus tumors. Real-time PCR analysis was performed using total RNA extracted from corpus tumors and adjacent nontumorous tissue specimens from Klf4+/+ and Klf4−/− mice. Total RNA obtained from colonic mucosa in Klf4+/+ and Klf4−/− mice was used as a control (C1). DNA was extracted from Klf4−/− corpus tumors, and PCR analysis was performed for genotyping of Klf4 alleles using DNA from TT and CC as a control (C2).
To further characterize the specific cell populations with Klf4 deletions in the Villin-Cre+;Klf4fl/fl tumors, we extracted DNA from three sources (Figure 4B1): 1) matched corpus mucosal tissue, 2) an in vitro cell culture of half of an antral tumor to eliminate stromal cells (Figure 4B2), and 3) the other half of the antral tumor. Genotypic analysis showed complete deletion of the Klf4 gene in the gastric tumor cells (Figure 4B3).
Additionally, we measured the KLF4 expression in gastric corpus tumors obtained from both Villin-Cre+;Klf4fl/fl and Villin-Cre−;Klf4+/+ mice using quantitative real-time PCR analysis. Our results showed that the KLF4 expression was significantly lower in all of the tumors than in adjacent corpus mucosal tissues (Figure 4C1). Finally, we sought to determine the integrity of the Klf4 gene in the corpus tumors obtained from Villin-Cre+;Klf4fl/fl mice and found that some but not all of the tumors exhibited Klf4 deletions (Figure 4C2).
Histopathology of the Stomach in Villin-Cre+;Klf4+/+ Mice
We also systematically examined preneoplastic changes in the gastric mucosa. Without MNU-based treatment, the preneoplastic lesions were primarily located in the antrum and progressed as the mice aged. Treatment with MNU promoted the formation of preneoplastic changes in these lesions, including hyperplasia (sFigure 3). As described above, neoplastic lesions formed in Villin-Cre+;Klf4+/+ mice that did not receive MNU (spontaneous tumors). However, we did not observe these lesions in control mouse strains, including Villin-Cre+;Klf4+/+, Villin-Cre−;Klf4+/fl, and Villin-Cre−;Klf4+/+ mice without MNU treatment, suggesting that gastric transformation was not impacted by the presence of floxed Klf4 alleles or the Cre transgene.
Aberrant FoxM1 Overexpression in Gastric Tumors
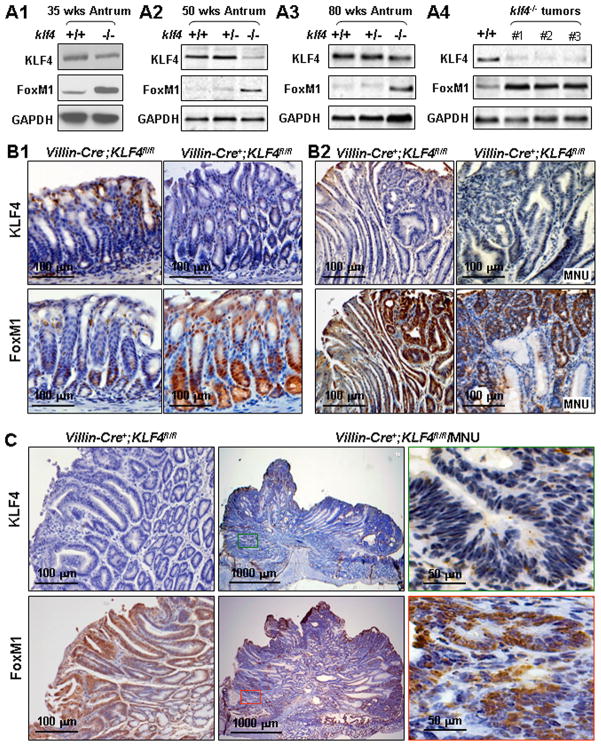
To characterize the underlying mechanism by which deletion of the Klf4 gene contributes to gastric transformation and tumorigenesis, we measured the KLF4 and FoxM1 expression in gastric antral tumors and normal gastric mucosal tissues obtained from Villin-Cre+;Klf4fl/fl, Villin-Cre+;Klf4+/fl, and Villin-Cre+;Klf4+/+ mice. In both Western blot and immunohistochemical analyses, lost KLF4 expression correlated with increased FoxM1 expression (Figure 5). Specifically, we found FoxM1 expression in the lower third and KLF4 expression in the upper third of the antral mucosa in Villin-Cre−;Klf4fl/fl mice, whereas we observed KLF4 underexpression and FoxM1 overexpression in the antral mucosa in Villin-Cre+;Klf4fl/fl mice (Figure 5B1). Concomitant underexpression of Klf4 and overexpression of FoxM1 were evident in intramucosal neoplastic lesions (Figure 5B2; sFigure 4) and tumors (Figure 5C) in Villin-Cre+;Klf4fl/fl mice. These results suggested that FoxM1 is a prominent downstream gene whose expression is negatively regulated by KLF4.
