Abstract
We describe a crowd-sourcing based solution for handling large quantities of data that are created by e.g., emerging digital imaging and sensing devices, including next generation lab-on-a-chip platforms. We show that in cases where the diagnosis is a binary decision (e.g., positive vs. negative, or infected vs. uninfected), it is possible to make accurate diagnosis by crowd-sourcing the raw data (e.g., microscopic images of specimens/cells) using entertaining digital games (i.e., BioGames) that are played on PCs, tablets or mobile phones. We report the results and the analysis of a large-scale public BioGames experiment toward diagnosis of malaria infected human red blood cells (RBCs), where binary responses from approximately 1000 untrained individuals from more than 60 different countries are combined together (corresponding to more than 1 million cell diagnoses), resulting in an accuracy level that is comparable to those of expert medical professionals. This BioGames platform holds promise toward cost-effective and accurate tele-pathology, improved training of medical personnel, and can also be used to manage the “Big Data” problem that is emerging through next generation digital lab-on-a-chip devices.
The internet revolution can be traced back to the invention of the transistor. It was the integration of transistors into electronic chips, and subsequently into computational systems that led to the development of computers and the internet. For over three decades, the number of transistors in integrated circuits has been doubling approximately every 18 months, a trend that is dubbed as the Moore’s Law. Quite interestingly, a similar trend seems to exist for the number of pixels installed on mobile phone cameras. In other words the megapixel count of mobile phones has been following Moore’s Law since the wide-scale introduction of camera phones in 2002 with 0.3 megapixels, reaching 41 megapixels in 2012.1 The massive volume of these consumer electronics components, reaching billions, has brought a phenomenal reduction in cost and unprecedented levels of access to such advanced digital devices despite their sophisticated hardware and software capabilities.
As a result of this revolution in digital electronics, we are now seeing a paradigm shift in imaging and sensing technologies with ultra-portable, cost-effective and high-throughput lab-on-a-chip platforms providing innovative solutions for e.g., point-of-care and telemedicine applications, among others.2–18 Cost-effectiveness and portability of these emerging digital imaging and sensing technologies through lab-on-a-chip platforms will allow wide-scale generation of large amounts of biomedical and environmental data even in traditionally resource-limited and remote parts of the world. The richness of this data will surely provide new opportunities for better understanding various phenomena such as global spatio-temporal patterns of various diseases and health conditions, helping us relate such patterns to their causes, potentially also influencing our global health and environmental policies. On the other hand, such massive amounts of data that will be continuously created by the next generation digital imaging, sensing, and lab-on-a-chip technologies will also add to our “Big Data” problem, which can fundamentally be related to the fact that the performance, training, and the number of available human experts (e.g., medical personnel, data analysts) do not scale as fast as Moore’s Law.
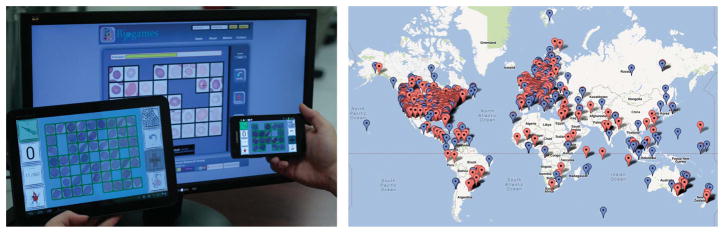
In this work, we introduce a platform that aims to tackle this emerging Big Data problem through entertaining games that crowd-source the digital data of interest (e.g., microscopic images of specimens) to minimally trained humans/gamers for diagnostic decision making or labelling. These digital games (termed BioGames – see Fig. 1) can be played all around the world on PCs, tablets or mobile phones. The individual responses of the gamers are collected using secure servers for analysis and decoding of the final decisions and the labelling of the original data.
Fig. 1.
Left: The designed game can be played on multiple platforms. The user is asked to kill or bank infected and healthy cells, respectively. Right: Geographic locations of gamers that have generated the diagnoses so far; each balloon indicates an individual gamer. A blue balloon indicates a gamer with more than 100 submitted cell diagnoses, and a red balloon indicates a gamer with less than 100 submitted cell diagnoses. Since its public launch in May 2012, we have had more than 2,150 gamers from 77 countries, who registered on our servers generating more than 1.5 million individual cell diagnoses.
