Abstract
The cbb3-type cytochrome c oxidases are members of the heme-copper proton pumping respiratory oxygen reductases. The structure of the cbb3-type oxidase from Pseudomonas stutzeri reveals that, in addition to the six redox-active metal centers (two hemes b, three hemes c and CuB), the enzyme also contains at least one Ca2+. The calcium bridges two propionate carboxyls at the interface between the low-spin heme b and the active-site heme b3 and, in addition, is ligated to a serine in subunit CcoO and by a glutamate in CcoN. The glutamate that is ligated to Ca2+ is one of a pair of glutamic acid residues that has previously been suggested to be part of a proton exit pathway for pumped protons. In the current work, mutants in these glutamates are investigated in the cbb3-type oxidases from Vibrio cholerae and from Rhodobacter sphaeroides. Metal analysis shows that each of these wild type enzymes contains Ca2+. Mutations of the glutamate expected to ligate the Ca2+ in each of these enzymes (E126 in V. cholerae; E180 in R. sphaeroides) result in the loss of activity as well as loss of Ca2+. Mutations in the nearby glutamate (E129 in V. cholerae; E183 in R. sphaeroides) also resulted in loss of oxidase activity and loss of Ca2+. It is concluded that the Ca2+ is essential for assembly of the fully functional enzyme and that neither of the glutamates is likely to be part of a pathway for pumped protons within the cbb3-type oxygen reductases. A more likely role for these glutamates is the maintenance of the structural integrity of the active conformation of the enzyme.
Keywords: cbb3–type Cytochrome c oxidase, Calcium binding, Rhodobacter sphaeroides, Vibrio cholerae
Introduction
The heme-copper superfamily includes respiratory oxygen reductases, which are proton pumps, as well as NO reductases, which are not pumps (1–4). The superfamily is defined by sequence homology within the integral membrane subunit which contains the catalytic bimetallic center where either O2 is reduced to water or NO is reduced to N2O. In the case of the oxygen reductases, the active site is comprised of a high spin heme and a nearby copper, called CuB. For the NO reductases, the bimetallic center is a high spin heme and a nearby Fe, called FeB. All of the members of the superfamily also contain a second, low spin heme within the same subunit which is required for electron transfer into the active site.
The respiratory oxygen reductases can be further subdivided into three major families, denoted the A, B and C families (1–3, 5–7). The two families of NO reductases that have been biochemically and structurally characterized, called the cNOR and qNOR families (1, 4, 8–11), are phyllogenetically most closely related to the C-family of oxygen reductases (12–16). The qNOR and cNOR enzymes are very closely related to each other (4, 8, 9, 17), but the cNOR enzymes use a cytochrome c as an electron donor (4), whereas qNORs use a quinol substrate (10).
It has been noted that in sequence alignments, the cbb3-type oxygen reductases, along with the cNOR and qNOR families have in common a pair of conserved glutamic acid residues that were predicted, prior to the recently available X-ray structures (10, 11), to be near the periplasmic surface of the proteins (18, 19). Site-directed mutagenesis on the cNOR from Paracoccus denitrificans showed that these glutamates are each necessary for function but not for assembly of the enzyme (18, 20, 21). Similar experiments with the cbb3-type oxidases from both Vibrio cholerae and Rhodobacter sphaeroides also indicate that these glutamates are important for function (7). It was suggested that these glutamates might function as part of a proton pathway in the NO reductases to deliver protons to the active site from the periplasm and, in the cbb3-type oxidases, to deliver pumped protons in the opposite direction, from the active site to the periplasm.
X-ray structures are now available for the cbb3-type oxygen reductase from Pseudomonas stutzeri (22), the cNOR from Pseudomonas aeruginosa (11) and the qNOR from Geobacillus stearothermophilus (10). The qNOR is in an inactive form with Zn2+ bound in place of the non-heme FeB. In each case, there is a single Ca2+ that bridges between propionates of the low spin and high spin heme components and, in addition, is ligated to one of the glutamates (e.g., E122 in the P. stutzeri cbb3; E126 in the V. cholerae cbb3; E180 in the R. sphaeroides cbb3; E135 in the P. aeruginosa cNOR; E429 in the G. stearothermophilus qNOR). In each of the structures, the second glutamate of each pair, located 3 residues on the C-terminal side, is not ligated to the Ca2+ but is engaged in a network of hydrogen bonds, more clearly shown in the higher resolution structures of the NO reductases. In the structure of the cbb3-type oxygen reductase, there is extra electron density near (but not adjacent to) the second glutamate which has been suggested as a possible second Ca2+ bound to the enzyme, but there is no indication of a second Ca2+ in the qNOR or cNOR structures.
