Summary
The ~300 human Cullin-RING ligases (CRLs) are multisubunit E3s in which a RING protein, either RBX1 or RBX2, recruits an E2 to catalyze ubiquitination. RBX1-containing CRLs also can bind Glomulin (GLMN), which binds RBX1’s RING domain, regulates the RBX1-CUL1-containing SCFFBW7 complex, and is disrupted in the disease Glomuvenous Malformation. Here we report the crystal structure of a complex between GLMN, RBX1, and a fragment of CUL1. Structural and biochemical analyses reveal that GLMN adopts a HEAT-like repeat fold that tightly binds the E2-interacting surface of RBX1, inhibiting CRL-mediated chain formation by the E2 CDC34. The structure explains the basis for GLMN’s selectivity toward RBX1 over RBX2, and how disease-associated mutations disrupt GLMN-RBX1 interactions. Our study reveals a mechanism for RING E3 ligase regulation whereby an inhibitor blocks E2 access, and raises the possibility that other E3s are likewise controlled by cellular proteins that mask E2-binding surfaces to mediate inhibition.
Introduction
Ubiquitin (Ub) ligation is highly regulated through the action of specific E1, E2, and E3 enzymes. Via a series of E1-dependent reactions, Ub’s C-terminus becomes covalently linked by a thioester bond to the E2’s active site cysteine. The resulting labile E2~Ub (~ denotes covalent bond) intermediate interacts with an E3 ligase, which recruits the substrate protein and promotes Ub transfer from the E2 to the substrate. Given the importance of ubiquitination in determining the fates of modified targets, it is of great interest to understand structural mechanisms by which E3 ligase activities are controlled.
Much knowledge of E3 regulation derives from investigations of the cullin-RING ligases (CRLs), which account for nearly half the predicted human E3s (Deshaies and Joazeiro, 2009). CRLs are multisubunit enzymes, nucleated by a cullin-RING catalytic core (Zimmerman et al., 2010). A cullin’s N-terminal domain (NTD) assembles with substrate-binding receptors (SRs), such as F-box proteins in the prototypic “SCF” (SKP1-CUL1-F-box protein) CRLs. Crystal structures have revealed how phosphorylation regulates binding of substrates such as CyclinE and p27 to SCFFBW7 and SCFSKP2-CKSHS1, respectively (Hao et al., 2007; Hao et al., 2005; Orlicky et al., 2003; Wu et al., 2003), and how other modifications or co-association with small molecules can activate substrate binding to other SRs (reviewed in (Duda et al., 2011)). Although mechanisms restraining substrate interactions with CRLs are less understood, structures revealed how the Mitotic Checkpoint Complex inhibitor blocks substrate binding to the highly divergent CRL, the Anaphase Promoting Complex (Buschhorn et al., 2011; Chao et al., 2012; da Fonseca et al., 2011; Herzog et al., 2009).
A cullin’s C-terminal domain (CTD) binds a catalytic RING-containing protein, which for cullins 1–4 is RBX1 and for CUL5 is RBX2 (Huang et al., 2009; Kamura et al., 1999b; Kamura et al., 2004; Ohta et al., 1999; Seol et al., 1999; Skowyra et al., 1999; Tan et al., 1999; Zheng et al., 2002b). The CTD comprises: a 4-helix bundle (4HB) linked to the cullin NTD; an α/β subdomain that forms an intermolecular β-sheet with a strand from RBX; and the extreme C-terminal WHB subdomain, which packs against and autoinhibits the RBX protein’s catalytic RING domain (Duda et al., 2008; Yamoah et al., 2008; Zheng et al., 2002b). CRLs are activated by autocatalytic covalent ligation of the Ub-like protein NEDD8 (Calabrese et al., 2011; Duda et al., 2008; Kamura et al., 1999a; Saha and Deshaies, 2008; Yamoah et al., 2008). This is thought to stimulate orientational flexibility of the RBX RING domain so that an associated Ub E2, such as CDC34, can be placed in multiple positions associated with processive polyubiquitination of substrate (Duda et al., 2008; Pierce et al., 2009; Saha and Deshaies, 2008; Wu et al., 2010; Yamoah et al., 2008). Notably, numerous factors have evolved to counteract neddylation: (1) the inhibitor CAND1 binds RBX1-CUL1 both at the SKP1/F-box protein-binding site and at the CUL1 WHB-RBX1 RING junction, simultaneously blocking both cullin-SR interactions and NEDD8 ligation (Goldenberg et al., 2004; Liu et al., 2002; Zheng et al., 2002a); (2) the deneddylating enzyme CSN removes NEDD8 thereby deactivating CRLs (Lyapina et al., 2001); and (3) the Gram-negative pathogenic bacterial effector Cif (cyclin inhibiting factor) inactivates neddylated cullins by deamidating Gln40 on NEDD8 (Cui et al., 2010; Jubelin et al., 2010; Morikawa et al., 2010).
Although structural data have provided important insights into mechanisms activating and inhibiting CRL-substrate binding, assembly, and neddylation (reviewed in (Duda et al., 2011; Zimmerman et al., 2010)), relatively less is known about structural mechanisms by which cellular proteins can attenuate Ub ligase activities of CRLs. We address this through studies of Glomulin (GLMN), a protein principally expressed in vascular smooth muscle cells (McIntyre et al., 2004). In humans, mutations of the GLMN gene cause the disorder Glomuvenous Malformation (GVM), which presents with nonmetastatic cutaneous vascular lesions resembling glomus tumors (Borroni et al., 2011; Brouillard et al., 2002; Brouillard et al., 2005). Prior work showed that the GLMN protein binds RBX1 but not RBX2-associated cullins in cells, that GLMN binds the isolated RBX1 RING domain, and that GLMN inhibits SCFFBW7-mediated ubiquitination of a CyclinE substrate in vitro (Tron et al., 2012). As with some other known CRL regulators, loss of GLMN influences only a subset of CRL activities in vivo (Arai et al., 2003; Tron et al., 2012). Knocking out GLMN in mice results in embryonic lethality due to severe developmental defects that partially mimic effects of ablating FBW7 (Tron et al., 2012). Decreased GLMN activity in cells leads to a striking CRL- and proteasome-dependent reduction in the level of FBW7, and a concomitant increase in levels of SCFFBW7 substrates CyclinE and c-Myc including in GVM patient samples (Tron et al., 2012). In order to gain insights into mechanisms by which GLMN can control SCF activities, we performed crystallographic, biochemical, and biophysical analyses of GLMN interactions with RBX1. Our data collectively reveal that GLMN binds and masks the E2 binding surface of RBX1’s RING domain to inhibit E3 ligase activity.
Results and Discussion
GLMN – a helical repeat protein with a C-terminal platform for binding RBX1’s RING domain
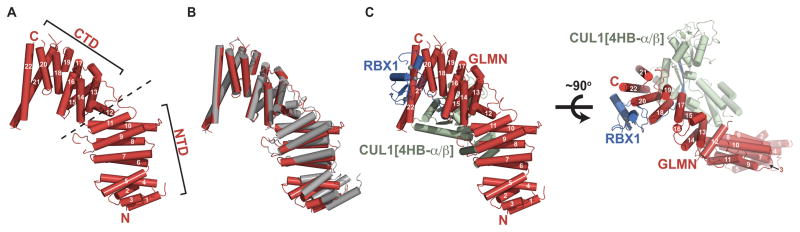
To understand the basis for GLMN-RBX1 interactions, we determined the 3.0 Å crystal structure of a complex between GLMN, RBX1, and a truncated form of CUL1 containing residues 411–690 that spans from the 4HB to the α/β subdomains (hereafter referred to as CUL1[4HB-α/β]), with two complexes per asymmetric unit (Table 1, HFig. 1, Fig. S1). The GLMN structure consists primarily of α-helices arranged as tandem antiparallel pairs, which form a double layer resembling HEAT (untingtin-Elongation-A subunit-TOR) repeats (Fig. 1A). Although HEAT-like repeats are often continuous and form superhelical solenoid structures (Groves and Barford, 1999), in GLMN the continuity is interrupted by a central helix lying roughly perpendicular to both adjacent repeats. As a result, the GLMN HEAT-like repeats are arranged into N-terminal and C-terminal domains, each containing 5 α-helical pairs plus some additional flanking helices. Despite no apparent sequence homology, GLMN’s two domains superimpose on each other with ~3 Å root-mean-square deviation. The connection between GLMN’s N- and C-terminal domains is somewhat flexible, as revealed by a ~10° difference in their relative orientations in the two molecules in the asymmetric unit (Fig. 1B). This agrees with our observations in solution using SAXS described below. Nonetheless, in both cases GLMN adopts a ~110 Å elongated structure, resembling a slightly bent finger.
