Abstract
The telomere end-protection problem is defined by the aggregate of DNA damage signaling and repair pathways that require repression at telomeres. To define the end-protection problem, we removed the whole shelterin complex from mouse telomeres through conditional deletion of TRF1 and TRF2 in nonhomologous end-joining (NHEJ) deficient cells. The data reveal two DNA damage response pathways not previously observed upon deletion of individual shelterin proteins. The shelterin-free telomeres are processed by microhomology-mediated alternative-NHEJ when Ku70/80 is absent and are attacked by nucleolytic degradation in the absence of 53BP1. The data establish that the end-protection problem is specified by six pathways [ATM (ataxia telangiectasia mutated) and ATR (ataxia telangiectasia and Rad3 related) signaling, classical-NHEJ, alt-NHEJ, homologous recombination, and resection] and show how shelterin acts with general DNA damage response factors to solve this problem.
Aspects of the end-protection problem have been revealed in yeast, plant, and mammalian cells based on adverse events at telomeres lacking certain telomeric proteins (1). However, the fate of telomeres devoid of all protective factors is unknown, and hence the end-protection problem remained undefined. Mammals solve the end-protection problem through the agency of shelterin (2), a multisubunit protein complex anchored onto duplex telomeric DNA by the TTAGGG repeat binding factors TRF1 and TRF2 (fig. S1). Both TRF1 and TRF2 interact with TIN2 (TRF1-interacting nuclear factor 2), which in turn links the heterodimer formed by TPP1 (TINT1/PTOP1/PIP1) and POT1 (protection of telomeres 1; POT1a and POT1b in mouse) to telomeres. TPP1/POT1 interacts with the single-stranded TTAGGG repeats present at mammalian chromosome ends in the form of a 50 to 400 nucleotide (nt) 3′ overhang. The sixth shelterin subunit, Rap1, is a TRF2-interacting factor. Deletion of each of the individual shelterin proteins revealed that the end-protection problem minimally involves the repression of ATM (ataxia telangiectasia mutated) and ATR (ataxia telangiectasia and Rad3 related) signaling as well as inhibition of double-strand break (DSB) repair by nonhomologous end-joining (NHEJ) and homology-directed repair (HDR). However, the possibility of redundant repression of additional DNA damage response (DDR) pathways has prevented a definitive description of the end-protection problem in mammalian cells.
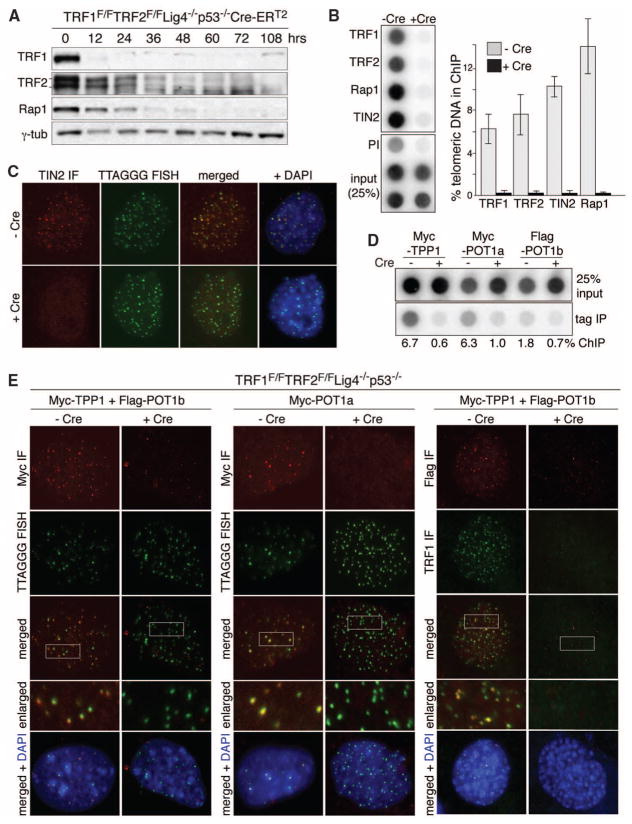
We sought to finalize the tally of telomere-threatening pathways by generating telomeres devoid of all shelterin proteins and their associated factors. We set out to remove both TRF1 and TRF2, which is predicted to lead to complete loss of shelterin (fig. S1). In this TRF1/2 double-knockout (DKO), NHEJ of telomeres devoid of TRF2 thwarts detection of potential novel pathways acting on deprotected chromosome ends. We therefore created conditional TRF1/2 DKO mouse embryo fibroblasts (MEFs) with additional deficiencies in DNA ligase IV (Lig4), Ku80, or 53BP1, which are predicted to minimize telomere fusion (3–5). Cre was expressed from a self-deleting Hit-and-Run (H&R-Cre) retrovirus or from a genetically introduced tamoxifen (4-OHT)–inducible Cre (Cre-ERT2 in the Rosa26 locus). TRF1F/FTRF2F/FLig4−/−p53−/−Cre-ERT2 MEFs rapidly lost TRF1, TRF2, and Rap1 when treated with 4-OHT and telomeric chromatin immunoprecipitation (ChIP) and immunofluorescence (IF) established that TRF1, TRF2, Rap1, and TIN2 disappeared from telomeres (Fig. 1, A to C). Furthermore, using tagged alleles to facilitate analysis, IF and ChIP documented loss of TPP1 and POT1a/b from the telomeres (Fig. 1, D and E, and fig. S2, A and B). Thus, the TRF1/2 DKO generates shelterin-free telomeres. However, the telomeric DNA remained packaged in nucleosomal chromatin (fig. S2C).