At the centre of the BioGames platform lies crowd-sourcing, which has recently emerged as a powerful strategy for tackling computationally difficult scientific problems.19–24 The basic idea behind a crowd-sourcing platform is to first break the task of interest into smaller pieces that can each be completed in a relatively short amount of time; and second to distribute these pieces of the problem to individual humans through a convenient medium, such as the internet; and finally to combine the collective responses of the individual participants to yield an optimal solution. A promising approach to distributing the data and finding human volunteers has been the use of entertaining computer games.21,25,26 In this approach the scientific or computational problem is embedded into a digital game and then distributed to gamers. The individual gamers may then cooperate, compete, or play independently to solve parts or the entire problem of interest.
Along the same lines, we have recently proposed a crowd-sourcing-based solution for diagnosing or labelling bio-medical images.24 In this small scale experiment (involving ~30 gamers), we showed that it is conceivable to achieve accurate diagnosis results by asking multiple individuals to label human RBC images that are potentially infected with the malaria parasite Plasmodium falciparum. In this manuscript, we expand on our earlier work and report the results and the related analysis of a large-scale public BioGames experiment toward diagnosis of malaria infected human RBCs, where binary responses from approximately 1000 minimally trained individuals from 6 different continents that have collectively diagnosed more than 1 million RBCs are combined, resulting in a diagnostic accuracy level that is comparable to those of expert medical professionals examining the same set of images (See Fig. 1).
We chose malaria for this public BioGames experiment since it is a disease that still afflicts a large number of people around the globe, and is most prevalent in impoverished and remote locations of the world. Furthermore, the binary nature of malaria diagnosis makes it suitable for the proposed BioGames approach. According to the World Health Organization, there were an estimated 174 million cases of malaria in 2010 resulting in 655 000 deaths, where 81% of the cases and 91% of the deaths occurred in Africa.27 Furthermore, approximately 86% of the global death toll has been in children under the age of 5.
Toward diagnosis of malaria using BioGames, we have designed a computer game in which a gamer is presented with multiple game frames containing a grid of RBC images (see Fig. 1). These images are taken from Giemsa stained thin blood smear samples using traditional bright-field microscopes with 100× objective-lenses. The Giemsa stain typically causes the malaria parasite to appear with a bluish colour, helping its diagnosis. Each gamer is given a brief online tutorial of what malaria infection looks like and is asked to kill or bank what s/he thinks are infected and healthy cells respectively in the frames that are presented throughout the game. Within each frame, we also include a set of control cells, i.e., RBC images whose labels (infected or healthy) are known to the game. The purpose of these control images is to allow us to determine the performance and error rates of each individual gamer as s/he is going through the frames. We use these performance numbers when combining and weighing the responses coming from multiple gamers. Stated differently, those gamers who are found to be highly accurate through these control images will be given more weight in the decision process compared to other gamers who perform poorly during the same games.
In our decoding strategy, we take a tele-communications analogy and start by assigning the labels 0 and 1 to correspond to healthy and infected cells, respectively. Using the control images, we compute the error probabilities associated with each gamer as where is the label that gamer k has provided for the ith cell image, xi is the true label of the image, and l ∈ {0,1}. Assuming we have M gamers and a prior , then we can compute the maximum a posteriori probability (MAP)24 estimate zi for the ith RBC as:
| (1) |
Given that we do not necessarily know the parasitemia level of the RBCs presented to the gamers, we assume equal prior probabilities for the infected and healthy cells, thus making the first term in the maximisation argument in eqn (1) irrelevant. Based on this decoding scheme, each gamer is essentially treated as a “repeater” in a noisy tele-communication channel, and using a MAP scheme we estimate the correct label for each unknown image in the game.24
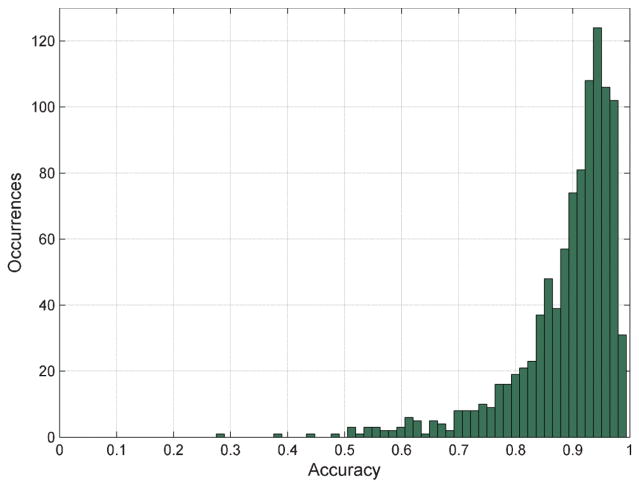
We made our BioGames interface available to the public on May 3, 2012, and as of August 4, 2012 we have had more than 2150 gamers from 77 countries, who registered on our servers generating more than 1.5 million individual cell diagnoses. Our image database contained approximately 8500 individual RBC images taken with different optical microscopes under various imaging conditions. Since labelling the full dataset (i.e., 8500 images) takes more time than what most gamers are willing to commit on average, it was expected that not all the gamers will diagnose the full set of images. For the purposes of data analysis and diagnosis of unknown RBC images, we only used the data collected from the gamers who managed to at least label 100 cells. This threshold is mainly to have sufficient statistics for estimating the gamer performance levels, but it also helps us to identify and select the “committed” gamers, yielding a total of 989 committed gamers (from 63 countries) with varying accuracy levels as shown in Fig. 2.