Table 1.
Crystallographic data and refinement statistics
| Data collection | Native | SeMet |
|---|---|---|
| Wavelength (λ) | 0.979 | 0.979 |
| Space Group | P21 | P21 |
| Unit cell parameters | ||
| a, b, c (Å) | 53.3, 193.9, 142.1 | 53.4, 195.0, 142.0 |
| α, β, γ (°) | 90, 98.8, 90 | 90, 98.9, 90 |
| Resolution (Å) | 50.0-3.0 (3.11-3.00) | 30.0-3.5 (3.62-3.50) |
| No. of measured reflections | 387,866 | 750,853 |
| No. of unique reflections | 56,830 | 35,888 |
| Overall Rsym (%) | 6.9 (68.4) | 15.2 (79.5) |
| Completeness (%) | 99.1 (99.1) | 99.5 (99.3) |
| Overall I/σI | 16.9 (1.3) | 19.1 (2.0) |
| Mean Redundancy | 2.5 | 6.3 |
|
| ||
| Refinement | ||
|
| ||
| Rwork/Rfree | 0.219/0.288 | |
| r.m.s.d. bond lengths (Å) | 0.008 | |
| r.m.s.d. bond angles (°) | 1.284 | |
| Subunits in asymmetric unit | 2 | |
| No. of atoms | ||
| Protein | 13,896 | |
| Zinc | 6 | |
| Ramachandran statistics | ||
| Residues in most favored regions (%) | 92.66 | |
| Residues in allowed regions (%) | 7.28 | |
| Residues in disallowed regions (%) | 0.06 | |
Highest resolution shell is shown in parenthesis. Rwork=Σ|Fo−Fc|/ΣFo. Rfree is the cross validation of R-factor, with 5% of the total reflections omitted in model refinement.
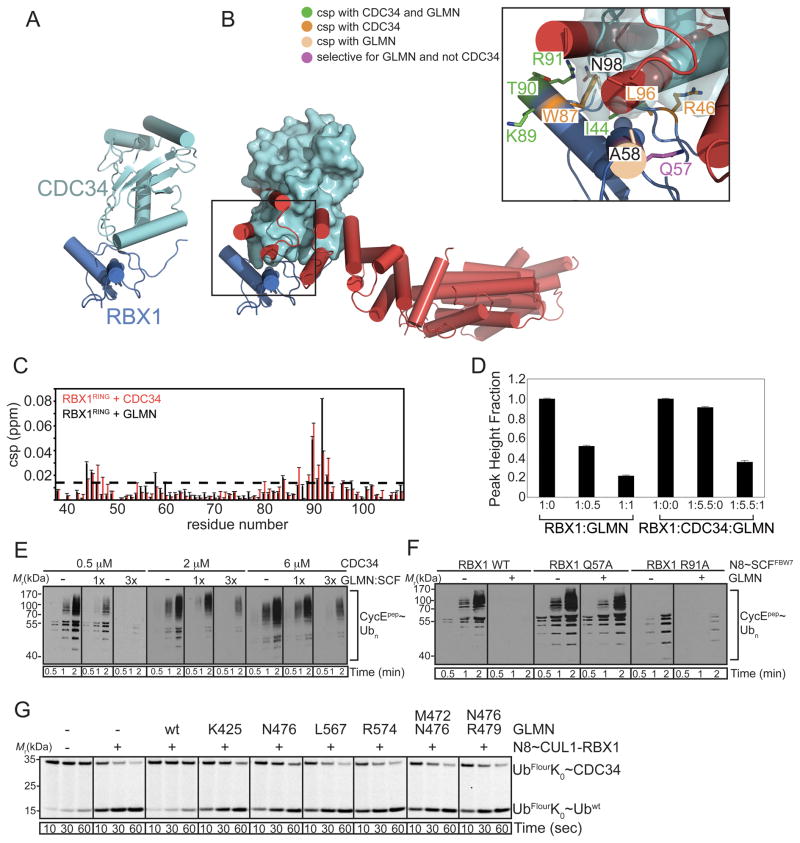
Fig. 1. Structure of a GLMN-RBX1-CUL1[4HB-α/β] complex.
A. The GLMN structure is comprised of helices arranged into tandem HEAT-like repeats, organized into N- and C-terminal domains (NTD and CTD).
B. The two molecules of GLMN in the asymmetric unit superimposed over their CTDs reveal structural flexibility as shown by differences between relative orientations of the NTD and CTD.
C. Structure of one GLMN (red)-RBX1 (blue)-CUL1[4HB-α/β] (green) complex from the asymmetric unit, shown in two orientations rotated ~90° around the x-axis.
The two RBX1-CUL1 portions of the asymmetric unit superimpose on each other. CUL1[4HB-α/β] and RBX1’s β-strand (residues 19–25), embedded in an intermolecular β-sheet within the CUL1 α/β-subdomain, superimpose on all prior CRL structures (Fig. S1H) (Angers et al., 2006; Calabrese et al., 2011; Duda et al., 2008; Fischer et al., 2011; Goldenberg et al., 2004; Zheng et al., 2002b). In isolation, RBX1’s RING domain also superimposes on all previous structures (Fig. S1H). However, in comparison to prior structures, the relative orientations differ for RBX1’s RING domain with respect to RBX1’s strand embedded in the CUL1 α/β-subdomain (Fig. S1I). This is consistent with the previously observed potential for RBX1 RING domain mobility (Calabrese et al., 2011; Duda et al., 2008; Fischer et al., 2011).
A 25 Å × 35 Å surface roughly centered on Ile44 of RBX1’s RING domain is essentially entirely covered by a platform formed by one face of the three C-terminal helical repeats that constitute GLMN’s “fingertip” (Fig. 1C). This agrees with our systematic deletion mutational analysis of GLMN, which mapped this identical region as required for RBX1 RING-binding (Fig. S2). The interaction buries ~880 Å2 from RBX1’s RING domain and ~825 Å2 from GLMN.
High-affinity interactions with the RING domain mediate stoichiometric GLMN binding to RBX1 complexes
In order to interpret functional roles of the GLMN-RBX1-CUL1 interactions, we wished to model GLMN bound to RBX1-CUL1 complexed with other components of SCF E3 complexes. However, in the crystals, GLMN not only contacts RBX1’s RING domain, but also makes some contacts with the linker between RBX1’s β-strand and RING, and also portions of CUL1 (Fig. 1C, S1J). It is not possible to model GLMN interacting with larger SCF complexes simultaneously via all the crystallographically-observed contacts due to differences in relative orientations of RBX1’s RING between prior structures and the complex with GLMN (Angers et al., 2006; Calabrese et al., 2011; Duda et al., 2008; Fischer et al., 2011; Goldenberg et al., 2004; Zheng et al., 2002b). Thus, to determine interactions in solution, we performed several experiments that reveal stoichiometric recruitment of GLMN to a variety of RBX1-containing complexes via direct interactions with the RING domain (Fig. 2).