Fig. 1.
Shelterin-free telomeres. (A) Immunoblots for TRF1, TRF2, and Rap1 after 4-OHT−induced TRF1/2 DKO from Lig4−/−p53−/−Cre-ERT2 MEFs. (B) ChIP for telomeric DNA associated with shelterin proteins in TRF1F/FTRF2F/Fp53−/−Lig4−/−MEFs (day 5 after H&R-Cre). Bars average percentage of telomeric DNA recovered in two independent experiments, ± SEMs. (C) IF-FISH for TIN2 at telomeres in TRF1F/FTRF2F/Fp53−/−Lig4−/−MEFs day 5 after H&R-Cre. TIN2 IF (red); telomeric PNA probe [fluorescein isothiocyanate (FITC), green]. (D) ChIP for telomeric DNA associated with Myc-TPP1, Myc-POT1a, and Flag-POT1b in TRF1F/FTRF2F/Fp53−/− Lig4−/−cells, with (+) and without (−) H&R-Cre. (E) IF for the telomeric localization of Myc-TPP1, Myc-POT1a, and Flag-POT1b (red, MYC or Flag antibodies) in TRF1F/FTRF2F/Fp53−/−Lig4−/− MEFs (5 days after H&R-Cre). Green, telomeric PNA probe or TRF1 IF.
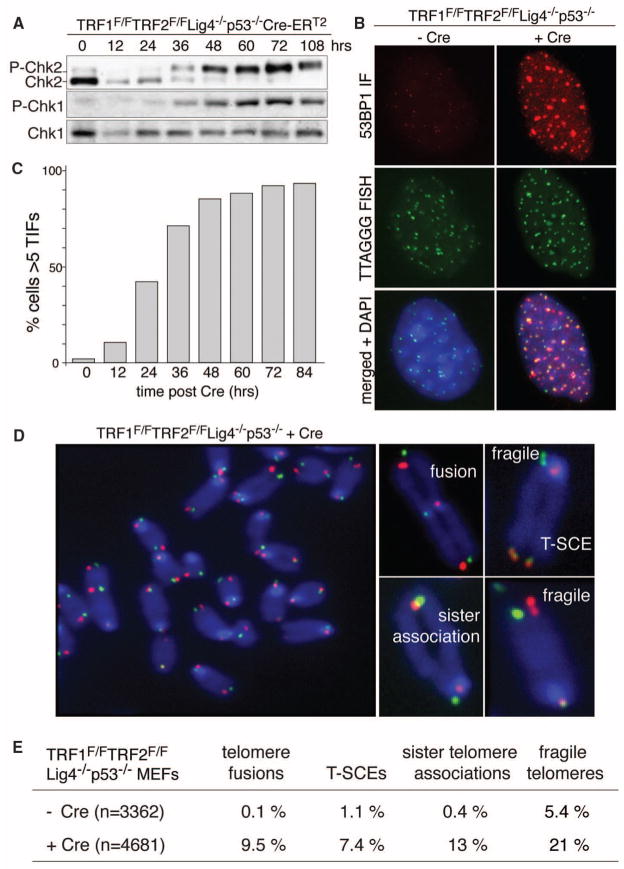
As expected from the ATM/ATR signaling elicited by removal of TRF2 and POT1a, respectively (6), cells with shelterin-free telomeres showed phosphorylation of Chk2 and Chk1, accumulated telomeric 53BP1 foci, and underwent polyploidization (Fig. 2, A to C, and fig. S2, D and E). Telomeric chromosome-orientation fluorescence in situ hybridization (CO-FISH) revealed a cornucopia of telomeric aberrations in metaphase spreads (Fig. 2, D and E). Telomeres often displayed the fragile telomere phenotype typical of the replication defect induced by TRF1 loss (7, 8). There were frequent sister telomere associations, which were previously noted in cells lacking TRF1, TIN2, TPP1, or POT1a/b (7, 9–11), and ~7.5% of the telomeres showed sequence exchanges between sister telomeres [telomere sister chromatid exchanges (T-SCEs)], indicative of the HDR activated upon loss of either Rap1 or POT1a/b (12, 13).