Figure 5.
Lost KLF4 expression and FoxM1 overexpression in gastric tumors. (A) Western blot analyses were performed using total protein lysates extracted from antral mucosa in mice at the ages of 35 (A1), 50 (A2), and 80 (A3) weeks and from antral tumors (A4). KLF4+/+, Villin-Cre−;Klf4fl/fl; KLF4+/−, Villin-Cre+;Klf4+/fl; KLF4−/−, Villin-Cre+;Klf4fl/fl. (B) Immunohistochemical staining for KLF4 (upper panels) and FoxM1 (lower panels) protein. From left to right were: normal, hyperplastic, intramucosal neoplastic, and neoplastic tissue from mice at age of 80 weeks. (C) Immunohistochemical staining of two gastric tumors for KLF4 and FoxM1 protein from mice at age of 80 weeks. The right panels are views of the middle panels at higher magnification.
Inverse Correlation between KLF4 and FoxM1 Expression in Human Gastric Tumors
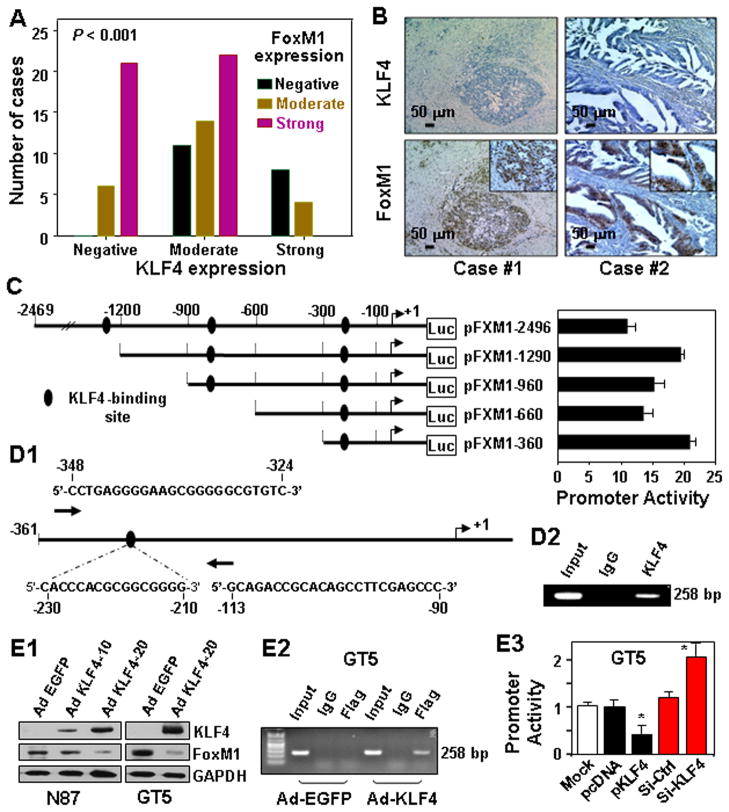
To validate our observation in our mouse model, we examined KLF4 and FoxM1 expression in human gastric tumor specimens. Analysis of FoxM1 and KLF4 expression in all 86 gastric tumors revealed a significant inverse correlation between FoxM1 expression and KLF4 expression (sTable 1; Figure 6A). In most of the gastric tumor specimens, KLF4 expression was significantly decreased or lost, whereas FoxM1 overexpression was strikingly evident (Figure 6B). These findings also suggested that expression of FoxM1 is negatively regulated by KLF4.
Figure 6.