Fig. 2.
Histogram of the overall accuracy levels of the 989 individual gamers who managed to label more than 100 cell images.
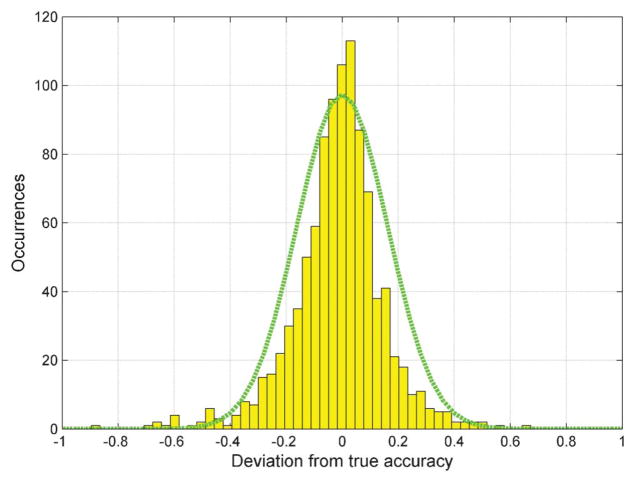
The ground truth labels for the images used in BioGames were obtained by asking a set of 9 medical experts to digitally label them and then taking their consensus responses. Based on this, we were able to quantify the individual performances of our gamers as well as their collective MAP decision estimates. We monitored the performance of the individual gamers using every 25th cell in this database as part of our control images. Fig. 3 shows the deviation of these accuracy estimates (based on control images) from the “true” accuracies of the gamers (based on all 8500 images). The distribution of these deviations is super-Gaussian and is concentrated close to zero, which implies that our control images (even though they are quite rare within the entire image set) provide sufficiently accurate sampling to represent each gamer’s overall error probability.
Fig. 3.
Distribution of the deviation from the true accuracies of those estimated using the control images; the mean is at −0.014 and the variance is 0.026. The green curve is that of a Normal distribution with the same variance as the deviations and a mean of 0.
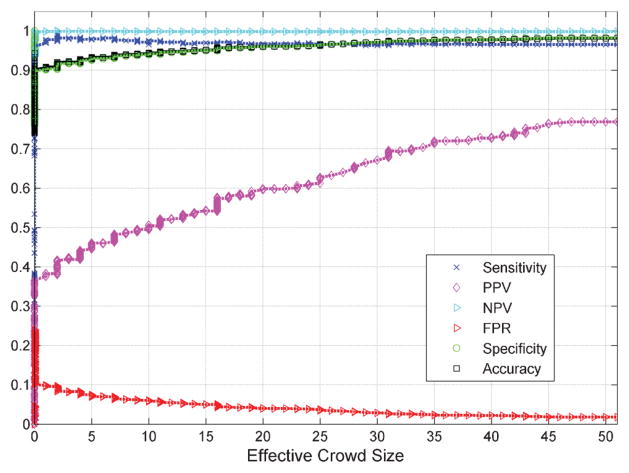
Combining the responses of these 989 gamers using MAP estimation,24 we were able to achieve an accuracy of 98.13% when compared to the ground truth data (generated by the consensus of 9 medical experts). This number on its own, however, can be misleading due the fact that there are significantly more uninfected cells in the database as compared to infected ones (which is naturally expected since typical parasitemia for a malaria infected patient is less than 5%). Some other important metrics include: the Positive Predictive Value (PPV), the Negative Predictive Value (NPV), and the False Positive Rate (FPR) (see Fig. 4 for definitions). In our BioGames experiments, the PPV was 76.85%, meaning that more than three quarters of the cells that were labelled as infected, were indeed infected. We also achieved an NPV of 98.78%, such that almost all of the cells labelled as negative are correctly labelled as such.
