Abstract
The extent to which evolutionary changes have impacted the phenotypic relationships among human diseases remains unclear. In this work, we report that phenotypically similar diseases are connected by the evolutionary constraints on human disease genes. Human disease groups can be classified into slowly or rapidly evolving classes, where the diseases in the slowly evolving class are enriched with morphological phenotypes and those in the rapidly evolving class are enriched with physiological phenotypes. Our findings establish a clear evolutionary connection between disease classes and disease phenotypes for the first time. Furthermore, the high comorbidity found between diseases connected by similar evolutionary constraints enables us to improve the predictability of the relative risk of human diseases. We find the evolutionary constraints on disease genes are a new layer of molecular connection in the network-based exploration of human diseases.
The recent rapid accumulation of functional-genomics and proteomics data provides insight into establishing the evolutionary relationship between the genotypes and phenotypes of human diseases1. We now know that sequence evolution correlates with diverse, genome-wide variables, including gene expression levels, connectivity of gene and protein interactions, and gene-knockout effects2,3,4. These correlations imply that there is an evolutionary relationship between genotypes and phenotypes, though their weakness makes it hard to establish functionally meaningful associations4.
Diseases are often thought of as rare mutations that trigger the loss of a function in an organism5. Sometimes, certain disease phenotypes are reported to be beneficial to the organism's survival and reproduction6,7, meaning that they might have undergone positive selection and evolved faster in response to particular environmental challenges8. While these findings have improved our understanding of the evolutionary constraints on human diseases, the true impact of the evolutionary connections between disease genes and the phenotypic similarities among human diseases needs to be elucidated more fully.
Disease progression is not dictated solely by a mutation in a single gene, but by the totality of the molecular connections in disease modules9,10,11,12. A disease module represents a group of disease genes that share phenotypic similarities as well as molecular connections such as co-expression, protein interactions, metabolic pathways, and co-localizations12,13,14. The disruptions of the connections within disease modules are known to cause particular disease phenotypes12. Thus, it is important to consider the phenotypic connections of diseases to characterize the mechanisms of disease progression9.
We investigated the evolution of human disease genes and found that genes in the same disease class share evolutionary constraints, indicating that the molecular evolution of genes is a novel factor in explaining the phenotypic connections between human diseases. Based on the evolutionary connections of human disease genes, we were able to identify slowly or rapidly evolving disease modules. Furthermore, disease pairs connected by similar evolutionary rates showed a nearly 2-fold increase in comorbidity in comparison with unconnected disease pairs, thus improving the predictability of the relative risk of human diseases. Our findings reveal, for the first time, the relationship between evolutionary connections and comorbidity among diseases, offering a new insight into previously unexplained mechanisms of disease progression.
Results
Diverse evolutionary rates of human disease genes
The evolution of the human genome is believed to have profoundly affected human phenotypes. Accordingly, the evolutionary history of human disease genes has been examined in the hope of gaining insights into the natural selection and adaptive evolution of disease-causing mutations15. To the best of our knowledge, however, a systematic exploration of the evolutionary connections between human disease genes, which may establish a novel molecular connection to human disease classes and phenotypes, is lacking. Hence, we analyzed the evolutionary history of 1,777 human disease genes from the Online Mendelian Inheritance in Man (OMIM) database16 within the framework of disease modules9. For the analysis of the evolution of disease modules, we grouped human diseases into 21 classes based on the affected physiological systems13.