KLF4 regulation of FoxM1 expression in human gastric tumors. (A) Tissue sections were prepared from 86 formalin-fixed, paraffin-embedded human gastric tumor specimens. Immunostaining of the sections was performed using specific anti-KLF4 and -FoxM1 antibodies. FoxM1 expression levels were inversely correlated with KLF4 expression levels (P<001; χ2 test). (B) Representative photographs of two cases showing KLF4 underexpression and FoxM1 overexpression. (C) Deletion mutants of FoxM1 promoter reporters were transfected into SK-GT5 cells in triplicate, and the relative promoter activities were measured 24 hours after transfection. (D) Schematic diagram of the FoxM1 proximal promoter. The nucleotide positions and sequences of the putative KLF4 binding site and PCR forward and reverse primers for ChIP analysis are shown (D1). Chromatin was extracted from SK-GT5 cells, and the ChIP assay was performed using a specific anti-KLF4 antibody and oligonucleotides flanking the FoxM1 promoter region containing the KLF4-binding site (D2). (E) N87 and SK-GT5 cells were transduced with a control Ad-EGFP (Neo) or Ad-KLF4 at an MOI of 10 or 20 for 24 hours. Total protein lysates were harvested from the cell cultures, and the levels of KLF4 (exogenous, as determined using an anti-FLAG antibody) and FoxM1 expression were determined using Western blot analysis (E1). SK-GT5 cells were transduced with Ad-KLF4 or Ad-EGFP at an MOI of 10 for 24 hours. Chromatin fragments were prepared for ChIP analysis using control IgG and an anti-Flag antibody (E2). The pFXM1-360 proximal promoter was transfected into SK-GT5 cells in triplicate with pcDNA3.1, a KLF4 expression vector, nontargeting control siRNA, or FoxM1-siRNA. The relative promoter activities were assessed 24 hours after transfection (E3).
Negative Regulation of FoxM1 Expression by KLF4
To determine whether KLF4 represses FoxM1 expression, we initially used a computer-bases software program to identify KLF4-binding sites on the FoxM1 promoter (Figure 6C). We found a putative KLF4-binding site between −230 and −210 bp in the FoxM1 proximal promoter (Figure 6D1). A ChIP assay indicated that KLF4 could bind to this region of the FoxM1 promoter in vivo (Figure 6D2). Consistently, increased KLF4 expression repressed FoxM1 expression in N87 and SK-GT5 cells (Figure 6E1), and increased KLF4 expression clearly bound to the target sequence of the FoxM1 promoter (Figure 6E2). Furthermore, transfection of a KLF4 expression vector repressed the activity of the pFXM1-360 proximal promoter, whereas knockdown of KLF4 expression by KLF4 small interfering RNA (siRNA) significantly increased the promoter activity (Figure 6E3). These results supported our hypothesis that KLF4 binds to the putative region of the FoxM1 promoter in vivo and represses FoxM1 transcription.
Gastric Antral Tumor Formation in Foxa3-Cre+;Klf4fl/fl Mice
A prior study has demonstrated that global deletion of Klf4 in stomach caused preneoplastic changes.27 However, whether those mice are prone to chemical carcinogenesis is unknown. In our final set of experiments, we bred and genetically verified Foxa3-Cre+;Klf4fl/fl mice according to previously published procedures.27 Consistent with that previous study, we observed extensive preneoplastic changes, including hyperplasia in gastric mucosa in both the corpus and antrum; furthermore, treatment with MNU promoted the formation of both preneoplasia and neoplasia (sFigure 5A & sFigure 6). Interestingly, extensive hyperplasia formed in the corpus, and no tumors had formed in mice by the age of 35 weeks without MNU-based treatment. However, MNU treatment promoted gastric tumor formation in all of the mice and gastric tumors were located predominantly in the antrum (sFigures 5B and 5C).
Finally, we compared the effects of Klf4 deletion in the corpus mucosa in Foxa3-Cre+;Klf4fl/fl and Villin-Cre+;Klf4fl/fl mice. We observed extensive Klf4 deletion in Foxa3-Cre+;Klf4fl/fl mice, whereas we found no discernible deletions in Villin-Cre+;Klf4fl/fl mice (sFigure 5D). These findings suggested that the extent of Klf4 deletion was directly correlated with that of gastric mucosal transformation.