Fig. 2.
Telomere dysfunction upon shelterin loss. (A) Induction of P-Chk1 and P-Chk2 after TRF1/2 codeletion. (B) IF-FISH assay for TIFs (telomere dysfunction-induced foci) in TRF1F/FTRF2F/FLig4−/−p53−/−Cre-ERT2 MEFs (5 days after Cre). FISH for telomeres (green), IF for 53BP1 (red), and 4′,6-diamidino-2-phenylindole (DAPI) as DNA counterstain (blue). (C) Time course of TIF response as in (B). TIFs were scored in TRF1F/FTRF2F/FLig4−/−p53−/−Cre-ERT2 cells at the indicated time points after 4-OHT. Cells with ≥5 telomeric 53BP1 foci were scored as TIF positive (n > 100 nuclei per time point). (D) Metaphase spread from TRF1F/FTRF2F/FLig4−/− p53−/− cells at 108 hours after Cre treatment, analyzed by telomeric CO-FISH using a FITC-OO-[CCCTAA]3 PNA probe (green) and a Tamra-OO-[TTAGGG]3 PNA probe (red). Blue, DAPI. Examples of fragile telomeres, chromosome- and chromatid-type fusions, sister telomere associations, and T-SCEs are on the right. (E) Quantification of aberrant telomeres in Cre-treated TRF1F/FTRF2F/FLig4−/−p53−/− MEFs analyzed as in (D).
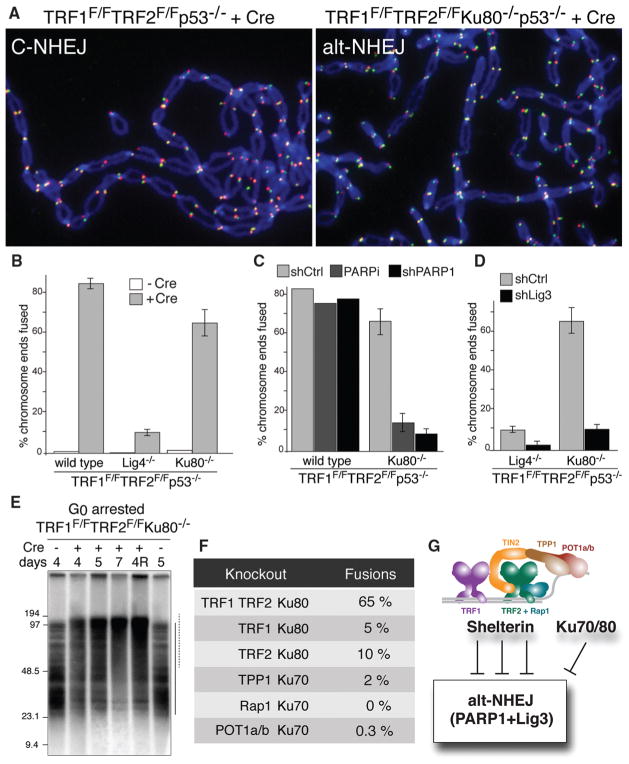
Because these Lig4 cells were NHEJ deficient, it was unexpected that nearly 10% of the telomeres became fused (Fig. 2E and Fig. 3). Furthermore, TRF1/2 DKO in Ku80-deficient MEFs resulted in fusions involving 65% of telomeres (Fig. 3, A and B, and fig. S3A). These results suggested that the shelterin-free telomeres are processed by alt-NHEJ, which is repressed by Ku70/80 and, to a lesser extent, by Lig4 (14–18). Consistent with alt-NHEJ, which is known to be promoted by poly (adenosine diphosphate ribose) polymerase 1 (PARP1) (16, 19), repression of PARP1 with a short hairpin RNA (shRNA) or olaparib (20) significantly reduced the fusion of shelterin-free telomeres in Ku-deficient cells (Fig. 3C and fig. S3B). ShRNA knockdown also implicated Lig3 in the alt-NHEJ of telomeres (Fig. 3D and fig. S3C), pointing to microhomology-mediated end-joining (21), possibly facilitated by the 2 A-T base pairs per telomeric repeat in annealing 3′ overhangs. Analysis of G0-arrested cells revealed that the alt-NHEJ pathway also operates in nonproliferating cells (Fig. 3E and fig. S3, D and E). Although most telomeres were processed by alt-NHEJ when shelterin was removed in toto, individual deletion of shelterin components from Ku null