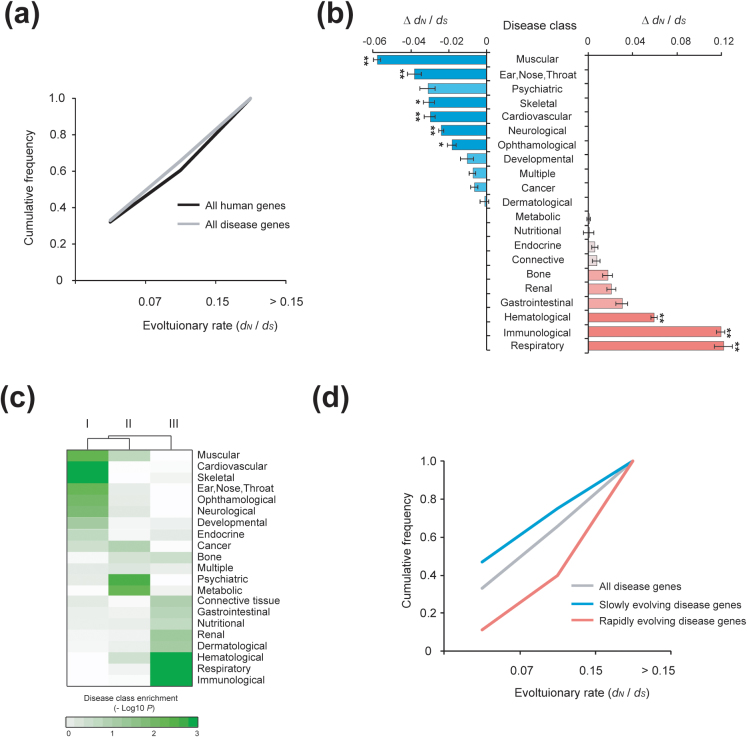
The evolution of human disease genes was examined through the ratios of the non-synonymous nucleotide substitution rate (dN) to the synonymous nucleotide substitution rate (dS) among orthologs of human disease genes; the evolutionary rate (dN/dS) represents the global mutation rate across genomes as well17. We observed a modest difference in the evolutionary rates between disease genes and all human genes (mean of dN/dS for human disease genes = 0.14, for all human genes = 0.16, P = 0.003, Mann-Whitney U test; Fig. 1a and Supplementary Fig. S1). When a gene shows a lower dN/dS than other genes, it has been under purifying selection due to a strong evolutionary constraint, whereas, a gene with a higher dN/dS than other genes indicates faster evolution via positive selection18.
Figure 1. Evolutionary divergence among disease genes in various disease classes.
(a) Cumulative frequency of the evolutionary rate (dN/dS) of human disease genes compared with all human genes (P = 0.003; Mann-Whitney U test). (b) Evolutionary rate of human disease genes classified by the phenotypic system affected; Δ dN/dS values indicate the difference in average evolutionary rates (dN/dS) between all human disease-associated genes and each disease class-associated gene (*P < 0.01, **P < 0.001; Mann-Whitney U test). (c) Enrichment of disease genes within each group quantified by the hypergeometric distribution. Each disease group was divided by the same number of disease genes. The intensity of the green color indicates the significance of enrichment (hypergeometric P-values). (d) Cumulative frequency of evolutionary rate (dN/dS) for all human disease genes and slowly and rapidly evolving human disease genes (P < 1.10 × 10−4; Mann-Whitney U test).
A careful examination of the genes in various disease classes revealed that they possess diverse evolutionary rates. In fact, most disease genes showed significantly different dN/dS values compared with the average (Fig. 1b). In particular, the evolutionary rates of the genes in the muscular, ear/nose/throat, skeletal, cardiovascular, neurological, and ophthalmological disease classes were significantly lower than the average (P < 0.01; Mann-Whitney U test). The means of dN/dS for these disease classes were about 50% smaller than mean value for all human genes, indicating that their sequences are under purifying selection. Meanwhile, the genes in the hematological, immunological, and respiratory disease classes showed significantly higher rates of evolution (P < 0.001; Mann-Whitney U test). Their mean dN/dS values were twice as large as the average for all human genes, indicating that their sequences experienced strong, positive selection. The current understanding of disease genes is that they are mutant alleles responsible for disorders and thus are subjected to purifying selection. Alleles associated with new functions, however, are subjected to positive selection and can be considered human-specific functional adaptations19,20. Notably, the genes of respiratory diseases showed the highest rates of evolution; the high rates may be the result of pathogens of intracellular physiology or positive selection on the proteins involved in respiratory functions that transduce environmental factors21.