Discussion
In this study, we observed selective inactivation of the klf4 gene in a distinct putative progenitor cell population at the bases of the pyloric glands in the antrum and subsequent development of spontaneous gastric tumors in the lesser curvature of the antrum but not in the corpus in Villin-Cre+;Klf4fl/fl mice. We also found that treatment with MNU accelerated gastric tumorigenesis in Villin-Cre+;Klf4fl/fl mice. Moreover, loss of KLF4 expression resulted in upregulation of FoxM1 expression and contributed to gastric tumor formation in Villin-Cre+;Klf4fl/fl mice. Consistently, KLF4 expression was inversely correlated with FoxM1 expression in human gastric tumors and downregulated FoxM1 expression in gastric cancer cells. Therefore, we provide clinical, experimental, and mechanistic evidence that loss of KLF4 expression in putative GPCs is an important step in gastric tumorigenesis and tumor progression. Because approximately 60–80% of intestinal-type gastric tumors form in the antrum, most often in the lesser curvature,3,28,29 our present mouse model is a close recapitulation of human gastric cancer and a novel tool for further investigation into the underlying molecular basis for gastric carcinogenesis.
The unique locations of both human intestinal-type gastric tumors and experimental gastric tumors in animals has spurred extensive investigations into their cellular origin.29,30 Reported evidence supports the existence of gastric stem cells, which are considered prime candidates as cells of origin for cancer.7,29,30 Studies tracing the in vivo lineage revealed that each gastric gland unit is functionally monoclonal in both the murine and human stomach, with all cell progeny arising from a single stem cell.6–7 However, the identities of these putative gastric stem cells and their causal link to gastric carcinogenesis are not clear. At least three putative types of gastric stem cells exist, and they may all be found in the antrum. Also, gastric cancer can originate from bone marrow-derived cells in a mouse model of chronic H. pylori infection.31 Interestingly, the biological characteristics of the pyloric glands in the antrum are developmentally similar to those in the small intestinal epithelium.31 Researchers have identified Lgr5+ cells as stem cell markers in both the intestine and stomach in mice, and activation of a Wnt signal by conditional deletion of APC gene in Lgr5+ cells efficiently initiates tumor formation in the distal stomach.7 Those researchers also recently observed that more active Lgr5+ stem cells affected the daily steady-state self-renewal of the pyloric epithelium in adults and both the corpus and pylorus in neonates.7 Even more recently, investigators identified Villin-positive cells as prospective progenitor cells in the bottom third of the pyloric gland. These cells are normally quiescent and multiply with multilineage potential via both symmetric and asymmetric division in response to inflammatory insults.6 All of these putative stem cell types appear to be capable of self-renewal and differentiation, with the cells’ statuses ranging from pluripotent to multipotent to tissue-specific adult stem cells.29 In the present study, selective inactivation of Klf4 in Villin-positive progenitor cells led to the development of tumor phenotypes in the lesser curvature of the antrum in old mice. The fact that the tumors were located exclusively at the pyloric junction was also consistent with the notion that the lesser curvature of the antrum is the predominant location of GPCs.6–7 Paradoxically, the GPCs marked by the villin-Cre transgene do not express the endogenous villin locus.6 Thus, to confirm efficient deletion of KLF4 in those cells will require performing lineage tracing experiments.
Furthermore, we speculated that bone marrow-derived cells (BMDCs), Lgr5+ cells, and Villin-positive cells all may contribute to gastric tumor formation, playing different roles linked with the differentiation status. BMDCs may be the most primitive uncommitted adult stem cells that can be recruited to tumor transformation by chronic inflammation. Lgr5+ cells are located exclusively in the bases of pyloric glands and are active stem cells responsible for the daily self-renewal of the pyloric mucosa. Villin-positive cells are quiescent stem cells that require injurious insults such as inflammation (by interferon-γ) for their activation. Villin is not normally expressed in the stomach; however, its expression can be induced in atrophic human and murine stomachs in response to chronic H. pylori infection via extracellular signal-regulated kinase signaling.32 Data from the present study also support this notion. Specifically, villin expression likely was induced by MNU-based treatment, which in turn drove Klf4 gene deletion and contributed to increased gastric tumor incidence. Also, the extent of Klf4 deletion varied in the antrum of mice at different ages or with exposure to