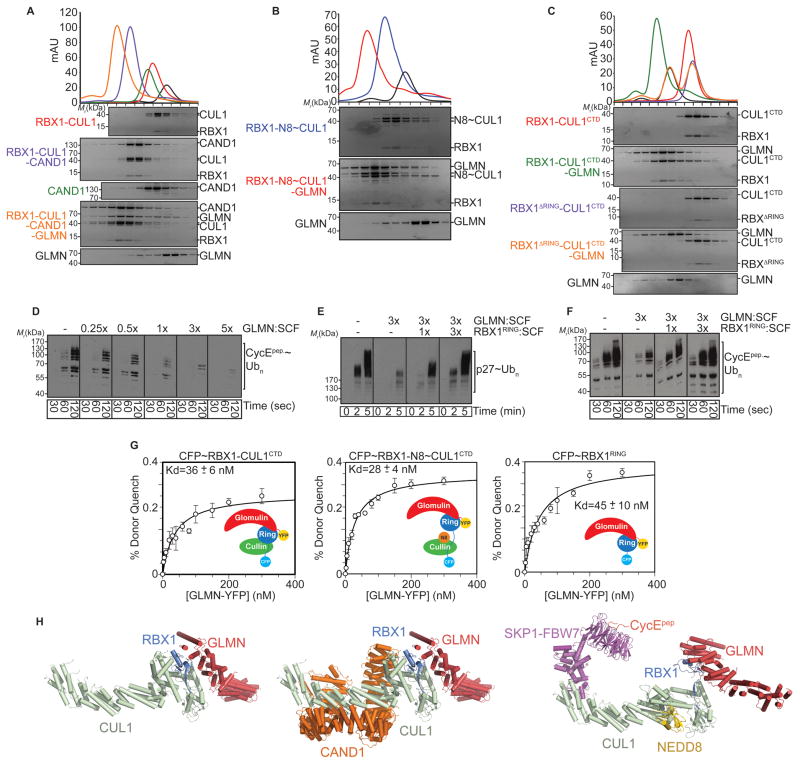
Fig. 2. Stoichiometric incorporation of GLMN into numerous SCF assemblies via RBX1’s RING domain.
A. Gel filtration chromatography elution profiles of GLMN, RBX1-CUL1, and CAND1 alone (black, red, and green, respectively), mixtures of RBX1-CUL1 and CAND1 (purple), and RBX1-CUL1 with CAND1 and GLMN (orange). RBX1-CUL1 corresponds to the “split ‘n coexpress” version in which full-length CUL1 is generated by coexpression of the N- and C-terminal domains as two noncovalently-associated polypeptides that migrate closely on SDS-PAGE (Zheng et al., 2002b). Sypro-stained SDS-PAGE gels of fractions from gel filtration peaks are shown below chromatograms.
B. Gel filtration chromatography elution profiles and Sypro-stained SDS-PAGE gels of fractions for GLMN and RBX1-CUL1~NEDD8 alone (black and blue, respectively), and of a mixture of RBX1-CUL1~NEDD8 and GLMN (red) using “split ‘n coexpress” RBX1-CUL1. These experiments were performed at the same time as those in panel A, with the same data for GLMN shown in both panels for reference.
C. Gel filtration chromatography elution profiles and Sypro-stained SDS-PAGE gels of fractions for GLMN, RBX1-CUL1 C-terminal domain (CTD), and a version lacking the RING domain (RBX1ΔRING) alone (black, red, and blue, respectively), and mixtures of RBX1-CUL1CTD and RBX1ΔRING-CUL1CTD with GLMN (green and orange, respectively).
D. Anti-biotin western blots showing polyubiquitination time-courses of a biotinylated CyclinE phosphopeptide in reactions containing 500 nM CDC34, 200 nM SCFFBW7, and the indicated ratio of GLMN:SCF (1× = 200 nM GLMN).
E. Anti-p27 western blots showing time-courses of SCFSKP2-CKSHS1-mediated polyubiquitination of phospho-p27, in the presence of 3x GLMN:SCF, or with GLMN and in competition assays also containing the indicated ratios of the isolated RBX1 RING domain:SCF.
F. Anti-biotin western blots showing time-courses of SCFFBW7-mediated polyubiquitination of a biotinylated CyclinE phosphopeptide, in the presence of 3x GLMN:SCF and in competition assays containing the indicated ratios of the isolated RBX1 RING domain:GLMN.
G. Quantification of dissociation constant (mean ± SD) from FRET-based binding assay between GLMN-YFP and CFP-RBX1-CUL1CTD, CFP-RBX1-CUL1CTD~NEDD8, or CFP-RBX1RING. Error bars represent standard error from 3 independent experiments (raw data shown in Fig. S3).
H. Structural models of GLMN-RBX1-CUL1, GLMN-RBX1-CUL1-CAND1, and GLMN-RBX1-NEDD8~CUL1-SKP1-FBW7-CyEphosphopeptide based on superimposing GLMN-RBX1 (RING) on the RBX1 RING domain of the prior RBX1-CUL1 (Zheng et al., 2002b) and RBX1-CUL1-CAND1 (Goldenberg et al., 2004) structures, and on the structural model of RBX1-CUL1~NEDD8 (Duda et al., 2008) also modeled with SKP1-FBW7- CyEphosphopeptide docked onto the N-terminal domain of CUL1 (Hao et al., 2007).
First, we considered that RBX1’s RING domain favors different relative orientations in different SCF complexes. Two extremes of RBX1 RING conformation are observed for RBX1-CUL1-CAND1, in which CAND1 rigidifies RBX1’s RING domain and locks it against CUL1’s WHB subdomain, and RBX1-cullin~NEDD8, in which RBX1’s RING domain is apparently more flexibly tethered and rotating (Duda et al., 2008; Goldenberg et al., 2004). Analysis of complex formation by gel filtration chromatography showed that GLMN associates stoichiometrically with the two very distinctive RBX1-complexes that also contain the “split ‘n coexpress” (Zheng et al., 2002b) version of full-length CUL1 (Fig. 2A, 2B).
Second, in order to test dependence of GLMN-cullin interactions on RBX1’s RING domain, we performed gel filtration chromatography of 1:1 mixtures of GLMN with RBX1 complexed with CUL1’s isolated C-terminal domain, or a mutant complex deleted for RBX1’s RING domain. The data show that GLMN forms a stoichiometric complex with RBX1-CUL1CTD, and that the RBX1 RING is necessary for this interaction (Fig. 2C).
Third, we used enzymatic competition assays to qualitatively compare GLMN’s interactions with RBX1’s isolated RING domain versus RBX1 assembled into active CRL complexes. We previously showed that addition of excess GLMN inhibits in vitro ubiquitination of a CyclinE phosphopeptide by SCFFBW7 (Tron et al., 2012). To gain insights into the ratio of GLMN:SCFFBW7 required for inhibition, we performed a titration experiment. Some inhibition is observed at a 1:1 ratio of GLMN:SCF, with more GLMN nearly completely squelching ubiquitination (Fig. 2D). If GLMN functions by binding to RBX1’s RING domain, then in vitro, in the absence of any cellular factors influencing regulation, GLMN would be expected to exert similar effects on other SCFs. Indeed, addition of GLMN also inhibited in vitro SCFSKP2-CKSHS1-mediated ubiquitination of phospho-p27 (Fig. 2E). To probe whether access to RBX1’s RING is important for GLMN-mediated SCF inhibition, we added increasing amounts of the isolated RBX1 RING domain to the assays. This relieves the inhibition, presumably by competing GLMN away from SCFs (Fig. 2E, 2F). This raised the possibility that GLMN binds equally well to RBX1’s RING domain in isolation and in active SCF complexes.