We also confirmed that phenotypically connected disease genes are linked by similar evolutionary constraints. Specifically, genes in the same disease class exhibit significant enrichment within a particular evolutionary group (Fig. 1c; P < 0.01, Mann-Whitney U test). The slowly evolving disease genes are significantly enriched in the muscular, cardiovascular, skeletal, ear/nose/throat, ophthalmological, and neurological disease classes; whereas the rapidly evolving disease genes are significantly enriched in the immunological, hematological, and respiratory disease classes. Therefore, from this point onward, we will refer to the diseases as either slowly or rapidly evolving. We also found significant differences in the evolutionary rates between the slowly and rapidly evolving disease genes (Fig. 1d; the mean dN/dS = 0.22 for rapidly evolving disease genes and 0.11 for slowly evolving disease genes, P < 1.10 × 10−4, Mann-Whitney U test).
Phenotypic similarity implies similar evolutionary history
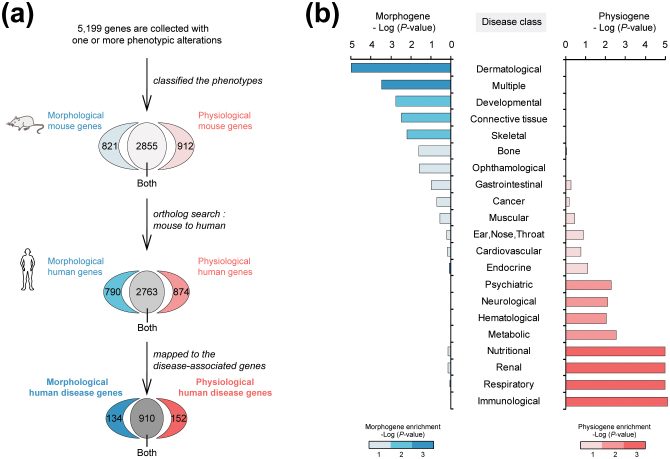
We discovered that disease genes associated with different phenotypes have distinct evolutionary rates; disease genes affecting morphological traits such as anatomical structures evolve more slowly than those affecting physiological traits such as immune responses (Fig. 2). To systematically analyze the relationship between molecular evolution and disease phenotypes, we utilized the mutational phenotypes in the Mouse Genome Database (MGD)22, which contains nearly 4,000 well-annotated genotype-phenotype associations identified through gene knockout, knockdown, trapping, or point mutation. We obtained 134 human morphological disease genes, 152 human physiological disease genes, and 910 human genes with both phenotypes via the ortholog mapping of mouse mutant phenotypes (Fig. 2a, see Methods). Each mouse mutant phenotype was categorized in the Mammalian Phenotype (MP) ontology and classified into three phenotypes: morphogenes which exclusively affect morphological traits; physiogenes which exclusively affect physiological traits; and morpho-physiological genes which affect both traits17.
Figure 2. Morphological- and physiological-disease genes enriched in different disease classes.
(a) Morphological and physiological human disease genes were classified by mapping mouse phenotypic alterations taken from the MGI database; the database describes 5,199 mouse genes that have one or more phenotypic alterations; morphological, physiological, and both; caused by knockout, knockdown, or other mutations. (b) Morphological and physiological human disease genes are enriched differently in various human disease classes. The intensity of color indicates the significance of enrichment (hypergeometric P-values).
We observed that morphological and physiological disease genes showed significantly disparate enrichment patterns across diverse disease classes (Fig. 2b; P < 0.001 in the hypergeometric distribution); for instance, morphogenes are significantly enriched in dermatological diseases and skeletal diseases, whereas physiogenes are grossly enriched in immunological disease and hematological diseases. Acheiropodia, a common skeletal disease that features bilateral congenital amputations of extremities and aplasia of the hands and feet23, shows enrichment of morphological disease genes. In contrast, Thrombocytopenia, a well-known hematological disease characterized by the destruction of platelets and the suppression of platelet production24, shows enrichment of physiological disease genes.