chemical insults, further supporting that the size of Villin-positive cell subpopulation may change. Interestingly, these rare villin-positive cells are similar in many aspects (rarity, location, slow proliferation and morphology) to the controversial and recently identified rare DCAMKL1-positive intestinal cells33 that are believed to be progenitor cells,34 that were later identified as tuft cells not progenitor cells.35 These cells are positive for villin, DCAMKL1, Cox1 and alpha tubulin. Further studies are clearly warranted to determine whether these rare villin-positive cells in antrum are also positive for either DCAMKL1, Cox1 and alpha tubulin or all of them; or whether these rare villin-positive gastric cells are in fact gastric tuft cells.36
Numerous genes are implicated in regulation of preneoplastic lesion and gastric tumor formation and progression in mouse models, including K-Ras,37 cdx2,38 gastrin,39,40 Wnt,7,41 and IKKβ/NF-κB.42 A prior study showed that loss of KLF4 expression in Foxa3-Cre+;Klf4fl/fl mice led to gastric hypertrophy, mucus cell hyperplasia, glandular distortion, and polypoid lesion development,27 and authors have reported reduced KLF4 expression in various types of human tumors,19,20 supporting KLF4’s tumor suppressor role. In the present study, we observed causal evidence of the critical role of loss of KLF4 expression in gastric carcinogenesis. Moreover, DNA analysis revealed Klf4 gene deletion from a significant majority of the tumors that formed in MNU-treated Villin-Cre+;Klf4fl/fl mice, whereas all of the tumor cells derived from Villin-Cre+;Klf4fl/fl mice exhibited Klf4 gene deletion. These results strongly suggest that these Klf4-deleted putative progenitor cells contribute to tumor formation in the gastric antrum in Villin-Cre+;Klf4fl/fl mice and give rise to hyperplastic lesions in the early stages of gastric carcinogenesis.
Finally, we observed that loss of KLF4 expression correlated directly with FoxM1 overexpression in a consistent manner in both human and murine gastric tumors. Several previous studies showed that FoxM1 expression is regulated by posttranslational modifications, degradation, and interaction with other transcription factors.43 Our experiments established FoxM1 as a novel downstream target of KLF4 in that KLF4 negatively regulates FoxM1 transcription, providing a novel molecular basis that causally links frequent FoxM1 overexpression with loss of KLF4 expression in various types of cancer, including gastric cancer.20,22 Given that FoxM1 positively regulates and KLF4 negatively regulates the progression of cell cycle, loss of expression of KLF4 and overexpression of FoxM1 should lead to dysregulated cell growth and cellular transformation as shown in the gastric antrum in Villin-Cre+;Klf4fl/fl mice and gastric antrum and corpus in Foxa3-Cre+;Klf4fl/fl mice. Thus, targeting this aberrant pathway may be an effective strategy for cancer prevention and treatment.
In summary, we derived mice with selective deletion of Klf4 in GPCs and showed that loss of Klf4 expression in these cells results in spontaneous gastric neoplasia and rapid development of gastric tumors in the setting of treatment with MNU, mostly in the lesser curvature of the antrum. The rapid progression to neoplasia observed in the Villin-Cre+;Klf4fl/fl mice was associated with overexpression of FoxM1 protein. In addition, we showed that KLF4 inhibited FoxM1 expression in part through its binding site. Therefore, KLF4 plays an important role in the homeostasis of normal gastrointestinal tissue, and inactivation of Klf4 in GPCs promotes carcinogenesis. Collectively, our findings strengthen the causative role gastric stem cells play in gastric carcinogenesis and the gastric tumor-suppressive role of Klf4 in response to carcinogen exposure.
Supplementary Material
Acknowledgments
The authors thank Mr. Don Norwood for editorial comments, Dr. Timothy C. Wang and Dr. James R. Goldenring for their critical reading and suggestions, and Dr. Julie Segre from the National Institutes of Health for Klf4loxP/loxP mice. Supported in part by Research Scholar Grant CSM-106640 from the American Cancer Society; grants 1R01-CA093829, R01-CA129956, R01-CA148954, and R01-CA152309 (to K.X.); and P01-CA130821 (to L.M.) from the National Cancer Institute, National Institutes of Health and the MD Anderson Development Fund (to D.T.). While this manuscript is in revision, Meiping Xie (1967-2011) tragically passed away after battling gastric cancer for 4 years. The work is dedicated to her memory.
Abbreviations used in this paper
- GPC
Gastric progenitor cell
- KLF4
Krüppel-like factor 4
- MNU
N-methyl-N-nitrosourea
- PCR
polymerase chain reaction
- EGFP
enhanced green fluorescent protein
- MOI
multiplicity of infection
- DAPI
4′6-diamidino-2-phenylindole
- ChIP
chromatin immunoprecipitation
Footnotes
Disclosure
Conflict of interest disclosures: no conflict of interest to disclose for all authors.
