Abstract
N-acetylglucosamine (GlcNAc), the monomer of chitin and constituent of bacterial peptidoglycan, is a preferred carbon and nitrogen source for streptomycetes. Recent studies have revealed new functions of GlcNAc in nutrient signaling of bacteria. Exposure to GlcNAc activates development and antibiotic production of Streptomyces coelicolor under poor growth conditions (famine) and blocks these processes under rich conditions (feast). Glucosamine-6-phosphate (GlcN-6P) is a key molecule in this signaling pathway and acts as an allosteric effector of a pleiotropic transcriptional repressor DasR, the regulon of which includes the GlcNAc metabolic enzymes N-actetylglucosamine-6-phosphate (GlcNAc-6P) deacetylase (NagA) and GlcN-6P deaminase (NagB). Intracellular accumulation of GlcNAc-6P and GlcN-6P enhanced production of the pigmented antibiotic actinorhodin. When the nagB mutant was challenged with GlcNAc or GlcN, spontaneous second-site mutations that relieved the toxicity of the accumulated sugar phosphates were obtained. Surprisingly, deletion of nagA also relieved toxicity of GlcN, indicating novel linkage between the GlcN and GlcNAc utilization pathways. The strongly enhanced antibiotic production observed for many suppressor mutants shows the potential of the modulation of GlcNAc and GlcN metabolism as a metabolic engineering tool toward the improvement of antibiotic productivity or even the discovery of novel compounds.
Keywords: metabolism, aminosugar, signaling, glycolysis, secondary metabolism, sporulation
Introduction
The alarming emergence of multiply antibiotic-resistant pathogens coupled with the lack of effective antimicrobials has revealed an urgent requirement for discovery of new antibiotics. Streptomycetes are Gram(+), soil-dwelling bacteria that have a complex life cycle that starts with the germination of a spore, which then grows out to form a branched vegetative mycelium. When morphological differentiation is initiated, a so-called aerial mycelium is produced, followed by the formation of long chains of unigenomic spores.1,2 Streptomycetes are best known for their ability to produce a broad range of secondary metabolites, including around 50% of all known antibiotics, as well as many antifungal, anticancer and immunosuppressant agents.3 The control of antibiotic production is complex, with typically many different transcriptional regulators dictating the expression of a single gene cluster, allowing the producing organism to correctly time biosynthesis, for example in response to growth phase or nutrient availability.4,5
As saprophytic soil bacteria, streptomycetes are able to grow on a wide range of organic compounds due to production of numerous extracellular enzymes, such as amylases, cellulases, chitinases, proteases and lipases. Since organic matter is also a predominant source of nutrition for other microorganisms, bacteria had to develop mechanism of predation leading to elimination of competing organisms. It is suggested that in natural habitat of bacteria, this mechanisms is stimulated by production of antibiotics leading to thwart of growth of competing organisms.3
Sequencing of the genomes of many Streptomyces and other actinomycetes revealed that most likely all filamentous actinomycetes possess a large arsenal of secondary metabolites, and only part of these are produced under standard laboratory conditions.6 Therefore, novel strategies not only for the improvement of antibiotic production, but also to wake up sleeping (also referred to as silent or cryptic) biosynthetic clusters are required to harness this potential. Current methods and technological advances regarding the discovery of new antibiotics have been reviewed elsewhere.7-9 Such approaches include engineering primary metabolism and regulatory cascades controlling antibiotic production.8,10,11
Role of GlcNAc in Control of Development and Antibiotic Production
N-acetylglucosamine (GlcNAc), the monomer or chitin and constituent of peptidoglycan, and the related amino acid glutamate are preferred nutrients as they serve as highly favorable carbon and nitrogen sources for streptomycetes.12,13 Besides as nutrient, GlcNAc also serves as a signaling molecule in streptomycetes. Accumulation of GlcNAc around the colonies at higher concentrations controls both morphological differentiation (sporulation) and antibiotic production; under poor nutritional conditions (famine) the accumulation of GlcNAc promotes development and antibiotic production, while under rich growth conditions (feast) these processes are inhibited.14 We previously proposed a complete signaling cascade from GlcNAc uptake to the onset of antibiotic production under famine conditions. This pathway is transmitted via the master regulator DasR, whose regulon includes genes for GlcNAc transport and metabolism as well as antibiotic production. A key molecule in this signaling cascade is GlcN-6P, which acts as an allosteric effector of DasR and therefore prevents its DNA binding ability.15 This results in the relief of transcriptional repression of the pathway-specific regulatory genes for antibiotic production, including actII-ORF4 and redZ, the pathway-specific activator genes for actinorhodin and prodiginine biosynthesis, respectively. DasR also represses the cpk gene cluster for the cryptic polyketide Cpk, although this interaction may be mediated via the control of scbA and scbR,5 which encode the synthetase for the γ-butyrolactone Scb1 and the Scb1-responsive transcriptional activator for Cpk biosynthesis, respectively.16,17 The molecular mechanism of GlcNAc signaling under rich nutritional conditions, which results in the blockage of sporulation and antibiotic production, is still unknown.