Evolutionary constraints and comorbidity
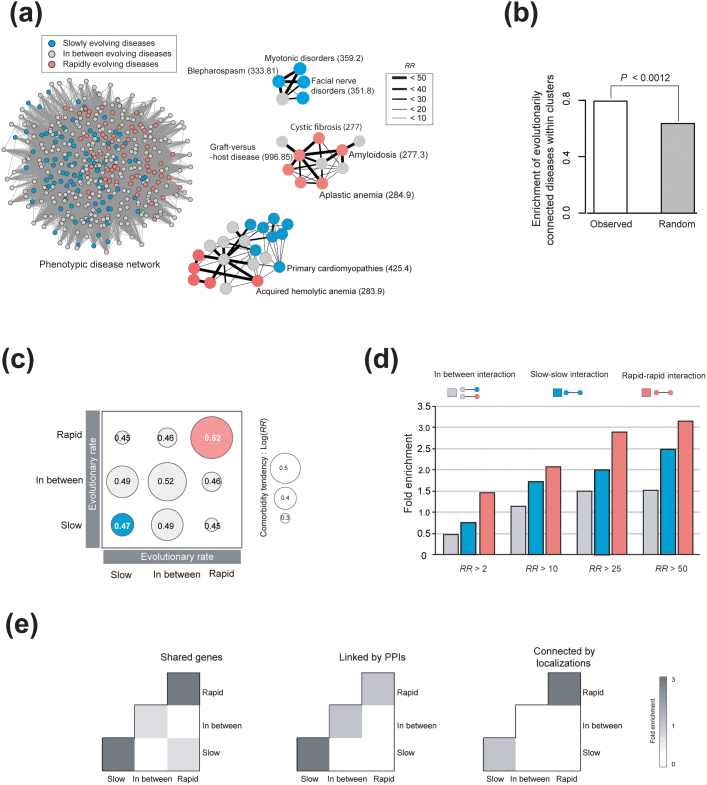
Given the strong correlation between the evolution of disease genes and disease phenotypes, we explored the possibility that the evolutionary connection could manifest itself at the population level via comorbidity25. To evaluate whether evolutionarily connected diseases are more comorbid than disconnected diseases, we measured the relative risks (RRs) of disease pairs with their evolutionary rates and represented them in a phenotypic disease network (see equation (1)) (Fig. 3a), where the nodes are diseases and links connect nodes that are significantly comorbid. We only used the disease pairs with relative risks based on 99% confidence intervals26,27 (see equations (2), (3) and (4)). The network consists of 515 diseases, of which 165 are slowly evolving, 91 are rapidly evolving, and 424 have in-between evolutionary rates.
Figure 3. Molecular evolution of disease genes connects disease comorbidity tendency.
(a) Network representation of 515 comorbid diseases (nodes) connected by 9,230 comorbidity links (comorbidity tendency, RR). Links with 99% confidence intervals are presented. Examples of evolutionarily connected diseases are shown with ICD-9 codes. (b) Comorbid disease pairs connected by similar evolutionary constraints are compared with a random control. (c) Average comorbidity tendencies (Log RR) in various links are indicated by the size of the circle. (d) Fold enrichment of phenotypic interactions according to link types: similarly evolving diseases (diseases that have similar evolutionary rates) and dissimilar disease pairs (diseases that have different evolutionary rates). Adjusted relative risk using 99% confidence intervals was used. (e) Comorbid disease pairs connected by similar evolutionary rates had shared genes linked by protein-protein interactions and connected by subcellular localizations. The enrichment ratio is proportional to the dark gray scale; fold-enrichment was calculated as the ratio of the observed to expected comorbid disease pairs in the evolutionary rate.
At first glance, the phenotypic disease network was largely divided into two obvious subnetworks of slowly evolving (blue) and rapidly evolving (red) diseases (Fig. 3a). To quantify the enrichment of slowly and rapidly evolving diseases, we used the Markov CLustering algorithm (MCL; Supplementary Fig. S2). We observed that disease pairs connected by similar evolutionary rates tended to enrich within clusters compared with the random control (Fig. 3b; P < 0.0012, Mann-Whitney U test). Moreover, we found that disease pairs that were connected by similar evolutionary rates tend to have high comorbidity tendency (Fig. 3c). For example, the pair of Blepharospasm (ICD-9-CM 33.81) and Facial Nerve Disorders (ICD-9-CM 351.8), both slowly evolving diseases, had high comorbidity (RR = 96.51), whereas the pair of Acquired Hemolytic Anemia (ICD-9-CM 283.9) and Primary Cardiomyopathies (ICD-9-CM 425.4), one rapidly evolving and the other slowly evolving, had low comorbidity (RR = 2.19).