cells failed to result in frequent telomere fusions (Fig. 3F). The finding that deletion of TPP1 does not elicit alt-NHEJ at telomeres in Ku null cells (Fig. 3F) contrasts with a previous suggestion that TPP1/POT1a/b are required to repress alt-NHEJ at telomeres (15). Possibly, the different method used to remove TPP1/POT1a/b in that study had additional effects. We conclude that Lig3/PARP1-dependent alt-NHEJ, is blocked by multiple shelterin components (or their interacting factors) as well as Ku70/80 (Fig. 3G).
Fig. 3.
Lig3- and PARP1-dependent alt-NHEJ in the absence of shelterin. (A) Metaphase chromosomes of the indicated MEFs analyzed (as in Fig. 2D) at 108 hours after Cre. (B) Quantification of telomere fusions in the indicated MEFs at 108 hours after H&R-Cre. Bars and not error bars means of three independent experiments, ± SDs. (C) Quantification of telomere fusions induced by deleting TRF1 and TRF2 [as in (A)] after treatment with PARP1 shRNA or 0.5 μM olaparib. (D) Quantification of telomere fusions [as in (C)] in cells treated with Lig3 or control shRNA. (E) Alt-NHEJ in G0 arrested TRF1F/FTRF2F/FKu80−/−p53+/+Cre-ERT2 MEFs. MboI and AluI digested DNA resolved on a pulsed-field gel electrophoresis probed with end-labeled [AACCCT]4. Dashed and solid lines: fused and unfused telomeres, respectively. Day 4R: cells released on day 4 and analyzed on day 5. (F) Percentage of fused telomeres in Ku-deficient MEFs lacking the indicated shelterin subunit(s). Cells were analyzed at 108 hours after Cre-mediated deletion of the floxed alleles of shelterin. (G) Summary of the repression of Lig3- and PARP1-dependent alt-NHEJ by shelterin and Ku70/80.
We anticipated that fully deprotected, un-fused telomeres would be subject to nucleolytic degradation, which is a marked outcome of telomere dysfunction in yeast [reviewed in (1)]. However, there was no evidence for overt nucleolytic processing of the shelterin-free telomeres (fig. S4A). In addition, in the absence of Ku70/80, which represses resection at telomeres in other eukaryotes (22–25), the overhang signal at the shelterin-free telomeres increased by a factor of <3, even when telomere fusions were repressed by inhibiting PARP1 (fig. S4, A to E). This modest effect suggested that Ku70/80 does not play a major role in repressing 5′ end resection.