N-acetylglucosamine Metabolism
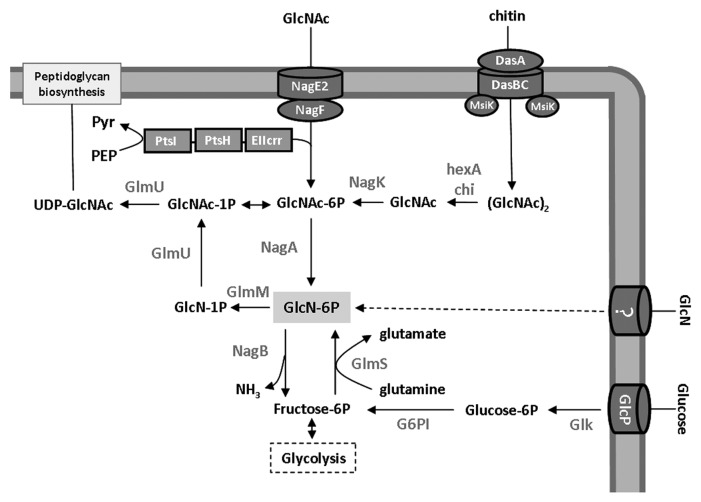
In S. coelicolor, the model organism of species, GlcNAc is transported via the phosphoenolpyruvate-dependent phosphotransferase system (PTS).18,19 During PTS-mediated carbon source uptake, a phosphoryl group is transferred from phosphoenolpyruvate (PEP) to the general phosphotransferase enzyme I (EI; encoded by ptsI), from there to HPr (encoded by ptsH), and then further onto enzyme IIA (EIIACrr; encoded by crr) and enzyme IIB (e.g., NagF).12,20 NagF phosphorylates incoming via the permease IIC (NagE2) GlcNAc to form N-acetylglucosamine-6-phosphate (GlcNAc-6P) (Fig. 1). Null mutants that have been constructed for any of the global PTS components (EI, EIIA, or HPr) locks streptomycetes in the vegetative growth phase.15
Figure 1. Glucose and aminosugar metabolism in S. coelicolor, with connecting pathways. Annotation is based on the KEGG database31 and experimental evidence. Arrows shows the direction of metabolism, and the responsible enzymes are presented in bold face. See text for further details.
Recently we characterized the metabolic enzymes in S. coelicolor, namely GlcNAc-6P deacetylase (NagA; SCO4284), GlcN-6P deaminase (NagB; SCO5236) and GlcNAc kinase (NagK; SCO4285).21 Similarly to the GlcNAc utilization pathway of other bacteria, intracellular GlcNAc-6P is deacetylated by NagA, which results in glucosamine-6-phosphate (GlcN-6P), the molecule which occupies a central position in the pathways toward cell-wall synthesis and glycolysis. GlcN-6P is converted by NagB to fructose-6-phosphate (Fru-6P), which enters the glycolytic pathway,22,23 but is also incorporated into murein following its conversion to UDP-GlcNAc by the action of GlmM (phosphoglucosamine mutase) and GlmU (N-acetylglucosamine-1-phosphate uridyltransferase) (Fig. 1). During growth on other carbon sources than aminosugars, GlcN-6P can be produced from Fru-6P by GlcN-6P synthase (glmS) (Fig. 1). NagK catalyzes the phosphorylation of intracellular GlcNAc to GlcNAc-6P (Fig. 1). In S. coelicolor, monomeric GlcNAc is transported via the PTS, resulting in intracellular GlcNAc-6P.12 Intracellular unphosphorylated GlcNAc is most likely internalized as N-N′-diacetylchitobiose [(GlcNAc)2] and/or other chito-oligosaccharides [(GlcNAc)n], such as those derived from chitin via the chitinolytic system.