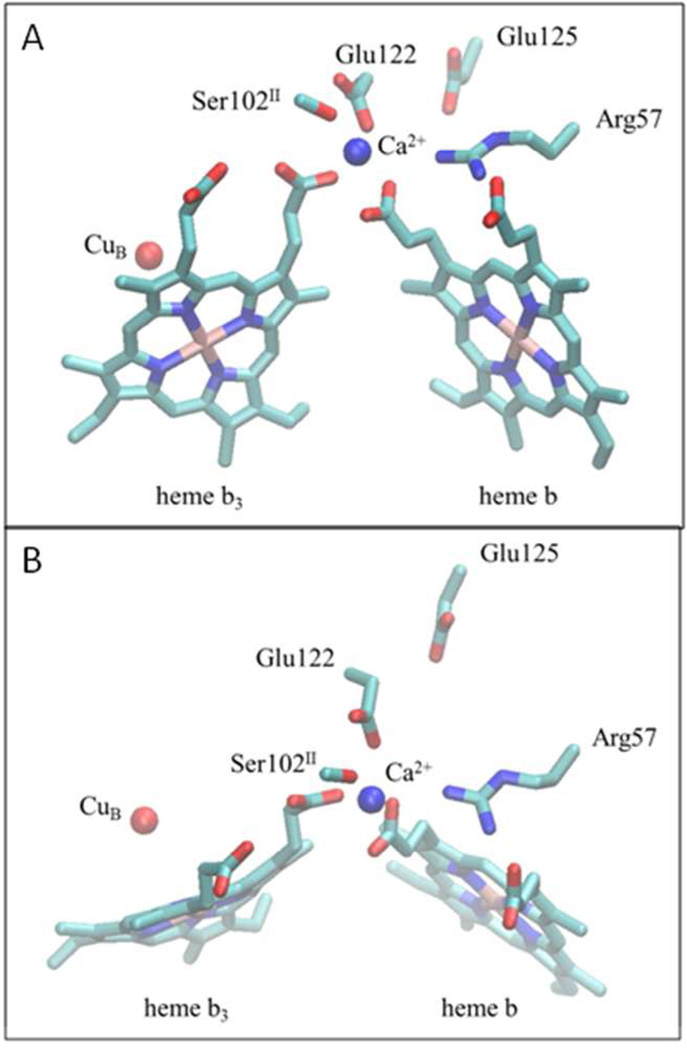
Figure 1 shows the Ca2+ binding site between the two hemes in the cbb3 oxygen reductase from P. stutzeri. In addition to the Ca2+ bridging the D-propionates of heme b and heme b3, an arginine (Arg57) is hydrogen bonded to the A- and D-propionates of the low-spin heme b.
Figure 1. Structure of the Ca+2 binding site in the C-family oxygen reductase from Pseudomonas stutzeri (PDB: 3MK7).
(A) The Ca+2 binding site, located at the interface between subunits I (CcoN) and II (CcoO), as seen from the side. The Ca2+ ion (blue) is ligated to the D-propionates of both the low-spin heme b and the active-site high spin heme b3, along with two conserved amino acid side chains, Glu122 in subunit I (CcoN) and Ser102 in subunit II (CcoO). The A- and D-propionates of the low-spin heme are both hydrogen bonded to a conserved arginine (Arg57) in subunit I. (B) A view of the same region of the protein as seen from the top (periplasmic side). The residue numbering corresponds to the P. stutzeri cbb3 oxygen reductase.
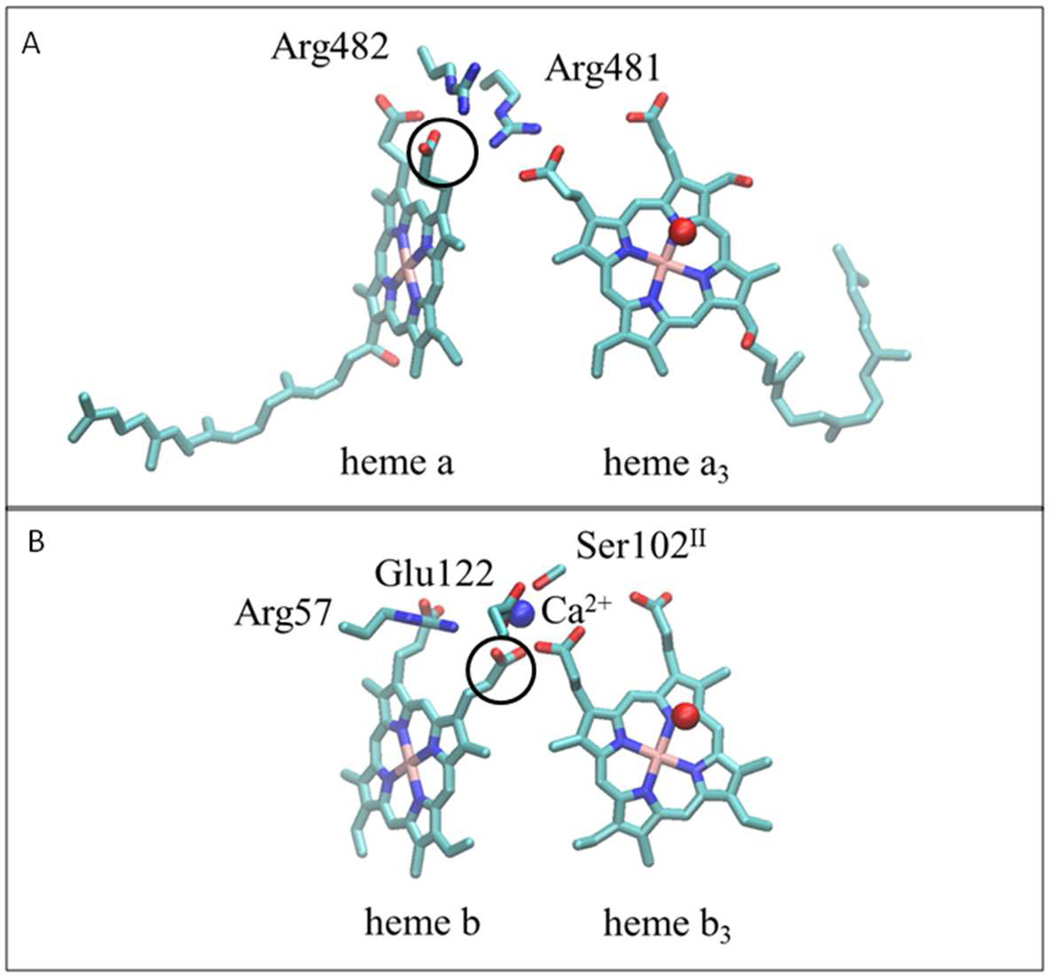
In the A- and B-family oxygen reductases, there is also an arginine which hydrogen bonds to both of the propionates of the low spin heme but, in place of the Ca2+ there is a second arginine which bridges the low-spin and active-site hemes by hydrogen bonding to two propionates (23, 24). Figure 2 shows the structure near the low-spin heme a and active-site heme a3 of the A-family cytochrome c oxidase from R. sphaeroides, and compares this to the corresponding region in the cbb3 oxygen reductase from P. stutzeri. Arg481 takes the place of the Ca2+ found in the cbb3 oxygen reductases, and the adjacent Arg482 bridges the two propionates of the low-spin heme. The low-spin heme propionate which is ligated to the Ca2+ in the C-family (cbb3) oxygen reductases (circled in Figure 2), as well as in the cNOR and qNOR families, has a different rotomeric configuration than the equivalent propionate which is hydrogen bonded to the adjacent arginine residues in the A- and B-family oxygen reductases. It is of interest that some of the A-family oxygen reductases, including the R. sphaeroides (25) and bovine (26) cytochrome c oxidases, also bind to a single Ca2+, but this cation is not ligated to the heme propionates. Mutagenesis studies have shown that this Ca2+ is not essential either for assembly or function (25, 27).