The tendency is widespread; pairs of similarly evolving diseases (slow-slow or rapid-rapid) have higher comorbidities than dissimilar pairs. The positive correlation between the similarity in evolutionary constraints and comorbidity raises an important question: would disease pairs with high comorbidity be strongly connected evolutionarily? To address this, we analyzed the increase in comorbidity tendency with regard to interaction types (Fig. 3d). Strongly comorbid disease pairs (RR > 50) showed a nearly 2-fold increase in similarly evolving groups compared with weakly comorbid pairs (RR > 2), suggesting that evolutionary constraints do indeed connect phenotypes and strengthen the relative risks of human diseases.
We further examined whether disease pairs with similar evolutionary rates also share other molecular connections. Because evolution is one of the important aspects of understanding molecular function, we expected that diseases connected by similar evolutionary constraints would share other molecular connections compared with non-connected disease pairs. Previous studies showed that comorbid diseases also share molecular connections such as genes, protein-protein interactions, subcellular localizations, and metabolic pathways12,14,25. Indeed, we discovered that diseases connected by similar evolutionary constraints tend to have other molecular connections as well (Fig. 3e). In particular, diseases in similarly evolving groups showed nearly a 2-fold increase in the strength of molecular connections. Our results strongly suggest that a deeper evolutionary analysis of disease genes is a frontier of research that can be useful in understanding the human disease network.
Discussion
We showed that evolutionary constraint is a new layer of molecular connection in the human disease network. Until now, evolution has not been studied quantitatively as a concrete link between phenotypes in the human disease network. Our analysis establishes for the first time that evolutionary connections definitely exist in disease modules, offering a new avenue for gaining insights into the etiology of human diseases and the mechanisms of disease progression.
We can ask whether evolution has also impacted phenotypic connections between complex diseases such as diabetes or bipolar disorder. It has been shown that both genetic and environmental factors contribute to the phenotypic connections between complex diseases. The genes associated with complex diseases are under more positive selection than genetic diseases, and they also display more recent evolutionary origins6,28. We analyzed the evolution of complex disease genes and found that various classes of complex diseases are also under diverse evolutionary constraints. The evolutionary rates of complex disease genes were analyzed based on the Genetic Association Database (GAD) which provides common, complex disease-gene relationships29. Specifically, complex diseases with similar evolutionary rates fall into the same classes, and complex diseases show a wide range of evolutionary rates, as we also observed for genetic diseases (Supplementary Fig. S3). For instance, the evolutionary rates of chemo-dependency and psychiatric diseases were low, whereas those of infectious and hematological diseases were high. Based on these observations, we conclude that evolution also affects the phenotypic connections of complex diseases.
Our findings on the evolution of disease classes are in good accord with the tissue specificity of human diseases, as various tissues and organs are associated with different disease types, and their features are reflected in the tissue-specific pathology of human diseases12,30. It was recently shown that tissue-specific gene expression varies according to different evolutionary pressures. Brawand et al. showed, from the comparison of mammalian transcriptomes, that the rate of gene evolution varied among mammalian organs31. In particular, the genes in neural tissues evolve much more slowly than the genes in other tissues, which is consistent with our finding that neurological disease genes evolve slowly. This presents a need for further exploration of the connections between tissue types and the evolution of human diseases.
We classified human disease genes by using mouse mutational phenotypes that relate to morphological or physiological traits. Among model organisms, the mouse is considered important for the study of human diseases because it offers extensive genotype-phenotype associations that allow us to overcome the present limitations on genotype-phenotype linkages in humans32. Although two different phenotypes, morphological and physiological traits, displayed distinct evolutionary patterns, it could be the result of a bias in the mouse mutational phenotypes. To further verify the relationship between disease phenotypes and evolutionary constraints, we analyzed the enrichment of human Gene Ontology (GO) biological processes according to evolutionary groups. We confirmed that each evolutionary group showed distinct human GO biological process enrichment patterns (Supplementary Fig. S4).