Fourth, to quantitatively compare binding between GLMN and RBX1’s RING domain either in isolation or when RBX1 is associated with a cullin, we developed a FRET-based binding assay, similar to that used previously to measure SCF binding to its cognate Ub E2, CDC34 (Kleiger et al., 2009). In this assay, GLMN-YFP binds to CFP-RBX1-CUL1CTD, CFP-RBX1-CUL1CTD~NEDD8, and CFP-RBX1RING with Kds of 36 ± 6 nM, 28 ± 4 nM, and 45 ± 10 nM, respectively (Fig. 2G). Thus, RBX1’s isolated RING domain is sufficient to bind GLMN with high-affinity, with similar interactions for RBX1-cullin complexes. Although the E2, CDC34, also interacts with RBX1-CUL1~NEDD8 with high affinity (Kleiger et al., 2009), the binding modes differ for GLMN, which requires RBX1’s RING domain (Fig. 2C), and CDC34, which does not (Fig. S3). While RBX1’s RING domain binds weakly to CDC34’s E2 core domain (Spratt et al., 2012), a CRL binds CDC34’s C-terminal acidic tail with high affinity even in the absence of CDC34’s E2 catalytic core domain and RBX1’s RING domain (Fig. S3).
Finally, we mutagenized GLMN residues mediating crystal contacts with CUL1 and observed no effect on inhibition (Fig. S1J, K). This is consistent with GLMN binding to RBX1’s RING domain in a range of orientations relative to CUL1, and with the notion that the observed contacts to CUL1 arose from packing during crystallization.
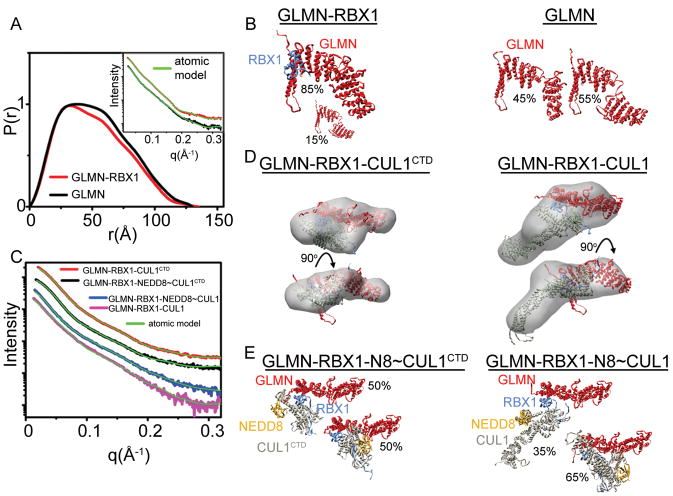
To gain insights into structures of GLMN-RBX1 complexes in solution in the absence of crystal contacts, we performed Small Angle X-ray Scattering (SAXS) studies (Putnam et al., 2007). The broadening in P(r) and Kratky plots indicate higher flexibility for GLMN alone as compared to in complex with RBX1’s RING domain (Fig. 3A, S4). Given the crystallographically-observed conformational flexibility of GLMN (Fig. 1), we reasoned that the coexistence of different GLMN conformations with a hinge between the N- and C-terminal domains could contribute to the experimental SAXS profile. Indeed, an ensemble of two GLMN conformers was required to successfully match the GLMN experimental data (Fig. 3A, 3B, and S4C) (Pelikan et al., 2009). For the GLMN complex with RBX1’s isolated RING domain, an ensemble predominantly corresponding to the GLMN-RBX1RING portion of the crystal and also containing 15% of free GLMN is sufficient to match experimental curve (Fig. 3A, 3B). These data are consistent with GLMN conformational flexibility (Fig. 1B) decreasing upon binding to the RBX1 RING. We also obtained SAXS data for GLMN bound to RBX1 complexes with unneddylated and neddylated “split ‘n coexpress” full-length CUL1 (Zheng et al., 2002b) and CUL1CTD. The data are consistent with GLMN binding the RBX1 RING domain as in our crystal structure, and with prior SAXS analyses of RBX1-CUL1CTD and RBX1-CUL1CTD~NEDD8, which indicated some conformational flexibility in the absence of neddylation, and substantial rotational flexibility of the RBX1 RING domain upon neddylation (Duda et al., 2008). For both unneddylated complexes the SAXS curves and envelopes matched with models obtained by docking GLMN onto the RING domain of RBX1 as in the original RBX1-CUL1 structures (Zheng et al., 2002b), in which the RBX1 RING approaches the CUL1 WHB subdomain (Fig. 2H, 3 and S4). For the neddylated structures, the SAXS data are consistent with a flexible hinge linking the RBX1-RING and GLMN to RBX1’s N-terminal strand and the remainder of CUL1 (Fig. 3C, 3E, and S4).
Fig. 3. SAXS analysis of structural assembly of GLMN with RBX1 complexes.
A. P(r) functions of SAXS data (shown in inset) for GLMN alone (black curve) or in complex with the isolated RBX1 RING domain (red curve). The broadening of the P(r) curve indicates more flexibility for free GLMN. Inset shows the matching of the experimental data and theoretical curves (green curve) obtained for MES models with goodness of fit values of χ2= 2.0 and χ2= 1.4 for GLMN-RBX1 and GLMN alone.
B. MES models for GLMN-RBX1 and GLMN shown with appropriate volume ratios.
C. Experimental SAXS curves with atomic models fits (green curves) for GLMN-RBX1-CUL1CTD : χ= 1.6, GLMN-RBX1-CUL1CTD ~NEDD8: χ= 0.9, GLMN-RBX1-CUL1: χ= 1.6 and GLMN-RBX1-CUL1 ~NEDD8: χ= 1.0. Unneddylated GLMN-RBX1-CUL1 models or MES conformers for neddylated GLMN-RBX1-CUL1 are shown in panels
D and E. D. GLMN-RBX1-CUL1 complexes, with the resulting SAXS envelopes. The SAXS envelopes were fit with models combining the GLMN-RBX1-CUL1[4HB-α/β] structure with the N-terminal domain of CUL1 from a prior structure (1LDJ.pdb) based on superimposing the CUL1 α/β domains from both structures.
E. Two selected conformers for neddylated GLMN-RBX1-CUL1- complexes are shown with appropriate volume ratios as defined by MES (Pelikan et al., 2009).
Taken together, our assembly, competitive inhibition/relief, quantitative binding, and solution structural data indicate that RBX1’s RING domain is both necessary and sufficient to recruit GLMN to numerous SCF ubiquitin ligase assemblies (Fig. 2H).
Details of GLMN-RBX1 RING interactions
The crystal structure reveals that GLMN-RBX1 RING interactions are anchored by two complementary hydrophobic surfaces from both proteins (Fig. 4A). One cluster involves GLMN’s Met472, Leu567, and Val571 packing against RBX1’s Ala43, Ile44, the aliphatic portion of Arg46, Trp87, Pro95, and Leu96, with support from Arg91. A second, smaller hydrophobic cluster lies at the distal end of the complex. Here, Tyr480 and Ile483 from the C-terminal end of the central helix in the GLMN tri-HEAT-like platform interacts with RBX1’s Ile54, Ala58, and hydrophobic portions of Glu55 and Gln57.