Figure 2. Comparison of heme propionate interactions in different heme-copper oxidoreductase families.
(A) In the A and B-families, represented by the structure of the A-family cytochrome c oxidase from Rhodobacter sphaeroides (PDB: 1M56), the heme propionates interact with a pair of conserved, adjacent arginine residues. The first arginine forms hydrogen bonds with propionates of the low-spin heme (heme a) and the active-site, high-spin heme (heme a3). The second arginine forms hydrogen bonds with the two propionates of the low-spin heme. (B) In the C-family oxidase from Pseudomonas stutzeri (PDB: 3MK7), the Ca+2 binding site plays an analogous role to one of the arginines (R481 in the top panel). Note the difference in the rotational configurations of the low-spin heme propionates (circled) in the two Panels. In the A and B-families the propionate is rotated out to form a hydrogen bond with the arginine, whereas in the C-family it is rotated in to ligate the Ca+2 and to allow room for the arginine to form hydrogen bonds with the low spin heme propionates. This structural feature is also found in the cNOR and qNOR families. The residue numbering in panel (B) is from the cbb3 oxygen reductase from P. stutzeri.
In the A- and B-family oxygen reductases, the two adjacent arginines which interact with the heme propionates (R481 and R482 in Figure 2) are nearly totally conserved, but there are no conserved acidic residues corresponding to the glutamate pair found in the C-family oxygen reductases and in the cNOR and qNOR enzymes.
In the current work, mutations of each of the glutamates of the conserved pair in the cbb3 oxygen reducatases from V. cholerae and from R. sphaeroides are further investigated, and it is shown that the loss of activity of the enzymes correlates with the loss of Ca2+. It is concluded that the mutagenesis data cannot be used to support the hypothesis that these glutamates are important for proton delivery to/from the active site in the cbb3 oxygen reductases. It is more likely that these residues are important for maintaining the structures of these enzymes in an active conformation.
Materials and methods
Mutagenesis of the cbb3-type oxidases from R. sphaeroides and V. cholerae
Site-directed mutagenesis and cloning were performed as previously reported (28)
Cell growth and purification of the His-tagged cbb3-Type oxidases
R. sphaeroides was grown semi-aerobically at 30° C in Sistrom media with 2 µg/mL tetracycline (Sigma). V. cholerae was grown aerobically at 37° C in modified M9 medium (0.1% glucose, 1mM MgSO4 and supplemented with, 10 mg/L thiamine, 10 µM CuSO4, 50 µM FeSO4) with 100 µg/mL ampicillin (Fisher Biotech) and 100 µg/mL streptomycin (Sigma). It was found that the expression of the V. cholerae cbb3 was equally good if the cells were grown in the minimal medium as in LB. However, protein purified from cells grown in minimal medium had fewer metal contaminants and more consistent metal analyses.
The wild type and mutant cbb3 proteins (with polyhistidine tags) from R. sphaeroides and V. cholerae were overexpressed and purified as previously described (28) using the detergent dodecylmaltoside (DDM). GeneMate Express SDS-PAGE gels from ISC BioExpress were used to evaluate the isolated protein complexes, using Coomassie Brilliant Blue staining. The CcoO and CcoP subunits were visualized by heme staining (29) since each of these subunits contains covalently bound heme c.
UV/Vis spectroscopic analysis
A Shimadzu UV/Vis-2101PC spectrophotometer was used to obtain the spectra of wild type and mutant enzymes. The concentrated protein samples were diluted with 10–20 mM Tris buffer and 0.05% DDM at pH 7.5–8.0. The enzymes were oxidized by air and reduced with sodium dithionite (Sigma). Spectra were measured from 300 nm to 800 nm and analyzed using Origin software.
Pyridine hemochrome assay
The concentrations of heme B and heme C were determined as previously described (30). 0.5ml of a stock solution containing 200 mM NaOH and 40% (by volume) pyridine and 3µl of 0.1M K3Fe(CN)6 were placed in a 1 mL cuvette. A 0.5 mL aliquot of the protein sample (~5 µM) was added with thorough mixing and spectrum of the oxidized hemes was recorded within 1 minute. Solid sodium dithionite (2–5mg) was then added and several successive spectra of the reduced pyridine hemochromes were recorded. The absorbance differences at selected wavelengths were used to obtain the concentrations of heme B and heme C. In some cases after reduction by dithionite, turbidity resulted in a distorted baseline. This was corrected by fitting the baseline to either an exponential or a 3rd order polynomial and subtracting this from the experimental spectrum.