Regardless of evolutionary origins, it is clear that evolutionary connections do exist in disease modules, offering a new insight into the etiology of human diseases and the mechanisms of disease progression. We demonstrated that diseases connected by similar evolutionary constraints tend to have similar phenotypes and comorbidity tendencies. The implications and applications of the evolutionary understanding of disease phenotypes are important in characterizing human disease phenotypes and contribute to a more robust foundation for network medicine.
Methods
Data set for the human disease genes
The gene-disease associations were collected from the OMIM database (http://www.ncbi.nlm.nih.gov/omim/) as described in Park et al.12 The OMIM database provides gene-disease associations between the 2,929 disease types in the Morbid Map and 1,777 disease-associated genes. The disease types were grouped into 1,340 distinct diseases by combing disease subtypes into single diseases based on their given disease names13. Finally, 2,161 disease terms were grouped into 1,228 unique diseases. For the evolutionary analysis of disease classes, diseases types were grouped into 21 classes based on the physiological systems they affected. For example, 215 disorders, such as HMG-CoA synthase-2 deficiency and CPT II deficiency, constituted the “Metabolic” disease class. This classification scheme reflects the phenotypic similarities among diseases in the same class and has been successfully used in the recent studies of systematic disease analyses12,14. To analyze phenotypically connected disease genes that are linked by similar evolutionary constraints, we grouped the disease genes according to their evolutionary rates. Each group contains roughly the same number of genes (type I, 687; type II, 659; and type III, 667). To analyze the evolutionary rates of non-Mendelian diseases, we used the GAD archive29 as described by Park et al.12 Among the 167 disease-associated genes, we collected 151 that have mouse-orthologs information.
Comorbidity information was obtained from the United States Medicare database recorded in the ICD-9-CM format (http://www.icd9data.com). This database contains the diagnoses of 13,039,018 elderly patients from 1990 to 1993 and has been successfully used in comorbidity analyses12,14,25,27. We manually mapped between ICD-9-CM codes and OMIM diseases14,25 to further analyze 83,924 comorbid pairs of hereditary diseases in this study.
Comparative genomics analysis
The dN/dS values of the human and mouse genes were computed. The human and mouse genomes were downloaded from National Center for Biotechnology Information NCBI36 and NCBIM36, respectively. The mouse orthologs, including sequences, of human genes were extracted using BioMart (http://www.biomart.org/). The human-mouse orthologs were grouped into 14,423 pairs, similarly to the work of Liao et al.33 Sequence alignments between the human and mouse genes were carried out through phylogenetic analysis (http://www.ensembl.org/info/docs/compara/homology_method.html). The dS and dN values between human and mouse orthologs were obtained from BioMart and estimated by the likelihood method33. Finally, the evolutionary rates of 1,662 disease genes were obtained from the 1,777 human disease genes (Supplementary Table S1).
Morphological and physiological disease genes
To obtain the morphological and physiological traits, the mutational phenotypes of the mouse genes were taken from the Mouse Genome Information (MGI) database (http://www.informatics.jax.org/) version 4.1122. The MGD provides a well-annotated list of 5,199 genes with one or more MP IDs identified from gene knockout, knockdown, trapping, or point mutations. The MP IDs were further classified into two groups: morphological (129 IDs), and physiological (183 IDs); similarly to the work of Liao et al.17 We analyzed 821 mouse morphogenes, 912 mouse physiogenes, and 2,855 mouse genes associated with both morphological and physiological IDs. Human orthologs of 4,588 mouse genes were obtained from MGI_MouseHumanSequence.rpt. Mouse-to-human ortholog mapping was downloaded from MGI; and finally, we obtained 4,379 human orthologs of mouse genes. In total, 790 human morphogenes, 874 human physiogenes, and 2,763 human genes with both phenotypes were obtained (Supplementary Table S2).