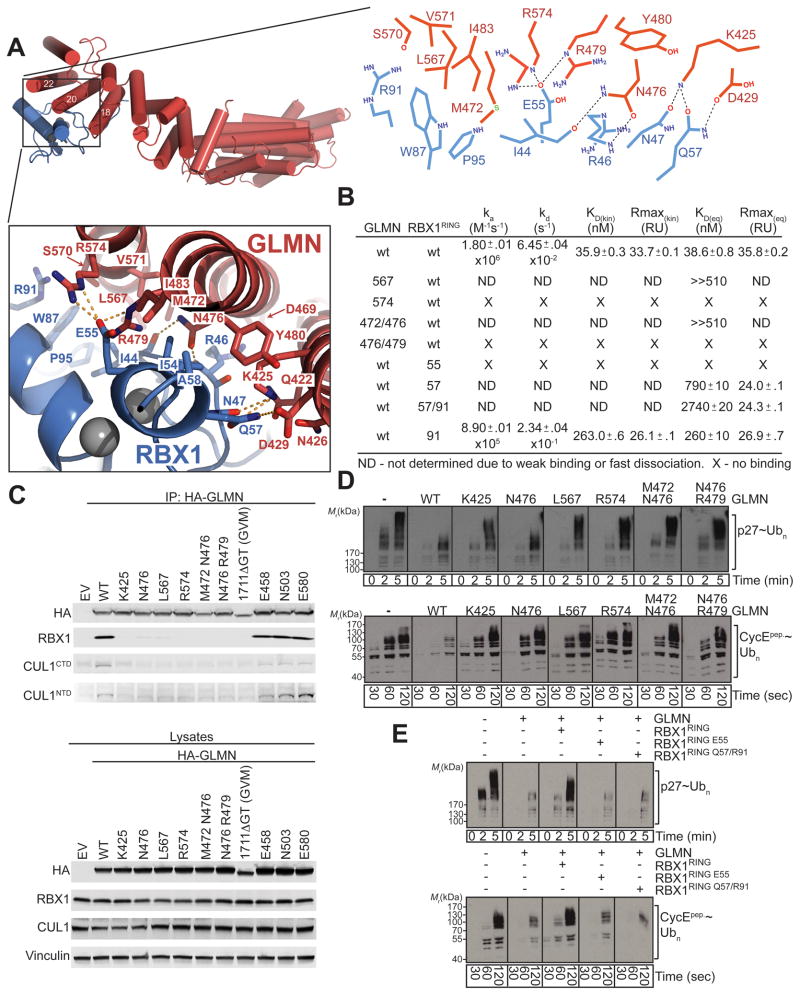
Fig. 4. Details of GLMN-RBX1 RING interactions.
A. Close-up (below) and schematic (right) views of indicated GLMN (red) - RBX1 RING (blue) interface from structure of GLMN-RBX1- CUL1[4HB-α/β] complex. Oxygens are shown in red, nitrogens in blue, sulfurs in green, and zinc atoms in grey.
B. Summary of kinetic and equilibrium data for binding constants measured by Biacore for interactions between wild-type and Ala substitutions for the indicated residues in GLMN and RBX1 RING domain. Standard deviations from curve fits using triplicate data are indicated. ND = Not determined due to weak binding or fast dissociation. X = no detectable binding.
C. Immunoblotting analysis, with antibodies specific for the indicated proteins, of immunoprecipitates (IP) (top) or whole-cell lysates (bottom) derived from HEK293T cells transfected with empty vector (EV), or the indicated HA-tagged GLMN constructs.
D. Top, anti-p27 western blots showing time-courses of SCFSKP2-CKSHS1-mediated polyubiquitination of phospho-p27, and bottom, anti-biotin western blots showing time-courses of SCFFBW7-mediated polyubiquitination of a biotinylated CyclinE phosphopeptide in the presence of wild-type or the indicated Ala substituted mutant versions of GLMN.
E. Top, Anti-p27 western blots showing time-courses of SCFSKP2-CKSHS1-mediated polyubiquitination of phospho-p27, and bottom, anti-biotin western blots showing time-courses of SCFFBW7-mediated polyubiquitination of a biotinylated CyclinE phosphopeptide in the absence or presence of GLMN, or with GLMN mixed 1:1 with either wild-type or mutant versions of the isolated RBX1 RING domain.
The GLMN-RBX1 complex is also stabilized by a vast network of electrostatic interactions, including between GLMN’s Gln422 and Lys425 and RBX1’s Asn47, between GLMN’s Lys425, Asn426, and Asp429 and RBX1’s Gln57, between GLMN’s Asn476 and the backbone oxygen and nitrogen respectively from RBX1’s Ile44 and Arg46, and between GLMN’s Arg479 and Arg574 and RBX1’s Glu55 (Fig. 4A).
We validated the functional importance of the structurally observed contacts between GLMN and RBX1 in four distinct assays. First, we measured binding between wild-type and mutant versions of GLMN and an isolated RBX1 RING domain using Biacore (Fig. 4B, S3). The Kd for wild-type GLMN binding to the RBX1 RING domain measured by Biacore (38.6 +/− 0.8 nM) is similar to that measured in our FRET assay. Ala subsitutions in place of key GLMN interface residues, either individually (Leu567 or Arg574), or pairwise substitutions in place of Asn476 and either Met472 or Asn479, impaired binding. Similarly, mutation of key contact residues from the RBX1 RING domain - Glu55, Gln57, or Arg91 - also decreased binding. Second, we tested the effects of mutations on the ability of HA-tagged GLMN to coimmunoprecipitate RBX1 when expressed in HEK293T cells. Indeed, the structure-based mutations diminished GLMN binding to endogenous RBX1 and its associated CUL1 in cells (Fig. 4C). As controls, the GVM disease-associated frameshift mutation with a 2-base deletion at position 1711 also eliminated the interaction (Tron et al., 2012), and Ala substitutions in GLMN at a variety of surface residues distal from the RBX1-binding surface (Glu458, Asn503, and Glu580) had no effect. Third, for our two different model Ub ligation systems, GLMN’s inhibitory activity was impaired by individual Ala substitutions for RBX1-contacting Lys425, Asn476, Leu567, and Arg574, or pairwise substitutions in place of Asn476 and either Met472 or Asn479 (Fig. 4D). Fourth, in our enzymatic competition assays, mutations in RBX1 that hinder binding to GLMN also impair relief from GLMN-inhibition of SCF E3 activities (Fig. 4E). Thus, the structurally-observed contacts mediate interactions between GLMN and the RBX1 RING domain in solution, and are critical for GLMN inhibition of RBX1-mediated E3 functions.
GLMN binds RBX1’s E2-binding site and prevents RBX1-mediated CDC34 Ub chain formation
Validated for GLMN-RBX1 interactions, the crystal structure allowed us to address how GLMN inhibits SCF activation of CDC34-mediated formation of Ub chains on substrates. We made a model of an RBX1-CDC34 complex by docking based on a homologous RING-E2 structure (Fig. 5A) (Ceccarelli et al., 2011; Zheng et al., 2000). We confirmed the model by examining NMR chemical shift perturbation (csp) upon adding CDC34 to 15N-labeled RBX1 RING domain in a 15N-1H TROSY experiment. In agreement with results published during our manuscript revision (Spratt et al., 2012), resonances displaying significant csp map to the canonical E2-binding site on RBX1 (Fig. 5B, 5C).
Fig. 5. GLMN binds the E2-binding surface of the RING domain and inhibits neddylated CUL1-RBX1-mediated ubiquitin chain synthesis.
A. Structural model of RBX1 (blue) -CDC34 (cyan) generated by superimposing the RING domain from the RING-E2 complex c-CBL-UbcH7 (Zheng et al., 2000) onto the RBX1 RING from RBX1-CUL1[4HB-α/β], and then CDC34 (Ceccarelli et al., 2011) onto UbcH7.
B. Same model, also showing GLMN (red), with surface of CDC34. Inset – close-up showing RBX1 residues for which 15N-1H resonances display chemical shift perturbation (csp) upon adding either CDC34 or GLMN (green), only upon adding CDC34 (orange), only upon adding GLMN (peach), or at the interface with GLMN but not CDC34 (purple).