Steady-state activity
A YSI model 53 oxygen monitor was used to polarographically measure steady-state oxidase activity at 25° C. To the sample chamber was added 20µL 1 M ascorbate, pH 7.4, 10µL of 0.1 M TMPD and 1.8 mL of 50 mM sodium phosphate buffer, pH 6.5 containing 100 mM NaCl and 0.05% DDM. Horse heart cytochrome c (Sigma) was added as the substrate to a final concentration of 40 µM in the case of R. sphaeroides cbb3. The reaction was initiated by adding 10 µL of a solution containing 1 µM enzyme, and the rate of oxygen consumption was monitored.
Metal analysis
Metal content of the protein was determined by inductively coupled plasma - optical emission spectroscopy (ICP-OES) using a Spectro Genesis spectrometer as previously described (31). The buffer containing the purified V. cholerae cbb3 protein was exchanged using a 100 kD cut-off Amicon concentrator (Millipore). The buffer was exchanged 3 times with 10 mM Tris buffer, pH 8, containing 0.05% DDM, 5% glycerol, 0.25 mM EDTA and 0.1 mM EGTA, followed by another 3 times with the same buffer without EDTA and EGTA. Purified R. sphaeroides cbb3 protein samples were incubated with 1 mM EDTA and 0.1 mM EGTA and then dialyzed against 20 mM Tris buffer, pH 7.5, containing 30 mM KCl, 1 mM EDTA and 0.05% DDM. After dialysis, the protein was concentrated using the 100 kD cut-off Amicon concentrator. Metal concentrations were calculated from regression lines of element standards run as samples rather than the regression lines produced by the Spectro software to avoid a 10% underestimation of element concentrations at low levels of the metals.
Results
Conservation of the glutamate residues at or near the Ca2+ binding site
The one Ca2+ clearly observed in the structure of the cbb3-type oxidase from P. stutzeri is ligated to the carboxyl of E122 (CcoN subunit) and to a serine, S102, located in the CcoO subunit, in addition to the two D-propionates from the low-spin heme b and the active-site heme b3, which are located in the CcoN subunit. Sequence alignments show that E122 (CcoN) is >99% conserved, replaced by a glutamine in the few exceptions, and S102(CcoO) is completely conserved. The nearby glutamate, E125 (CcoN) is not ligated to the Ca2+ but is also very highly conserved (>99%). These data are included in Table 1. Also shown is the pattern of conservation of the equivalent residues in the qNOR and cNOR families. In the cNOR enzymes, as in the C-family oxygen reductases, the Ca2+ has ligands that bridge two separate subunits (NorC and NorB), and it is the ligand from the subunit which contains cytochrome c (CcoO or NorC), which is most highly conserved. The two glutamates are less conserved in the cNOR family than in the cbb3 oxygen reductases, and even less conserved in the qNOR family.
Table 1. Conservation of residues important for calcium binding in the C-family of oxygen reductases and the cNOR and qNOR families of NO reductases.
The percent conservation of the residues is given, along with the variants for each family.
| Family | E1221 (in CcoN) Ca+2 ligand |
E1251 (in CcoN) | S1021 (in CcoO) Ca2+ ligand |
|---|---|---|---|
| C-family | E (Q) >99% | E (D, Q, K) 99% | S, T 100% (in CcoO) |
| cNOR | E (K, P, H, V, G) 85% | E (D) 95% | Y >99% (in NorC) |
| qNOR | E (A, S, D, P) 80% | D (E, S, Q, N, T) 45% | Y 90% |
The numbering of the residues refer to the cbb3 oxygen reductase from Pseudomonas stutzeri.
Metal and heme content of the wild type cbb3-type oxygen reductases
The metal contents of the cbb3 enzymes from V. cholerae and from R. sphaeroides are shown in Table 2. These enzymes contain 3 hemes c, 2 hemes b and 1 CuB, so the Fe/Cu ratio should be 5/1. The observed ratios are higher than this, Fe/Cu = 7.5 and 10.1 for the V. cholerae and R. sphaeroides enzymes, respectively. These high values could be due to loss of CuB or contaminating Fe, or both. The ratio of heme C/heme B should be 3/2=1.5, and the measured values are 1.9 and 1.5 for the V. cholerae and R. sphaeroides enzymes, respectively (Table 2).
Table 2. Characteristics of wild type and mutant cbb3-type oxygen reductases.
| Subunit | Strain | mutant | Activity (% WT) |
[C]/[B] ratio |
Ca/Cu | 5(Ca/Fe) |
|---|---|---|---|---|---|---|
| CcoN | R. sphaeroides | WT | 1001 | 1.5 | 2.4±0.3 (n = 3) |
1.44±0.5 (n = 3) |
| R. sphaeroides | E180D | <1 | 1.72 | 0 (n = 1) | ||
| R. sphaeroides | E180Q | 1 | 1.62 | 0 (n = 1) | ||
| R. sphaeroides | E180G | <1 | 2 | |||
| R. sphaeroides | E183D | 1 | 1.62 | 0 (n = 1) | ||
| R. sphaeroides | E183Q | 7 | 1.5 | 1.92±0.34 (n = 3) |
1.13±0.23 (n = 3) |
|
| R. sphaeroides | E183G | 1 | 4 | |||
| V. cholerae | WT | 1001 | 1.9 | 1.82±0.05 (n = 7) |
1.11±0.2 (n = 7) |
|
| V. cholerae | E126D | 2 | 1.8 | |||
| V. cholerae | E126Q | <1 | 1.2 | 0.55±0.33 (n = 7) |
||
| V. cholerae | E126G | <1 | 1.2 | |||
| V. cholerae | E129D | 11 | 1.3 | |||
| V. cholerae | E129Q | 18 | 1.7 | 0.63±0.4 (n = 8) |
||
| V. cholerae | E129G | 2 | 1.8 | |||
| V. cholerae | E129A | 2 | 1.5 | |||
| CcoO | R. sphaeroides | S105T | <1 | |||
| R. sphaeroides | S105A | <1 | ||||
| R. sphaeroides | S105G | <1 |
The activities of the R. sphaeroides and V. cholerae cbb3-type oxygen reductases were 800 e−/sec and 500 e−/sec, respectively. See Methods section for details.