Comorbidity measure (RR)
We used the RR as the quantitative measure of the comorbidity tendency of disease pairs25. The RR allows us to quantify the co-occurrence of disease pairs compared with the random expectation. It is calculated as
where N is the total number of Medicare patients (13,039,018), Ii is the incidence of disease i, Ij is the incidence of disease j, Cij is the number of patients who had both diseases i and j, and Cij* is equal to the random expectation IiIj/N.
To calculate the significance of the RR, we used the method of Katz et al. to estimate confidence intervals34. According to their estimation, the 99% confidence interval for the RR between two diseases i and j is calculated by:
where σ12is given by:
Disease pairs within the 99% confidence interval are only considered if the LB value is larger than 1 when RR is larger than 1, or if the UB value is smaller than 1 when RR is smaller than 1.
Clustering analysis in the phenotypic disease network
To identify clusters in the phenotypic disease network, we used the MCL with the default option. To quantify the enrichment of slowly and rapidly evolving diseases in the clusters, we measured the difference of enrichment between the slowly and rapidly evolving diseases within the clusters. To measure statistical significance, we randomly assigned disease classes to the diseases. After 10,000 randomizations, we calculated the P-value by the Mann-Whitney U test.
Gene ontology enrichment analysis
Each disease-associated gene was annotated with a GO biological process based on the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov)35,36. The P-values in the functional enrichment analyses were calculated using a hypergeometric distribution on each evolutionary group: slow, rapid, and in-between.
Subcellular localization analysis
Subcellular localization information was assigned to every disease-associated protein12. Briefly, the underlying data stems from the Swiss Prot annotation information and subcellular-localization predictors, ConLoc, and Proteome Analyst37,38, which have previously been shown to be effective for mapping subcellular-localization information to disease-associated proteins12. Subcellular-localization information was available for 1,168 disease-associated proteins from the “Cellular Component” field of Swiss Prot. The subcellular localizations of the remaining proteins were predicted by ConLoc and Proteome Analyst. ConLoc was designed to predict protein subcellular localization by optimizing the results of 13 predictors for five major localizations (cytosol, extracellular, mitochondria, nucleus, and plasma membrane)38. Other subcellular localizations (endoplasmic reticulum, Golgi, peroxisome, and lysosome) were predicted by Proteome Analyst37.
Protein-protein interaction network
The human protein interaction network was compiled by integrating eight existing interaction databases: the Biomolecular Interaction Network Database, the Human Protein Reference Database, the Molecular Interaction database, the Database of Interacting Proteins, IntAct, BioGRID, Reactome, and the Protein-Protein Interaction Database. Redundant interactions and low-confidence interactions were filtered out12. The final network comprises 65,135 interactions among 10,652 human proteins.
Author Contributions
SP designed the whole work, produced all the data, and wrote the paper. JSY, JK, YES, and JHH helped to analyze the data. JP wrote the paper and provided comorbid disease pairs. SKJ supervised the work. SK supervised the whole work and contributed to the manuscript preparation. All authors read and corrected the manuscript before the submission.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Drs. Jianzhi Zhang (University of Michigan) for providing mouse genes associated with phenotype traits. This work was supported in part by National Research Foundation grants of Korea (2012002568, 20110027840, and R312011000101 of the World Class University program), Seoul National University Cancer Research Institute (cri-10-2), and a POSTECH BSRI grant.