C. Csp of Rbx1RING resonances upon adding 1 equivalent of GLMN300-594 (black) or 5.5 equivalents of CDC34 (red). Dashed line delineates 2 standard deviations above the mean csp.
D. Normalized average signal intensity from [1H, 15N] TROSY spectra of 15N RBX1RING in isolation, or in the presence of the indicated molar ratios of GLMN300-594, CDC34, or both.
E. Anti-biotin western blots showing polyubiquitination time-courses of a biotinylated CyclinE phosphopeptide in reactions containing 200 nM SCFFBW7, 200 (1x) or 600 (3x) nM GLMN, and 500 nM (10 s exposure), 2 or 6 μM (2 s exposure) CDC34.
F. Anti-biotin western blots showing polyubiquitination time-courses of a biotinylated CyclinE phosphopeptide in the absence or presence of GLMN, and SCFFBW7 containing either wild-type or the indicated mutant versions of RBX1.
G. Phosphorimager data for pulse-chase assays of RBX1-CUL1~NEDD8 activation of CDC34-mediated free Ub chain formation in the absence or presence of the indicated versions of GLMN.
Next, we superimposed our RBX1-GLMN structure onto the RBX1-CDC34 model. The comparison suggests that GLMN completely masks the E2-binding site on RBX1 (Fig. 5B). Because GLMN is a large protein for NMR experiments, we added substoichiometric amounts of a minimal RBX1-binding domain from GLMN, corresponding to residues 300-594, and observed csp for several of the same RBX1 resonances as upon addition of CDC34 (Fig. 5C, S3). Addition of stoichiometric GLMN300-594 led to severe line broadening and decreased signal intensity for RBX1 resonances, presumably due to the elongated high-affinity >40 kDa RBX1RING-GLMN300-594 complex tumbling slowly on the NMR timescale. This occurred even when 1 molar equivalent of GLMN300-594 was added to a 1:5.5 RBX1RING:CDC34 mixture (Fig. 5D). In a related vein, some GLMN inhibition is observed even with a 1:10 ratio of GLMN to CDC34 in our assays (Fig. 5E). Together, the results suggest that GLMN can outcompete excess CDC34 for binding to the RBX1 RING domain.
Consistent with RBX1 using a common surface to bind GLMN and CDC34, an Ala substitution in place of Arg91 in this surface impairs SCF E3 activity, and the remaining activity is not completely eliminated upon adding GLMN (Fig. 5F). However, the structural data also suggest that RBX1’s Gln57 does not bind CDC34, but uniquely binds GLMN. Indeed, a Gln57Ala mutant RBX1 RING domain was impaired for binding to GLMN (Fig. 4B). In the context of a neddylated SCF complex, the RBX1 Gln57Ala mutant shows wild-type E3 activity in our assay, but is not inhibited by GLMN in agreement with the notion that this RBX1 surface binding to GLMN is essential for inhibition (Fig. 5F).
A key intrinsic E3 function of RBX1’s RING domain in the context of neddylated CUL1-RBX1 complexes is to stimulate CDC34’s innate ability to build a Ub chain even in the absence of substrate (Saha and Deshaies, 2008). Indeed, CDC34-mediated di-Ub chain formation is stimulated by addition of RBX1-CUL1CTD~NEDD8 in a pulse-chase assay, and GLMN inhibits this reaction (Fig. 5G). Inhibition by GLMN depends on binding to RBX1’s RING domain, because Ala substitutions in place of GLMN’s RBX1-binding residues hinder its inhibitory activity (Fig. 5G).
Structural basis for GLMN’s specificity for RBX1 over related RINGs
A prior study revealed GLMN as displaying absolute specificity for binding to RBX1 and its associated CUL1-CUL4, but not the nearly 50% identical RBX2 protein or its associated CUL5 in cells (Tron et al., 2012). Furthermore, domain swap experiments showed that GLMN binding in cells depends on the identity of the RBX RING domain. Indeed, we find that GLMN specifically recognizes the RBX1 RING domain, but not the RING domains from RBX2 or the more distal relative APC11 (Fig. S5). The GLMN-RBX1- CUL1[4HB-α/β] crystal structure shows key contact residues differing between RBX1 and RBX2, including RBX1’s Asn47, Glu55, and Arg91 (Fig. S5), which correspond to RBX2’s Val55, Arg63, and Asn96, respectively. Val55 and Asn96 would be unable to make the favorable contacts achieved by their RBX1 counterparts. Furthermore, RBX2’s Arg63 would repel GLMN’s Arg479 and Arg574, which make favorable contacts with RBX1’s Glu55.
GVM disease mutations and implications for GLMN functions in vivo
The GLMN-RBX1-CUL1[4HB-α/β] structure provides a rationale for understanding the effects of mutations associated with Glomuvenous Malformation disease (Borroni et al., 2011; Brouillard et al., 2002; Brouillard et al., 2005). To date, no missense mutations have been found. Instead, most disease mutations result in premature truncations that delete the entire RBX1-binding region (Fig. S2C). The structure explains why RBX1 cannot bind even to the mutant with the most minimal truncation, which deletes GT at position 1711 and generates a frameshift altering the distal 3 ½ turns of the C-terminal helix in GLMN. This both removes key RBX1-binding residues and would also likely decrease stability due to loss of hydrophobic residues that face the adjacent HEAT-like repeat helices. In a related vein, the only in-frame deletion, of Asn393, would lead to destabilization of GLMN’s C-terminal domain, thereby impairing binding to RBX1.
Implications for Ub ligase regulation
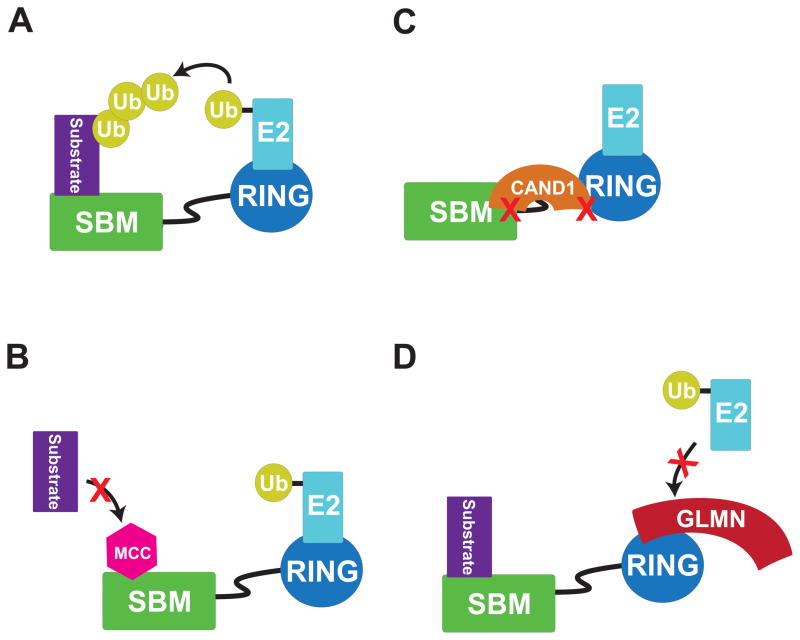
At a rough level, GLMN may be considered to resemble the other CRL inhibitor CAND1, as both proteins are composed of HEAT/HEAT-like repeats (Fig. 1, 2H). However, the two inhibitors bind CRLs differently, and function through entirely distinct mechanisms. CAND1 targets unassembled, unneddylated CRLs at two points, and inhibits CRL assembly and activation in 3 ways: (1) by preventing binding to SR complexes, (2) by blocking the NEDD8 acceptor Lys, and (3) by locking RBX1 against the cullin to prevent rotation of RBX1’s RING domain required for neddylation (Goldenberg et al., 2004). In contrast, a larger RBX1 surface distinct from the CAND1-binding site but encompassing the E2-interacting region binds GLMN (Fig. 5, 6, S6). Thus, unlike CAND1, GLMN binding is entirely compatible with CRL assembly and RBX1 RING domain conformational flexibility, but instead targets CRL catalytic function (Fig. 6).