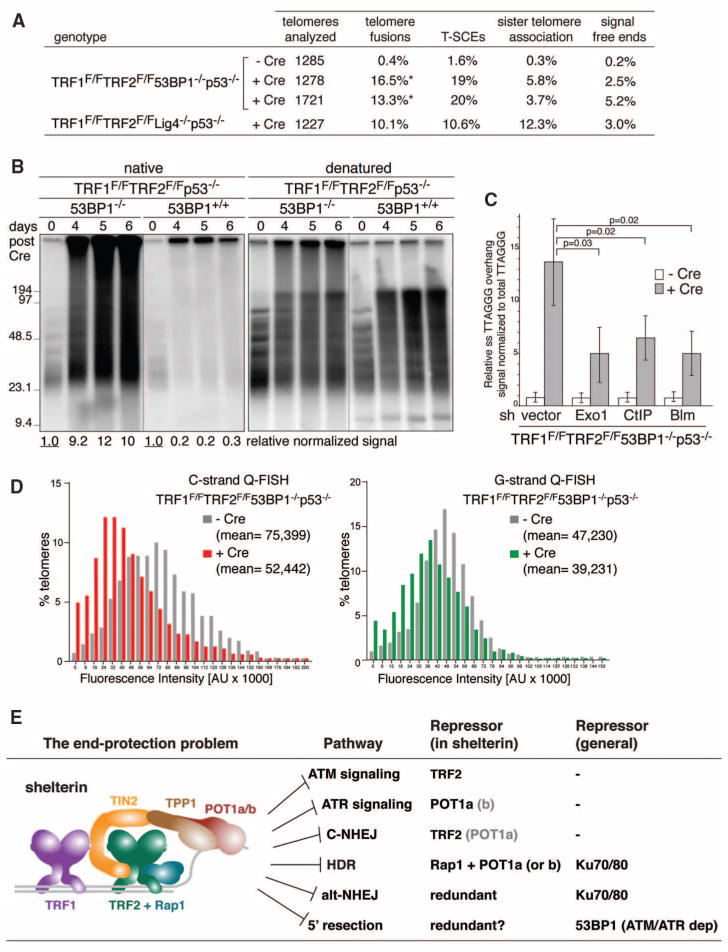
It was recently shown that 5′ end resection at DSBs is minimized by 53BP1, a DDR factor that binds near DSBs and at dysfunctional telomeres in response to ATM or ATR signaling (26, 27). To test the role of 53BP1 at shelterin-free telomeres, we generated TRF1F/FTRF2F/F53BP1−/−p53−/−MEFs. Neither classical nor alt-NHEJ is anticipated at the shelterin-free telomeres of these cells, because 53BP1 is required for Lig4-dependent telomere fusions (5) and Ku70/80 impedes alt-NHEJ (Fig. 3). Indeed, TRF1/2 DKO in 53BP1 null cells elicited a modest level of telomere fusions, mediated mainly by Lig3 (Fig. 4A and fig. S5, A and B), and infrequent sister telomere associations (Fig. 4A). The telomeric overhang signal increased by a factor of ~10 after the TRF1/2 DKO, but not when either TRF1 or TRF2 were deleted from 53BP1-deficient cells (Fig. 4, B and C, and fig. S5C). The excessive signal was due to single-stranded DNA at a 3′ end, as it was removed by the Escherichia coli 3′ exonuclease Exo1 (fig. S5D). The increase in the overhang signal was maximal in cycling cells, regardless of the cell cycle phase, but also occurred in G0 arrested cells (fig. S6, A to D). Because 5′ end resection at DSBs is mediated by CtIP, Blm, and Exo1 (28–30), we examined the role of these factors by shRNA knockdown. Depletion of CtIP, Blm, or Exo1 significantly reduced the overhang signal, establishing that 5′ end resection contributes to the phenotype (Fig. 4C and fig. S7, A to E). Furthermore, quantitative FISH (Q-FISH) recorded a 20 to 30% reduction in the length of the telomeric G-rich and C-rich strands, consistent with nucleolytic degradation (Fig. 4D). Thus, telomeres are in danger of excessive 5′ end resection by enzymes involved in DSB processing. This hyperresection is blocked by shelterin and, in the absence of shelterin, by 53BP1 (Fig. 4E and fig. S7F).
Fig. 4.
53BP1 blocks 5′ end resection and shortening of shelterin-free telomeres. (A) Quantification of telomere aberrations in Cre-treated (108 hours) TRF1F/FTRF2F/F53BP1−/− p53−/− and TRF1F/FTRF2F/FLig4−/−p53−/−MEFs. *, 93% of the cells had ~12% of chromosome ends fused, whereas 7% of the cells had more than 50% of the chromosome ends fused. (B) Representative in-gel 3′ overhang analysis of the indicated MEFs after Cre treatment. Relative overhang signals were normalized to total telomeric DNA (lanes without Cre set to 1). (C) Quantification of 3′ overhangs of TRF1F/FTRF2F/F53BP1−/−p53−/− MEFs (+ or − H&R-Cre, 108 hours) treated with Exo1, CtIP, and Blm shRNAs. The ss/total signal ratios of the +Cre samples are expressed relative to the −Cre samples for each shRNA treatment. Means of three independent experiments ± SDs. P values: two-tailed student’s t tests. (D) Q-FISH of telomeres in TRF1F/F TRF2F/F53BP1−/−p53−/−MEFs with or without H&R-Cre (day 5). (E) Summary of the end-protection problem.