For these mutants, the low spin heme b component of the oxidase, though present, was not reduced by dithionite in the assembled enzyme.
The Ca contents of the enzymes are presented in Table 2 as either a ratio to Cu or to Fe. If the Ca stoichiometry is taken as a ratio to the total iron content, the data indicate at Ca content of 1.1 and 1.4 equivalents per mol of enzyme (5 Fe per enzyme molecule) for the V. cholerae and R. sphaeroides cbb3 oxygen reductases, respectively. The Ca stoichiometry that is determined from the Ca/Cu ratio is higher in both cases, about 1.8 for the V. cholerae enzyme and 2.4 for the R. sphaeroides enzyme. It is noted that the possible presence of a second Ca2+ in the cbb3 from P. stutzeri is also unclear, both from the crystal structure itself and from the metal analysis (22).
At this point, with the current enzyme preparations, it is not possible to conclude with confidence whether these enzymes contain one or two equivalents of Ca2+. For convenience, the Ca2+ stoichiometries of various mutants are reported as the values determined based on the amount of Fe present (assuming 5 Fe per mol of enzyme).
Site-directed mutagenesis of the V. cholerae cbb3 oxygen reductase
E126 (Ca2+ ligand)
The E126D, E126Q and E126G mutations were characterized. As shown in Table 2, each of these mutations is virtually inactive (2%, <1%, <1%, respectively). The ratio of heme C to heme B of the E126D mutant is similar to that of the wild type, but both E126Q and E126G have a lower ratio. SDS-PAGE analysis shows that both of the heme C-containing subunits (22)., CcoO and CcoP, are present in each mutant, though this is qualitative and not quantitative. The Ca stoichiometry was measured for the E126Q mutant and is about half of the wild type. Hence, mutation of the Ca ligand results in both the loss of Ca and loss of activity.
E129
The E129D, E129Q, E129G and E129A mutants were characterized. The E129D and E129Q mutants have 11% and 18% of the wild type activity, whereas the E129G and E129A have less than 2% activity (Table 2). The heme C/heme B ratio ranges from 1.3 to 1.8 (expected is 1.5). The Ca stoichiometry was determined for the E129Q mutant and was shown to be about half of the wild-type value (Table 2), 0.63 vs 1.11 equivalents of Ca per mol of enzyme. Even though E129 is not directly ligated to the bound Ca, a mutation in E129 can still cause loss of this metal, apparently by an indirect interaction.
Site-directed mutagenesis of the R. sphaeroides cbb3 oxygen reductase
E180 (Ca2+ ligand)
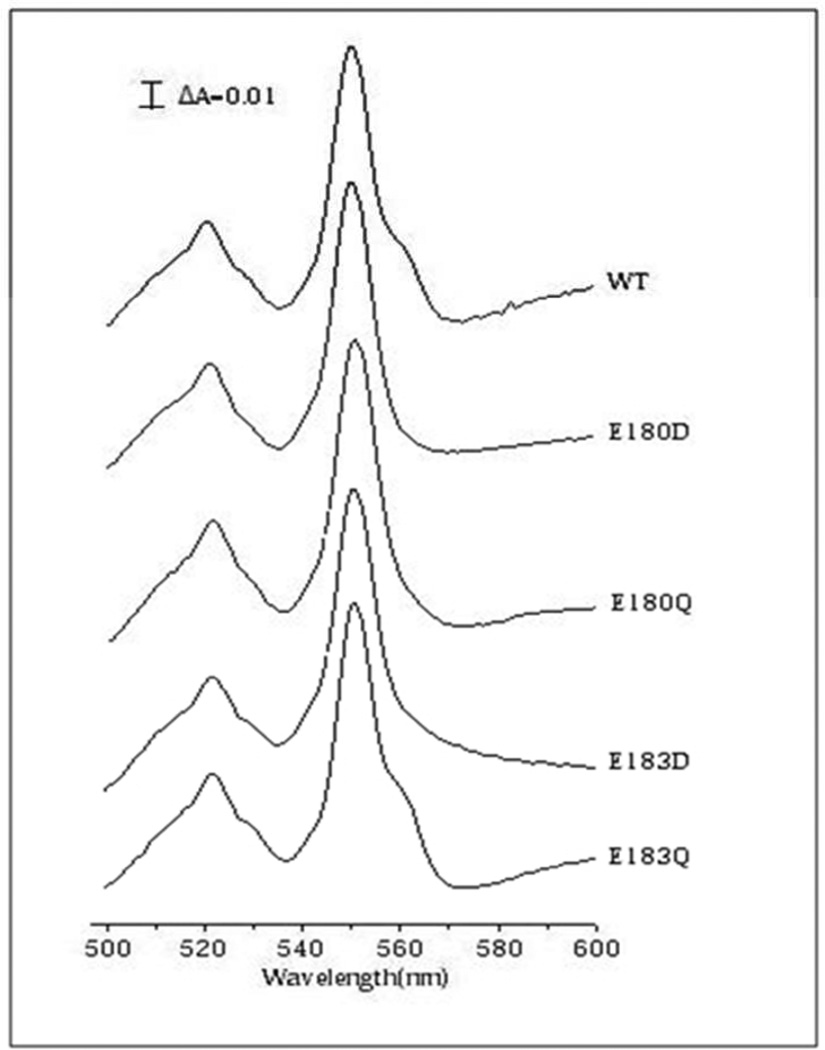
The E180D, E180Q and E180G mutants were examined. All were virtually inactive (Table 2) and the yield of E180G was very low, implying instability or an assembly problem. The E180G mutant was not examined further. Metal analysis of both E180D and E180Q showed that not only was there no Ca but also that there was no Cu. In addition, the dithionite reduced-minus-oxidized UV/visible spectra show that the loss of Cu in E180Q and E180D is coincident with the lack of reduction of heme b. The content of heme B in the mutant enzymes is similar to that of the wild type enzyme, as shown by the pyridine-hemochrome analysis (Table 2), but the low spin heme is not reduced by dithionite in the assembled enzyme. This indicates substantially more damage due to the alteration of the Ca ligand in the R. sphaeroides enzyme than in the V. cholerae enzyme. The nature of this perturbation to the low spin heme b was not explored further. It is noted, however, that this unusual mutant phenotype of a heme that is not reducible by dithionite has been previously observed in a mutant of the cytochrome bd quinol oxidase from E. coli (32).