Fig. 6. Models depicting distinct mechanisms of RING E3 ligase inhibition by cellular factors.
A. Schematic model of a RING E3 ligase, with a substrate-binding module (SBM) recruiting substrate and a RING domain recruiting an E2 for Ub ligation to the associated substrate. The RING is flexibly tethered to the SBM to allow positioning in multiple orientations associated with polyubiquitination.
B. Schematic view of inhibition of APC, a distal cullin-RING E3 family member, by the Mitotic Checkpoint Complex (MCC), which blocks substrate binding.
C. Schematic view of CRL inhibition by CAND1, which blocks RING assembly with the SBM, rotation of the RING domain, and NEDD8 ligation.
D. Schematic view of CRL inhibition by GLMN, which blocks the E2 binding site of the RBX1 RING domain.
Despite their different modes of inhibition, CAND1 and GLMN have some common properties. Both appear to exert broad biochemical effects on multiple RBX1-associated cullins: the presence of CAND1 leads to decreased cullin neddylation in vitro and in vivo (Bennett et al., 2010; Liu et al., 2002; Zheng et al., 2002a). Similarly, the presence of GLMN leads to decreased CRL-mediated neddylation and ubiquitination in vitro, and absence of GLMN leads to diminished levels of cullins 1–4 and RBX1 in vivo (Fig. 2 and (Tron et al., 2012)). Strikingly, loss of either CAND1 or GLMN in vivo appears to affect only selected CRL substrates in terms of biological function. The levels of many well-studied CRL targets appear uninfluenced by loss of CAND1 in either mammalian cells or worms (Bennett et al., 2010; Bosu et al., 2010; Chua et al., 2011). Indeed, only a small fraction of the total cellular pool of CRLs is CAND1-associated, and phenotypes upon CAND1 gene mutation in C. elegans reflect deregulation of SCFLIN-23, but not other CRLs (Bennett et al., 2010; Bosu et al., 2010; Chua et al., 2011). Similarly, GLMN binds only ~7 and 0.2 percent of cellular RBX1 and CUL1, respectively (Fig. S7), and GLMN’s known biological effects seem particularly directed toward SCFFBW7, whose levels are decreased in a CRL- and proteasome dependent manner in the absence of GLMN (Tron et al., 2012). At this point, the reasons why among F-box proteins FBW7 is particularly sensitive to loss of GLMN remain unknown. It is also possible that other F-box proteins are co-regulated by presently unidentified inhibitors that can compensate for loss of GLMN. Future studies will be required to expand our understanding of GLMN’s functions in vivo, whether other CRL substrates are also regulated by GLMN under various cellular settings, and how GLMN-mediated inhibition is overcome.
Although few other cellular inhibitors of E3 ligases have been described, there are many mechanisms maintaining E3s in an “off-state”. For example, a secondary substrate binding site in the RING E3, Ubr1p, is occluded until activation by peptide binding to a primary substrate binding site (Du et al., 2002). Also, both cIAP1 and c-CBL make intramolecular interactions that prevent dimerization and/or E2 binding by their RING domains, with c-CBL being activated by phosphorylation (Dou et al., 2012; Dueber et al., 2011; Kobashigawa et al., 2011; Lopez et al., 2011). Autoinhibition also regulates several HECT E3s, which function by a distinct catalytic mechanism from RING E3s. HECT E3s form a covalent, thioester-linked HECT~ubiquitin intermediate via a catalytic Cys from which Ub is transferred (reviewed in (Kee and Huibregtse, 2007)). Accessibility of the HECT domain active site Cys can be blocked by intra-E3 interactions with domains that mediate cellular localization or substrate binding for active forms of HECT ligases (Gallagher et al., 2006; Wiesner et al., 2007). Finally, in parkin, which is a member of the recently described RING-HECT hybrid class of E3 that contains both a RING domain for E2 binding and a catalytic Cys, an internal Ub-like domain binds near the parkin RING domain and prevents interaction with Ub (Chaugule et al., 2011; Wenzel et al., 2011). Thus, as with CRL autoinhibition that is overcome by neddylation, representative members of all three of the major known cellular E3 classes appear to normally exist in inhibited states, awaiting activation.
This wide-range of mechanisms mediating E3 autoinhibition suggests numerous potential means for inhibition of CRLs and other E3s by trans-acting cellular factors (Fig. 6). We anticipate that many other E3s will eventually be discovered to have modes of regulation that parallel GLMN binding to RBX1, with cellular proteins masking the E2 binding sites on other RING or HECT E3s (Fig. 6). Furthermore, it is an exciting possibility that GLMN may represent the tip of the iceberg in terms of E3 ligase inhibitors mutated in diseases. Recent reports of several small molecules that ultimately inhibit CRLs by diverse mechanisms - through blocking NEDD8 ligation, substrate binding, or CDC34 activity (Ceccarelli et al., 2011; Orlicky et al., 2010; Soucy et al., 2009) - inspire confidence that it may in the future be possible to therapeutically restrain CRL activities that are mis-activated due to mutation of cellular E3 inhibitors in diseases.
Experimental Procedures
X-ray crystallography
We obtained an initial electron density map from crystals of SeMet-labeled GLMN complexed with RBX1-CUL1CTD by MR-SAD after merging 9 different SAD datasets to obtain data nearly complete to 3.5 Å resolution. In addition to 11 of 14 SeMets in the GLMN sequence visible in the SAD map, data from 19 more site-specific SeMets guided structure building. CUL1’s helix 29 and WHB subdomain, which correspond to regions we previously found undergo rotations in CRLs Duda et al., 2008), were not observed in the electron density maps, and thus were deleted in the construct used for final structure refinement at 3 Å resolution. Details are in Supplemental Information.
Binding and enzyme assays
Detailed experimental procedures are in Supplemental Information. Briefly, to measure binding by FRET, equilibrium dissociation constants were obtained from titrations monitoring YFP quenching of a donor CFP signal. Interactions were examined between a C-terminal YFP fusion to GLMN and CFP-RBX1 purified alone, and in complexes with unneddylated and neddylated CUL1CTD, and between C-terminal YFP fusions to CDC34 or CDC34TAIL and RBX1 or RBX1 ΔRING complexed with split- ‘n-coexpress CUL1 C-terminally fused to CFP. The FRET efficiency (E) was determined by taking the maximum fluorescence intensity for the donor (CFP-conjugate) at 475 nm in the presence (FDA) and absence (FD) of acceptor (GLMN- or CDC34- or CDC34TAIL-YFP fusion protein) and solving for E=1−(FDA/FD). Kd was estimated by fitting of the titration curves to a hyperbolic equation assuming a single-site binding model.
For Biacore assays, RBX1’s RING domain was injected over minimally biotinylated GLMN (or mutant) captured on a CM-4 chip coated with covalently immobilized streptavidin. Triplicate injections were made for each concentration of RBX1’s RING domain (or mutant), and the data were processed, double-referenced and analyzed with the software package Scrubber2 (version 2.0b, BioLogic Software). The kinetic rate constants and/or equilibrium dissociation constants were determined by fitting the data to a 1:1 (Langmuir) interaction model.