Involvment with the manuscript:
Study concept and design: KEPING XIE, QIANG LI, SUYUN HUANG
Acquisition of data: QIANG LI, LI WANG, ZHILIANG JIA, KUN GUO, XIANGYU KONG, QI LI, KEPING XIE
Analysis and interpretation of data: KEPING XIE, QIANG LI, SUYUN HUANG, ZHILIANG JIA, XIANGYU KONG, KUN GUO, QI LI, DONGFENG TAN, LOPA MISHRA
Drafting of the manuscript: QIANG LI, KEPING XIE, SUYUN HUANG, LOPA MISHRA
Critical revision of the manuscript for important intellectual content: SUYUN HUANG
Statistical analysis: QIANG LI, ZHILIANG JIA, XIANGYU KONG, SUYUN HUANG, KEPING XIE
Obtained funding: KEPING XIE, SUYUN HUANG, LOPA MISHRA
Technical support: XIANGDONG LE, ZHILIANG JIA, DAOYAN WEI, QI LI
Material support: KEPING XIE, SUYUN HUANG
Study supervision: KEPING XIE, SUYUN HUANG
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest. 2007;117:60–69. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merchant JL. Inflammation, atrophy, gastric cancer: connecting the molecular dots. Gastroenterology. 2005;129:1079–1082. doi: 10.1053/j.gastro.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 4.Galluzzi L, Maiuri MC, Vitale I, et al. Cell death modalities: classification and pathophysiological implications. Cell Death Differ. 2007;14:1237–1243. doi: 10.1038/sj.cdd.4402148. [DOI] [PubMed] [Google Scholar]
- 5.Karam SM, Leblond CP. Dynamics of epithelial cells in the corpus of the mouse stomach. I. Identification of proliferative cell types and pinpointing of the stem cell. Anat Rec. 1993;236:259–279. doi: 10.1002/ar.1092360202. [DOI] [PubMed] [Google Scholar]
- 6.Qiao XT, Ziel JW, McKimpson W, et al. Gumucio DL. Prospective identification of a multilineage progenitor in murine stomach epithelium. Gastroenterology. 2007;133:1989–1998. doi: 10.1053/j.gastro.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker N, Huch M, Kujala P, van de Wetering M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 8.McDonald SA, Greaves LC, Gutierrez-Gonzalez L, et al. Mechanisms of field cancerization in the human stomach: the expansion and spread of mutated gastric stem cells. Gastroenterology. 2008;134:500–510. doi: 10.1053/j.gastro.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 9.Bjerknes M, Cheng H. Multipotential stem cells in adult mouse gastric epithelium. Am J Physiol Gastrointest Liver Physiol. 2002;283:G767–G777. doi: 10.1152/ajpgi.00415.2001. [DOI] [PubMed] [Google Scholar]
- 10.Madison BB, Dunbar L, Qiao XT, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 11.Craig SW, Powell LD. Regulation of actin polymerization by villin, a 95,000 dalton cytoskeletal component of intestinal brush borders. Cell. 1980;22:739–746. doi: 10.1016/0092-8674(80)90550-4. [DOI] [PubMed] [Google Scholar]
- 12.Maunoury R, Robine S, Pringault E, et al. Developmental regulation of villin gene expression in the epithelial cell lineages of mouse digestive and urogenital tracts. Development. 1992;115:717–728. doi: 10.1242/dev.115.3.717. [DOI] [PubMed] [Google Scholar]
- 13.Ezzell RM, Chafel MM, Matsudaira PT. Differential localization of villin and fimbrin during development of the mouse visceral endoderm and intestinal epithelium. Development. 1989;106:407–419. doi: 10.1242/dev.106.2.407. [DOI] [PubMed] [Google Scholar]
- 14.Braunstein EM, Qiao XT, Madison B, et al. Villin: A marker for development of the epithelial pyloric border. Dev Dyn. 2002;224:90–102. doi: 10.1002/dvdy.10091. [DOI] [PubMed] [Google Scholar]
- 15.Pinto D, Robine S, Jaisser F, et al. Regulatory sequences of the mouse villin gene that efficiently drive transgenic expression in immature and differentiated epithelial cells of small and large intestines. J Biol Chem. 1999;274:6476–6482. doi: 10.1074/jbc.274.10.6476. [DOI] [PubMed] [Google Scholar]
- 16.Robine S, Sahuquillo-Merino C, Louvard D, et al. Regulatory sequences on the human villin gene trigger the expression of a reporter gene in a differentiating HT29 intestinal cell line. J Biol Chem. 1993;268:11426–11434. [PubMed] [Google Scholar]