E183
The E183D, E183Q and E183G mutants were characterized. All the mutants have very low (or no) oxidase activity, with the highest activity (7%) observed with E183Q (Table 2). The E183G was obtained in very low yield and was not examined further. The E183D mutant had properties similar to those of the E180 mutants insofar as there was loss of both Ca and CuB, and the low spin heme b component was not reduced by dithionite. The E183Q mutant retains a small amount of oxidase activity (7%), and has partial loss of Ca2+ but no loss of Cu (Table 2). This is the only R. sphaeroides mutant of those examined in which loss of activity is not correlated with a massive loss of Ca2+.
S105(CcoO)
The S105T, S105A and S105G mutations were made but no expressed protein was obtained for any of the mutants. This Ca2+ ligand appears to be absolutely necessary for protein assembly/stability.
Discussion
Prior to the availability of X-ray structures, the conserved pair of glutamates in a putative periplasmic “loop” appeared to be reasonable candidates as participants in proton translocation to/from the active sites of the cbb3-type (C-family) oxygen reductases as well as the related cNOR and qNOR families of NO reductases. Models suggested that these glutamates might be located near the perisplasmic surface of the proteins (18, 19) and site-directed mutagenesis indicated that the glutamates are functionally important in both the cbb3 oxygen reductases and cNOR enzymes (18, 20, 21). In the case of the cNOR enzymes, protons required for the chemistry at the active site come from the periplasm (18, 33, 34) and the limited data were consistent with these glutamates playing a role to provide a pathway for proton transfer to the active site (34). For the cbb3 enzymes, protons for the chemistry at the active site must come from the opposite side of the membrane (2, 7), so the glutamates were proposed to provide a pathway for pumped protons to exit to the periplasm (7). The X-ray structures (10, 11, 22) as well as recent functional information, including the data presented in the current work, make these proposals much less likely.
The X-ray structures of the cbb3 oxygen reductase, the cNOR and the qNOR each reveal a single Ca2+ that bridges propionates on heme b and heme b3, and is also ligated to a glutamate carboxyl (10, 11, 22), E122 in the case of the P. stutzeri cbb3. There is extra electron density in the P. stutzeri cbb3 structure (22) that suggests the possibility of a second Ca2+ near E125. Metal analysis also indicates that there might be a second Ca2+ present in the P. stutzeri cbb3 oxidase (22). The current data on the V. cholerae and R. sphaeroides cbb3 enzymes are similarly ambiguous about the possibility of a second Ca2+ (Table 2). Metal impurities in the protein preparations, present even after three column chromatography steps, as well as possible loss of metals, contribute to the uncertainty.
The X-ray data (10, 11, 22) show that in each of these enzyme families (cbb3, cNOR, qNOR), the two glutamates are not near the surface in contact with the bulk aqueous phase, but are more interior. The upstream member of the pair (lower residue number) is, in each case, ligated to a Ca2+. The downstream glutamate is not ligated to the Ca2+ nor part of a salt bridge, but participates in a network of hydrogen bond interactions in each protein. The possible second Ca2+ in the cbb3 structure is located near this downstream glutamate (22). There is only one Ca2+ in the structures of the cNOR and qNOR (10, 11).
The very high (> 99%) degree of conservation of each of the two glutamates in the sequences of the cbb3 oxygen reductases makes it evident that they are both important in these enzymes (Table 1). This is confirmed by the site-directed mutagenesis presented in the current work (Table 2). Regardless of the uncertainty of whether there is one or two Ca2+ in the wild type enzymes, the pattern for both the V. cholerae and R. sphaeroides cbb3 mutants is that substitution for either of the glutamates results in substantial loss of activity as well as partial or complete loss of Ca2+ (Table 2). Changing the Ca2+ ligand, E126 (V. cholerae) or E180 (R. sphaeroides), results in virtually complete loss of function as well as partial or complete loss of Ca2+ (Table 2). In the R. sphaeroides mutants, the extent of evident damage is greater, including loss of CuB. Note that if the wild type enzyme is interpreted to contain 2 Ca2+, then a single mutation (E180D or E180Q) results in the loss of both Ca2+. Loss of function due to these mutations can be most easily interpreted as a consequence of gross conformational changes. The mutagenesis data do not rule out a role of this glutamate in proton translocation but, importantly, cannot be used in support of such a role. If E180 were to cycle through a state during catalysis in which it were protonated and no ligated to the Ca2+, one might not expect the E180Q mutation to cause so much damage since glutamine to some extent resembles a protonated glutamate.