Fig. 4.
The performance results obtained through combining responses of 989 gamers. The “effective crowd size” is the minimum number of times that every cell in the dataset has been diagnosed. Accuracy ≜ (TP + TN)/(TP + TN + FP + FN), Sensitivity ≜ TP/(TP + FN), Specificity ≜ TN/(TN + FP), PPV ≜ TP/(TP + FP), NPV ≜ TN/(TN + FN), FPR ≜ FP/(TN + FP), where TP, TN, FP, and FN correspond to the number of true positive, true negative, false positive, and false negative labels respectively.
In Fig. 4 we also report the change of the above discussed performance metrics as a function of the effective crowd size, which we define as the minimum number of times that all of the cells in the dataset have been diagnosed by the gamers. Since not all of the gamers diagnosed the complete set of images, the maximum effective crowd size that was achieved in our public BioGames experiment was 51 with some cells being diagnosed up to 450 times due to the overlap that existed among the ranges of cells diagnosed by different gamers. It is important to note that both the overall accuracy and the PPV are increasing as the effective size of the crowd increases. Furthermore, as desired, the FPR is consistently decreasing with larger crowd sizes (see Fig. 4). These experimental results validate the efficacy of our BioGames approach toward obtaining highly accurate diagnostics results and image labels with large-enough crowds of minimally trained individuals, that are physically scattered across several continents.
We should also emphasise that this distributed BioGames platform can be extended to scenarios where a binary diagnosis is no longer the case. It is possible to systematically combine results of n-ary decisions in an optimal fashion to produce a more accurate overall result/decision. However, a possible issue in such a more complicated n-ary scenario is the increased number of crowd decisions that may be required for accurate diagnosis, making certain tasks (especially the ones that demand real-time decisions) impractical for crowd-sourcing purposes.
Looking forward, the BioGames concept could help us better manage the Big Data problem emerging with the introduction of next generation imaging, sensing and lab-on-a-chip devices which all benefit from ubiquitous digital communications technologies (e.g., mobile phones, tablet PCs, etc) for wide-scale generation of massive amounts of biomedical and environmental data, even in resource poor settings and remote locations. Through intelligent crowd-sourcing and digital gaming strategies, we can potentially harness the power of human crowds and their innate visual pattern recognition and learning abilities to better sort, classify, diagnose, and manage this emerging Big Data.
Our grand vision is to further develop this BioGames interface into a generic telemedicine and telepathology platform, further extending it to other medical diagnostics tasks. In addition to generating remote biomedical diagnosis through engaging games, the presented platform can serve as an information hub for the global healthcare and environmental research community. This digital hub will allow for the creation of very large databases of microscopic images that can be used for the purposes of training experts and automated computer vision algorithms. It can also serve as an analysis tool for health-care and environment related policy makers toward better management and/or prevention of epidemics, pandemics and disasters.
In conclusion, here we described how the task of binary image-based diagnosis can be crowd-sourced to minimally trained individuals and yet yield accurate results. We reckon that under many circumstances this methodology is more practical than the use of automated computer algorithms for the same purpose since the human visual system offers a very low-cost and highly superior pattern recognition platform for such image understanding tasks. We demonstrated our BioGames results for images taken with traditional bright-field microscopes, but given that the task of diagnosis is independent of the imaging modality, as long as enough detail and resolution is present in the image, the proposed approach is applicable to other imaging modalities. This makes it a viable approach for managing the large quantities of data that will be created by our next generation lab-on-a-chip imaging and sensing devices that are rapidly emerging for especially point-of-care diagnostics and telemedicine applications through ubiquitous digital communications platforms such as mobile phones, tablet PCs etc.
Acknowledgments
A. Ozcan gratefully acknowledges the support of the Presidential Early Career Award for Scientists and Engineers (PECASE), ARO Young Investigator Award, NSF CAREER Award, the ONR Young Investigator Award 2009 and the NIH Director’s New Innovator Award DP2OD006427 from the Office of The Director, NIH.