In enzyme assays, GLMN was incubated with RBX1-CUL1~NEDD8 for 10 minutes prior to mixing with other SCF components and initiating reactions. Polyubiquitination assays were adapted from those described previously (Duda et al., 2008; Jubelin et al., 2010), with either 5 μM of a biotinylated CyclinE phosphopeptide or 0.2 μM phospho-p27 (complexed with CyclinA-CDK2) as substrates, 200 nM neddylated SCFFBW7 or SCFSKP2-CKSHS1, and 500 nM CDC34 for all figures except 5E, where CDC34 concentrations range from 500 nM-6 μM. GLMN was 50 nM-1 μM for Fig. 2D, 200 or 600 nM (1x or 3x relative to SCF) for Fig. 5E, and 600 nM in all other polyubiquitiation assays. SCFSKP2-CKSHS1 polyubiquitination assays also contained 250 nM UbcH5B. In the competition assays, 600 nM GLMN was incubated for 10 min on ice with 200 (1x in Fig. 2E and F) or 600 (3x in Fig. 2E and F and Fig. 4E) nM of the indicated version of the isolated RBX1RING, prior to incubation with RBX1-CUL1~NEDD8.
For pulse-chase di-Ub chain synthesis assays, a charging reaction containing 10 μM CDC34, 20 μM fluorescein-ubiquitin K0 and 150 nM UBA1 was quenched by 5-fold dilution into buffer containing 50 mM EDTA. This was diluted 10-fold into chase mixes containing 500 μM unlabeled native ubiquitin (SIGMA), and where indicated 500 nM RBX1-CUL1~NEDD8 and 2 μM GLMN. CDC34’s ability to synthesize a di-Ub chain was monitored by detecting transfer of fluorescein-K0 Ub from CDC34 to the free Ub.
Supplementary Material
Highlights.
Glomulin (GLMN) binds with ~40 nM Kd to RBX1 but not other related RING E3s
A crystal structure shows GLMN binding RBX1’s E2-interacting surface
Biochemical and NMR data indicate GLMN inhibits RBX1 E3 by blocking E2 access
The structure suggests GVM disease mutations would prevent GLMN binding to RBX1
Acknowledgments
This work was supported by ALSAC, PHS grants 5P30CA021765, R01GM069530 (BAS), RO1CA93804, R01CA63113 and P01CA050661 (JAD), and fellowships from the Pew Charitable Trust and the International Human Frontier Science Program (AET). BAS is an Investigator of the Howard Hughes Medical Institute. We are grateful to DC Scott, DW Miller, DJ Miller Polson, N Brown, P Soule, CO Rock, JW Harper, and EJ Enemark for advice and/or reagents; I Kurinov and C Ralston for synchrotron assistance; P Rodrigues and R Cassell for peptide synthesis and purification; S Bozeman and J Bollinger for administrative/computational support; and R Clubb for providing the sortase clone. NECAT is supported by NIH NCRR RR-15301, APS by US DOE W-31-109-ENG-38, ALS by US DOE DE-AC02-05CH11231. SAXS at the SIBYLS beamline BL12.3.1 of ALS is supported in part by the US DOE Integrated Diffraction Analysis Technologies (IDAT).
Footnotes
Accession code: Coordinates and structural data have been deposited in the Protein Data Bank with accession code 4F52.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Arai T, Kasper JS, Skaar JR, Ali SH, Takahashi C, DeCaprio JA. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni RG, Narula N, Diegoli M, Grasso M, Concardi M, Rosso R, Cerica A, Brazzelli V, Arbustini E. A novel mutation of the glomulin gene in an Italian family with autosomal dominant cutaneous glomuvenous malformations. Exp Dermatol. 2011;20:1032–1034. doi: 10.1111/j.1600-0625.2011.01387.x. [DOI] [PubMed] [Google Scholar]
- Bosu DR, Feng H, Min K, Kim Y, Wallenfang MR, Kipreos ET. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Developmental biology. 2010;346:113–126. doi: 10.1016/j.ydbio.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillard P, Boon LM, Mulliken JB, Enjolras O, Ghassibe M, Warman ML, Tan OT, Olsen BR, Vikkula M. Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations (“glomangiomas”) American journal of human genetics. 2002;70:866–874. doi: 10.1086/339492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillard P, Ghassibe M, Penington A, Boon LM, Dompmartin A, Temple IK, Cordisco M, Adams D, Piette F, Harper JI, et al. Four common glomulin mutations cause two thirds of glomuvenous malformations (“familial glomangiomas”): evidence for a founder effect. J Med Genet. 2005;42:e13. doi: 10.1136/jmg.2004.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nature structural & molecular biology. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese MF, Scott DC, Duda DM, Grace CR, Kurinov I, Kriwacki RW, Schulman BA. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nature structural & molecular biology. 2011;18:947–949. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. Autoregulation of Parkin activity through its ubiquitin-like domain. The EMBO journal. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua YS, Boh BK, Ponyeam W, Hagen T. Regulation of cullin RING E3 ubiquitin ligases by CAND1 in vivo. PLoS ONE. 2011;6:e16071. doi: 10.1371/journal.pone.0016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Yao Q, Li S, Ding X, Lu Q, Mao H, Liu L, Zheng N, Chen S, Shao F. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science (New York, NY. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nature structural & molecular biology. 2012 doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Current opinion in structural biology. 2011;21:257–264. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EC, Schoeffler AJ, Lingel A, Elliott JM, Fedorova AV, Giannetti AM, Zobel K, Maurer B, Varfolomeev E, Wu P, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science (New York, NY. 2011;334:376–380. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- Fischer ES, Scrima A, Bohm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, et al. The Molecular Basis of CRL4(DDB2/CSA) Ubiquitin Ligase Architecture, Targeting, and Activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1717–1722. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Groves MR, Barford D. Topological characteristics of helical repeat proteins. Current opinion in structural biology. 1999;9:383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Molecular cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science (New York, NY. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Molecular cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin G, Taieb F, Duda DM, Hsu Y, Samba-Louaka A, Nobe R, Penary M, Watrin C, Nougayrede JP, Schulman BA, et al. Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes & development. 1999a;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science (New York, NY. 1999b;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes & development. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochemical and biophysical research communications. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20579–20584. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Molecular cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Lopez J, John SW, Tenev T, Rautureau GJ, Hinds MG, Francalanci F, Wilson R, Broemer M, Santoro MM, Day CL, et al. CARD-mediated autoinhibition of cIAP1’s E3 ligase activity suppresses cell proliferation and migration. Molecular cell. 2011;42:569–583. doi: 10.1016/j.molcel.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science (New York, NY. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- McIntyre BA, Brouillard P, Aerts V, Gutierrez-Roelens I, Vikkula M. Glomulin is predominantly expressed in vascular smooth muscle cells in the embryonic and adult mouse. Gene Expr Patterns. 2004;4:351–358. doi: 10.1016/j.modgep.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Morikawa H, Kim M, Mimuro H, Punginelli C, Koyama T, Nagai S, Miyawaki A, Iwai K, Sasakawa C. The bacterial effector Cif interferes with SCF ubiquitin ligase function by inhibiting deneddylation of Cullin1. Biochemical and biophysical research communications. 2010;401:268–274. doi: 10.1016/j.bbrc.2010.09.048. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Molecular cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Molecular cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & development. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science (New York, NY. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Spratt DE, Wu K, Kovacev J, Pan ZQ, Shaw GS. Selective recruitment of an E2~ubiquitin complex by an E3 ubiquitin ligase. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M112.353748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Molecular cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Tron AE, Arai T, Duda DM, Kuwabara H, Olszewski JL, Fujiwara Y, Bahamon BN, Signoretti S, Schulman BA, DeCaprio JA. The Glomuvenous Malformation Protein Glomulin Binds Rbx1 and Regulates Cullin RING Ligase-Mediated Turnover of Fbw7. Molecular cell. 2012;46:67–78. doi: 10.1016/j.molcel.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Molecular cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Molecular cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Molecular cell. 2002a;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002b;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Current opinion in structural biology. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.