Changing the downstream glutamate, E129 (V. cholerae) or E183 (R. sphaeroides), also invariably results in loss of function, though the results are more varied and not as absolute as observed with the mutations of the Ca2+-ligating glutamate. The V. cholerae protein is more tolerant of mutations at this position than is the R. sphaeroides enzyme. E129D and E129Q in the V. cholerae enzyme retain 11% and 18% activity, respectively, whereas E183D and E183Q from the R. sphaeroides oxidases retain only 1% and 7% activity. In all cases, either partial or complete loss of Ca2+ is observed.
The following are reasonable conclusions from the data.
The cbb3 oxygen reductases contain a either one or two Ca2+. Our data do not resolve this uncertainly, raised initially from the work on the enzyme from the P. stutzeri enzyme (22).
The Ca2+ at the binding site that is apparent in the X-ray structure is essential for function of the cbb3 oxygen reductases. Mutations that result in the loss of Ca2+ or disruption of the Ca2+ binding site results in the loss of enzyme activity. It is likely that the Ca2+ is important to maintain the protein in a conformation in which the hemes and CuB are functional. The role of the Ca2+ to form a bridge between the CcoN and CcoO subunits is also critical, as evidenced by the lack of protein assembly when the Ca2+-liganding amino acid in the CcoO subunit (S105) is substituted by another amino acid. The essential nature of the Ca2+ in the cbb3 oxygen reductases is in contrast to the Ca2+ which binds near the low-spin heme in some of the A-family oxygen reductases, but which is not required for assembly or enzyme activity (25, 27).
Depending on the mutation and which enzyme is studied, mutating the non-liganding glutamate (downstream member of the pair), the cbb3 enzyme can be inactivated along with partial or complete loss of Ca2+. There is no reason to attribute the loss of function to blocking proton transfer of the pumped proton away from the cbb3 active site. It is not evident, in any event, that blocking the pumped proton would necessarily inhibit oxidase activity.
The conclusion that the role of the Ca2+ is primarily to stabilize the heme and protein for optimal activity has also been made for the related NOR enzymes (4, 8–10). Note that the two glutamates examined in the current work are not nearly as well conserved in the cNOR and qNOR sequences as in the sequences of the cbb3 oxygen reductases (Table 1). This is particularly true for the qNORs. When present, the Ca2+ ligands are important, as shown by the fact that mutants of each of the Ca2+ ligands (E429 and Y93) in the qNOR from Geobacillus stearothermophilus lose most of their activity as well as Ca2+ (10). It is possible that in the qNOR and cNOR variants lacking the equivalent glutamate, the Ca2+ has a different ligand or, alternatively, the role of the Ca2+ is filled by alternative structural features. There are credible hydrophilic pathways which bypass the two glutamates for delivering protons from the periplasm to the active site in the cNOR enzymes, so there is no need to propose any role of the glutamate pair in proton translocation in either the cNOR or qNOR enzymes (4, 9). Experimental tests will be required, however, to determine the input pathway(s) for protons to reach the active sites of the qNOR and cNOR enzymes.
Figure 3. Dithionite-minus-air oxidized difference spectra of the R. sphaeroides cbb3–type oxygen reductases.
The 551 nm peak is due to hemes c and 561 nm peak is mostly due to the low spin heme b. The data show the absence of the spectroscopic signature for reduced heme b in the E180D, E180Q and E183D mutants.
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL16101).
References
- 1.Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 2.Han H, Hemp J, Pace LA, Ouyang H, Ganesan K, Roh JH, Daldal F, Blanke SR, Gennis RB. Adaptation of aerobic respiration to low O2 environments. Proc Natl Acad Sci U S A. 2011;108:14109–14114. doi: 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira MM, Sousa FL, Verissimo AF, Teixeira M. Looking for the minimum common denominator in haem-copper oxygen reductases: Towards a unified catalytic mechanism. Biochim Biophys Acta. 2008;1777:929–934. doi: 10.1016/j.bbabio.2008.05.441. [DOI] [PubMed] [Google Scholar]
- 4.Hino T, Nagano S, Sugimoto H, Tosha T, Shiro Y. Molecular structure and function of bacterial nitric oxide reductase. Biochim Biophys Acta. 2012;1817:680–687. doi: 10.1016/j.bbabio.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 5.Pereira MM, Santana M, Teixeira M. A Novel Scenario for the Evoluation of Haem-copper Oxygen Reductases. Biochim. Biophys. Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 6.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc Natl Acad Sci U S A. 2009;106:16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemp J, Han H, Roh JH, Kaplan S, Martinez TJ, Gennis RB. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 oxidase) of heme-copper oxygen reductases. Biochemistry. 2007;46:9963–9972. doi: 10.1021/bi700659y. [DOI] [PubMed] [Google Scholar]
- 8.Shiro Y, Sugimoto H, Tosha T, Nagano S, Hino T. Structural basis for nitrous oxide generation by bacterial nitric oxide reductases. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:1195–1203. doi: 10.1098/rstb.2011.0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiro Y. Structure and function of bacterial nitric oxide reductases: Nitric oxide reductase, anaerobic enzymes. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2012 doi: 10.1016/j.bbabio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto Y, Tosha T, Pisliakov AV, Hino T, Sugimoto H, Nagano S, Sugita Y, Shiro Y. Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat Struct Mol Biol. 2012;19:238–245. doi: 10.1038/nsmb.2213. [DOI] [PubMed] [Google Scholar]