Footnotes
References
- 1.Nokia. Nokia 808 Pureview. 2012 http://europe.nokia.com/find-products/devices/nokia-808-pureview.
- 2.Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. PLoS One. 2009;4:e6320. doi: 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tseng D, Mudanyali O, Oztoprak C, Isikman SO, Sencan I, Yaglidere O, Ozcan A. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishara W, Su T-W, Coskun AF, Ozcan A. Opt Express. 2010;18:11181–11191. doi: 10.1364/OE.18.011181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller AR, Davis GL, Oden ZM, Razavi MR, Fateh A, Ghazanfari M, Abdolrahimi F, Poorazar S, Sakhaie F, Olsen RJ, et al. PLoS One. 2010;5:e11890. doi: 10.1371/journal.pone.0011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith ZJ, Chu K, Espenson AR, Rahimzadeh M, Gryshuk A, Molinaro M, Dwyre DM, Lane S, Matthews D, Wachsmann-Hogiu S. PLoS ONE. 2011;6:11. doi: 10.1371/journal.pone.0017150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isikman SO, Bishara W, Sikora U, Yaglidere O, Yeah J, Ozcan A. Lab Chip. 2011;11:2222–2230. doi: 10.1039/c1lc20127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Yaglidere O, Su T-W, Tseng D, Ozcan A. Lab Chip. 2011;11:315–322. doi: 10.1039/c0lc00358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Mavandadi S, Coskun AF, Yaglidere O, Ozcan A. Anal Chem. 2011;83:6641–6647. doi: 10.1021/ac201587a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenbaum A, Sikora U, Ozcan A. Lab Chip. 2012;12:1242–1245. doi: 10.1039/c2lc21072j. [DOI] [PubMed] [Google Scholar]
- 11.Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Lab Chip. 2012;12:2678. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 13.Sia SK, Linder V, Parviz BA, Siegel A, Whitesides GM. Angew Chem, Int Ed. 2004;43:498–502. doi: 10.1002/anie.200353016. [DOI] [PubMed] [Google Scholar]
- 14.Martinez AW, Phillips ST, Carrilho E, Thomas SW, Sindi H, Whitesides GM. Anal Chem. 2008;80:3699–3707. doi: 10.1021/ac800112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schatz B, Berlin R, Richard J, Berlin B. Healthcare Infrastructure: Health Systems for Individuals and Populations. Springer; 2011. [Google Scholar]
- 16.McKlernan G. Searcher: The Magazine for Database Professionals. 2010;18:48. [Google Scholar]
- 17.Adler R. California Healthcare Foundation. 2007. [Google Scholar]
- 18.Hurling R, Catt M, Boni MD, Fairley BW, Hurst T, Murray P, Richardson A, Sodhi JS. J Med Internet Res. 2007;9:e7. doi: 10.2196/jmir.9.2.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Von Ahn L, Maurer B, McMillen C, Abraham D, Blum M. Science. 2008;321:1465–1468. doi: 10.1126/science.1160379. [DOI] [PubMed] [Google Scholar]
- 20.Roman D. Commun ACM. 2009;52:12–12. [Google Scholar]
- 21.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, Leaver-Fay A, Baker D, Popovic Z, Players F. Nature. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper S, Baker D, Popovic Z, Treuille A, Barbero J, Leaver-Fay A, Tuite K, Khatib F, Snyder AC, Beenen M, et al. Proceedings of the Fifth International Conference on the Foundations of Digital Games - FDG ‘10; 2010. pp. 40–47. [Google Scholar]
- 23.Khatib F, Cooper S, Tyka MD, Xu K, Makedon I, Popovic Z, Baker D, Players F. Proc Natl Acad Sci U S A. 2011;108:18949–18953. doi: 10.1073/pnas.1115898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavandadi S, Dimitrov S, Feng S, Yu F, Sikora U, Yaglidere O, Padmanabhan S, Nielsen K, Ozcan A. PLoS One. 2012;7:e37245. doi: 10.1371/journal.pone.0037245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Ahn L, Dabbish L. Commun ACM. 2008;51:58–67. [Google Scholar]
- 26.EteRNA. EteRNA—Played by Humans, Scored by Nature. 2011 http://eterna.cmu.edu/content/EteRNA.
- 27.WHO. Who, World. 2011. p. 278. [Google Scholar]