References
- Streelman J. T. & Kocher T. D. From phenotype to genotype. Evol Dev 2, 166–73 (2000). [DOI] [PubMed] [Google Scholar]
- Herbeck J. T. & Wall D. P. Converging on a general model of protein evolution. Trends Biotechnol 23, 485–7 (2005). [DOI] [PubMed] [Google Scholar]
- Wolf Y. I. Coping with the quantitative genomics ‘elephant’: the correlation between the gene dispensability and evolution rate. Trends Genet 22, 354–7 (2006). [DOI] [PubMed] [Google Scholar]
- Koonin E. V. & Wolf Y. I. Evolutionary systems biology: links between gene evolution and function. Curr Opin Biotechnol 17, 481–7 (2006). [DOI] [PubMed] [Google Scholar]
- Blekhman R. et al. Natural selection on genes that underlie human disease susceptibility. Curr Biol 18, 883–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J. J., Borenstein E., Chen R. & Petrov D. A. Similarly strong purifying selection acts on human disease genes of all evolutionary ages. Genome Biol Evol 1, 131–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender E. J. & Lahn B. T. Positive selection on the human genome. Hum Mol Genet 13 Spec No 2, R245–54 (2004). [DOI] [PubMed] [Google Scholar]
- Sabeti P. C. et al. Positive natural selection in the human lineage. Science 312, 1614–20 (2006). [DOI] [PubMed] [Google Scholar]
- Barabasi A. L., Gulbahce N. & Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 12, 56–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage K. et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol 25, 309–16 (2007). [DOI] [PubMed] [Google Scholar]
- Wu X., Jiang R., Zhang M. Q. & Li S. Network-based global inference of human disease genes. Mol Syst Biol 4, 189 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. et al. Protein localization as a principal feature of the etiology and comorbidity of genetic diseases. Mol Syst Biol 7, 494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K. I. et al. The human disease network. Proc Natl Acad Sci U S A 104, 8685–90 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. S. et al. The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci U S A 105, 9880–5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Systemic determinants of gene evolution and function. Mol Syst Biol 1, 2005 0021 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A., Scott A. F., Amberger J. S., Bocchini C. A. & McKusick V. A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33, D514–7 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B. Y., Weng M. P. & Zhang J. Contrasting genetic paths to morphological and physiological evolution. Proc Natl Acad Sci U S A 107, 7353–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes A. et al. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci 14, 19–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Schreiber F., Thanintorn N. & Sonnhammer E. L. OrthoDisease: tracking disease gene orthologs across 100 species. Brief Bioinform 12, 463–73 (2011). [DOI] [PubMed] [Google Scholar]
- Barreiro L. B. & Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet 11, 17–30 (2010). [DOI] [PubMed] [Google Scholar]
- Mukherjee A. B. & Zhang Z. Allergic asthma: influence of genetic and environmental factors. J Biol Chem 286, 32883–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig J. T. et al. The Mouse Genome Database (MGD): from genes to mice--a community resource for mouse biology. Nucleic Acids Res 33, D471–5 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianakiev P. et al. Acheiropodia is caused by a genomic deletion in C7orf2, the human orthologue of the Lmbr1 gene. Am J Hum Genet 68, 38–45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuker A. & Cines D. B. Immune thrombocytopenia. Hematology Am Soc Hematol Educ Program 2010, 377–84 (2010). [DOI] [PubMed] [Google Scholar]
- Park J., Lee D. S., Christakis N. A. & Barabasi A. L. The impact of cellular networks on disease comorbidity. Mol Syst Biol 5, 262 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbahce N. et al. Viral perturbations of host networks reflect disease etiology. PLoS Comput Biol 8, e1002531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo C. A., Blumm N., Barabasi A. L. & Christakis N. A. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol 5, e1000353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N. & Kehrer-Sawatzki H. Exploring the potential relevance of human-specific genes to complex disease. Hum Genomics 5, 99–107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K. G., Barnes K. C., Bright T. J. & Wang S. A. The genetic association database. Nat Genet 36, 431–2 (2004). [DOI] [PubMed] [Google Scholar]
- Jiang X. et al. Modularity in the genetic disease-phenotype network. FEBS Lett 582, 2549–54 (2008). [DOI] [PubMed] [Google Scholar]
- Brawand D. et al. The evolution of gene expression levels in mammalian organs. Nature 478, 343–8 (2011). [DOI] [PubMed] [Google Scholar]
- Lee I., Blom U. M., Wang P. I., Shim J. E. & Marcotte E. M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res 21, 1109–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B. Y. & Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A 105, 6987–92 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D., J B., Azen S. P. & M C., P. . Obtaining confidence interval for the risk ratio in cohort studies. Biometrics 34, 469–474 (1978). [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafron D. et al. Proteome Analyst: custom predictions with explanations in a web-based tool for high-throughput proteome annotations. Nucleic Acids Res 32, W365–71 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Yang J. S., Jang S. K. & Kim S. Construction of functional interaction networks through consensus localization predictions of the human proteome. J Proteome Res 8, 3367–76 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information