- 11.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. Structural Basis of Biological N2O Generation by Bacterial Nitric Oxide Reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 12.Castresana J, Lübben M, Saraste M, Higgins DG. Evolution of Cytochrome Oxidase, an Enzyme Older Than Atmospheric Oxygen. EMBO. J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castresana J, Saraste M. Evolution of Energetic Metabolism: The Respiration-early Hypothesis. Trends-Biochem. Sci. 1995;20:443–448. doi: 10.1016/s0968-0004(00)89098-2. [DOI] [PubMed] [Google Scholar]
- 14.Gribaldo S, Talla E, Brochier-Armanet C. Evolution of the haem copper oxidases superfamily: a rooting tale. Trends in Biochemical Sciences. 2009;34:375–381. doi: 10.1016/j.tibs.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Ducluzeau A-L, Ouchane S, Nitschke W. The cbb3 Oxidases Are an Ancient Innovation of the Domain Bacteria. Molecular Biology and Evolution. 2008;25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- 16.Ducluzeau A-L, van Lis R, Duval S, Schoepp-Cothenet B, Russell MJ, Nitschke W. Was nitric oxide the first deep electron sink? Trends in Biochemical Sciences. 2009;34:9–15. doi: 10.1016/j.tibs.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Jones CM, Stres B, Rosenquist M, Hallin S. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol Biol Evol. 2008;25:1955–1966. doi: 10.1093/molbev/msn146. [DOI] [PubMed] [Google Scholar]
- 18.Reimann J, Flock U, Lepp H, Honigmann A, Adelroth P. A pathway for protons in nitric oxide reductase from Paracoccus denitrificans. Biochim Biophys Acta. 2007;1767:362–373. doi: 10.1016/j.bbabio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 19.Flock U, Reimann J, Adelroth P. Proton transfer in bacterial nitric oxide reductase. Biochem Soc Trans. 2006;34:188–190. doi: 10.1042/BST0340188. [DOI] [PubMed] [Google Scholar]
- 20.Thorndycroft FH, Butland G, Richardson DJ, Watmough NJ. A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem J. 2007;401:111–119. doi: 10.1042/BJ20060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butland G, Spiro S, Watmough NJ, Richardson DJ. Two conserved glutamates in the bacterial nitric oxide reductase are essential for activity but not assembly of the enzyme. J Bacteriol. 2001;183:189–199. doi: 10.1128/JB.183.1.189-199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 23.Mills DA, Geren L, Hiser C, Schmidt B, Durham B, Millett F, Ferguson-Miller S. An Arginine to Lysine Mutation in the Vicinity of the Heme Propionates Affects the Redox Potentials of the Hemes and Associated Electron and Proton Transfer in Cytochrome c Oxidase. Biochemistry. 2005;44:10457–10465. doi: 10.1021/bi050283d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puustinen A, Wikström M. Proton Exit from the Heme-copper Oxidase of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:35–37. doi: 10.1073/pnas.96.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee A, Kirichenko A, Vygodina T, Siletsky SA, Das TK, Rousseau DL, Gennis R, Konstantinov AA. Ca(2+)-binding site in Rhodobacter sphaeroides cytochrome C oxidase. Biochemistry. 2002;41:8886–8898. doi: 10.1021/bi020183x. [DOI] [PubMed] [Google Scholar]
- 26.Kirichenko A, Vygodina T, Mkrtchyan HM, Konstantinov A. Specific cation binding site in mammalian cytochrome oxidase. FEBS Lett. 1998;423:329–333. doi: 10.1016/s0014-5793(98)00117-3. [DOI] [PubMed] [Google Scholar]
- 27.Kirichenko AV, Pfitzner U, Ludwig B, Soares CM, Vygodina TV, Konstantinov AA. Cytochrome c oxidase as a calcium binding protein. Studies on the role of a conserved aspartate in helices XI–XII cytoplasmic loop in cation binding. Biochemistry. 2005;44:12391–12401. doi: 10.1021/bi050376v. [DOI] [PubMed] [Google Scholar]
- 28.Hemp J, Christian C, Barquera B, Gennis RB, Martinez TJ. Helix Switching of a Key Active-Site Residue in the Cytochrome cbb3 Oxidases. Biochemistry. 2005;44:10766–10775. doi: 10.1021/bi050464f. [DOI] [PubMed] [Google Scholar]
- 29.Thomas PE, Ryan D, Leven W. An Improved Straining Procedure for the Detection of the Peroxidase Activity of Cytochrome P-450 on Sodium Dodecyl Sulfate Polyacrylamide Gels. Anal. Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 30.Berry EA, Trumpower BL. Simultaneous Determination of Hemes a, b, and c from Pyridine Hemochrome Spectra. Anal. Bioch. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 31.Carrell CJ, Ma JK, Antholine WE, Hosler JP, Mathews FS, Davidson VL. Generation of novel copper sites by mutation of the axial ligand of amicyanin. Atomic resolution structures and spectroscopic properties. Biochemistry. 2007;46:1900–1912. doi: 10.1021/bi0619674. [DOI] [PubMed] [Google Scholar]
- 32.Belevich I, Borisov VB, Zhang J, Yang K, Konstantinov AA, Gennis RB, Verkhovsky MI. Time-resolved Electrometric and Optical Studies on Cytochrome bd Suggest a Mechanism of Electron-proton Coupling in the Di-heme Active Site. Proc Natl Acad Sci USA. 2005;102:3657–3662. doi: 10.1073/pnas.0405683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendriks JH, Jasaitis A, Saraste M, Verkhovsky MI. Proton and electron pathways in the bacterial nitric oxide reductase. Biochemistry. 2002;41:2331–2340. doi: 10.1021/bi0121050. [DOI] [PubMed] [Google Scholar]
- 34.Shapleigh JP, Payne WJ. Nitric oxide-dependent proton translocation in various denitrifiers. J Bacteriol. 1985;163:837–840. doi: 10.1128/jb.163.3.837-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]