The deleterious events at shelterin-free telomeres revealed that six pathways define the end-protection problem (Fig. 4E). Shelterin is the main armor of chromosome ends, providing protection against classical NHEJ and inadvertent activation of the ATM and ATR signaling. In addition to these primary threats, telomeres can fall victim to alt-NHEJ, HDR, and unmitigated resection. However, these pathways are also blocked by either Ku70/80 or 53BP1, providing a second layer of defense. Although 53BP1 can minimize hyperresection, it will only do so at telomeres that elicit a DNA damage signal. Therefore, the protective ability of 53BP1 is limited and shelterin must prevent hyperresection under most conditions. We speculate that the mechanism by which shelterin fulfills this task is related to how it governs the formation of the correct telomeric overhangs after DNA replication. In contrast to 53BP1, Ku70/80 should be available to blocks alt-NHEJ and HDR at telomeres independent of a DNA damage signal. Why, then, should shelterin also repress these pathways? The redundancy may ensure greater protection, or the repression of alt-NHEJ and HDR may be a secondary outcome of the mechanism by which shelterin executes one of its other functions. As the genetic deconstruction of telomeres has illuminated the full spectrum of processing reactions that threaten chromosome ends lacking proper protection, this study provides a framework for the understanding of the consequences of telomere dysfunction arising from telomere attrition in aging and cancer.
Supplementary Material
Acknowledgments
We thank D. White for exceptional dedication to the mouse husbandry involved in this project and members of the de Lange laboratory for comments on this manuscript. This work was supported by grants from the NIH to T.dL. (GM49046 and AG016642). T.dL. is an American Cancer Society Research Professor.
Footnotes
References and Notes
- 1.de Lange T. Science. 2009;326:948. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. Annu Rev Genet. 2008;42:301. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.Celli GB, de Lange T. Nat Cell Biol. 2005;7:712. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 4.Celli GB, Denchi EL, de Lange T. Nat Cell Biol. 2006;8:885. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 5.Dimitrova N, Chen YC, Spector DL, de Lange T. Nature. 2008;456:524. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denchi EL, de Lange T. Nature. 2007;448:1068. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 7.Sfeir A, et al. Cell. 2009;138:90. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez P, et al. Genes Dev. 2009;23:2060. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. Mol Cell. 2011;44:647. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kibe T, Osawa GA, Keegan CE, de Lange T. Mol Cell Biol. 2010;30:1059. doi: 10.1128/MCB.01498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hockemeyer D, Daniels JP, Takai H, de Lange T. Cell. 2006;126:63. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 12.Sfeir A. Science. 2010;327:1657. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palm W, Hockemeyer D, Kibe T, de Lange T. Mol Cell Biol. 2009;29:471. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boboila C, et al. J Exp Med. 2010;207:417. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rai R, et al. EMBO J. 2010;29:2598. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, et al. Nucleic Acids Res. 2006;34:6170. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simsek D, Jasin M. Nat Struct Mol Biol. 2010;17:410. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan CT, et al. Nature. 2007;449:478. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 19.Mansour WY, Rhein T, Dahm-Daphi J. Nucleic Acids Res. 2010;38:6065. doi: 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menear KA, et al. J Med Chem. 2008;51:6581. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 21.Simsek D, et al. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann P, Cech TR. Mol Biol Cell. 2000;11:3265. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravel S, Larrivée M, Labrecque P, Wellinger RJ. Science. 1998;280:741. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 24.Polotnianka RM, Li J, Lustig AJ. Curr Biol. 1998;8:831. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- 25.Riha K, Shippen DE. Proc Natl Acad Sci USA. 2003;100:611. doi: 10.1073/pnas.0236128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouwman P, et al. Nat Struct Mol Biol. 2010;17:688. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunting SF, et al. Cell. 2010;141:243. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mimitou EP, Symington LS. Nature. 2008;455:770. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Cell. 2008;134:981. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravel S, Chapman JR, Magill C, Jackson SP. Genes Dev. 2008;22:2767. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.