- 17.Feinberg MW, Cao Z, Wara AK, et al. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 18.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McConnell BB, Ghaleb AM, Nandan MO, et al. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei D, Gong W, Kanai M, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 21.Katz JP, Perreault N, Goldstein BG, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Zhang N, Jia Z, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 2011;10:396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai B, Pieper RO, Li D, et al. FoxM1B regulates NEDD4-1 expression, leading to cellular transformation and full malignant phenotype in immortalized human astrocytes. Cancer Res. 2010;70:2951–61. doi: 10.1158/0008-5472.CAN-09-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M, Dai B, Kang SH, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 26.Gartel AL. A new target for proteasome inhibitors: FoxM1. Expert Opin Investig Drugs. 2010;19:235–42. doi: 10.1517/13543780903563364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz JP, Perreault N, Goldstein BG, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–945. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 28.Tomita H, Yamada Y, Oyama T, et al. Development of gastric tumors in Apc(Min/+) mice by the activation of the beta-catenin/Tcf signaling pathway. Cancer Res. 2007;67:4079–4087. doi: 10.1158/0008-5472.CAN-06-4025. [DOI] [PubMed] [Google Scholar]
- 29.Quante M, Wang TC. Stem cells in gastroenterology and hepatology. Nat Rev Gastroenterol Hepatol. 2009;6:724–737. doi: 10.1038/nrgastro.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houghton J, Stoicov C, Nomura S, et al. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–1571. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]
- 31.Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003;3:592–600. doi: 10.1038/nrc1141. [DOI] [PubMed] [Google Scholar]
- 32.Rieder G, Tessier AJ, Qiao XT, et al. Helicobacter-induced intestinal metaplasia in the stomach correlates with Elk-1 and serum response factor induction of villin. J Biol Chem. 2005;280:4906–4912. doi: 10.1074/jbc.M413399200. [DOI] [PubMed] [Google Scholar]
- 33.Giannakis M, Stappenbeck TS, Mills JC, et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem. 2006:28111292–300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- 34.May R, Riehl TE, Hunt C, et al. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630–7. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- 35.Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology. 2009;137:2179–80. doi: 10.1053/j.gastro.2009.06.072. [DOI] [PubMed] [Google Scholar]
- 36.Saqui-Salces M, Keeley TM, Grosse AS, et al. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochem Cell Biol. 2011;136(2):191–204. doi: 10.1007/s00418-011-0831-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brembeck FH, Schreiber FS, Deramaudt TB, et al. The mutant K-ras oncogene causes pancreatic periductal lymphocytic infiltration and gastric mucous neck cell hyperplasia in transgenic mice. Cancer Res. 2003;63:2005–2009. [PubMed] [Google Scholar]
- 38.Mutoh H, Sakurai S, Satoh K, et al. Development of gastric carcinoma from intestinal metaplasia in Cdx2-transgenic mice. Cancer Res. 2004;64:7740–7747. doi: 10.1158/0008-5472.CAN-04-1617. [DOI] [PubMed] [Google Scholar]
- 39.Zavros Y, Eaton KA, Kang W, et al. Chronic gastritis in the hypochlorhydric gastrin-deficient mouse progresses to adenocarcinoma. Oncogene. 2005;24:2354–2366. doi: 10.1038/sj.onc.1208407. [DOI] [PubMed] [Google Scholar]
- 40.Kang W, Rathinavelu S, Samuelson LC, et al. Interferon gamma induction of gastric mucous neck cell hypertrophy. Lab Invest. 2005;85:702–715. doi: 10.1038/labinvest.3700260. [DOI] [PubMed] [Google Scholar]
- 41.Clements WM, Wang J, Sarnaik A, et al. beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62:3503–3506. [PubMed] [Google Scholar]
- 42.Shibata W, Takaishi S, Muthupalani S, et al. Conditional Deletion of IkappaB-Kinase-beta Accelerates Helicobacter-Dependent Gastric Apoptosis, Proliferation, and Preneoplasia. Gastroenterology. 2010;138:1022–1034. doi: 10.1053/j.gastro.2009.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388:1257–1274. doi: 10.1515/BC.2007.159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.