Interestingly, some Streptomyces species, including S. bingchenggensis, S. cattleya, and S. violaceusniger, do not have a nagB ortholog. Genome mining revealed the presence of an open reading frame, which appears to be a hybrid of nagB/glmS2 and shows similarity to the nagB-II genes present in Gram-positive bacteria. These NagB-II homologs contain the C-terminal domain of GlmS, but display only the catabolic activity of NagB.24,25 The fact that these three streptomycetes also contain the PTS genes for internalization and phosphorylation of GlcNAc, including nagE2 (SBI_06686, SCATT_19300, Strvi_8260) and nagF (SBI_06687, SCATT_19290, Strvi_8259), as well as nagA (SBI_04956, SCAT_2457, Strvi_8764) for the subsequent deacetylation of GlcNAc-6P to GlcN-6P, strongly suggests that indeed these organisms can catabolize GlcNAc using the NagB/GlmS2 hybrid enzyme instead of the canonical NagB.
Engineering of Aminosugar Metabolism as an Approach to Enhance Antibiotic Production
Better understanding of GlcNAc-mediated signaling in streptomycetes requires the creation of multiple mutants, single and in combination, to block metabolic routes so as to force the accumulation of specific metabolites. Thus new insight is obtained with regard to the flux of GlcNAc and related metabolites, and how this influences the global regulatory routes that control Streptomyces development and antibiotic production. In the absence of nagA, addition of GlcNAc results in the accumulation of GlcNAc-6P, while in nagB mutants GlcN-6P accumulates (Fig. 1). nagA deletion mutants showed strongly enhanced production of the blue-pigmented antibiotic actinorhodin in the presence of GlcNAc, on both rich (R2YE) and poor (MM) solid media.21 The likely accumulation of GlcN-6P in nagB mutants led to a strong increase in Act production on R2YE agar plates supplemented with GlcNAc, while the mutant failed to grow on MM agar plates supplemented with either GlcNAc or its deacetylated form, glucosamine (GlcN) (ref. 21 and Fig. 2). Toxicity of GlcN-6P was reported previously in Escherichia coli.26-28 In line with the idea that the accumulation of GlcN-6P was the cause of the toxicity, nagAB double mutants (which do not accumulate GlcN-6P for the lack of NagA activity; Fig. 1) were able to grow on GlcNAc, but the enhanced antibiotic production remained (Fig. 2). However, we could not immediately explain the surprising observation that also the toxicity of GlcN to nagB mutants was relieved by additional deletion of nagA, as involvement of NagA in GlcN metabolism was not anticipated (Fig. 2). This sheds new light on GlcN metabolism and suggests direct involvement of NagA in, or linkage between, both pathways.
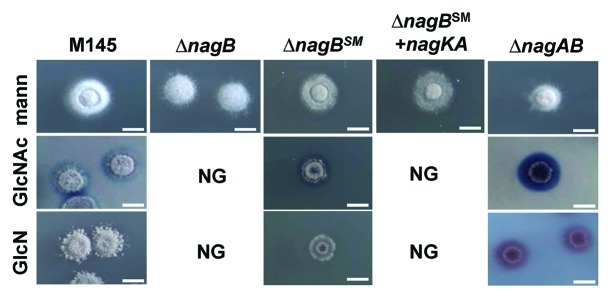
Figure 2. Phenotypic analysis of nag mutants. Stereomicrographs showing the phenotypes of S. coelicolor M145, its nagB and nagAB mutants, nagB suppressor mutant SMG4 (obtained on GlcN) that has a deletion in nagA, and the complemented suppressor mutant expressing nagKA. Strains were grown for 5 days on MM agar plates supplemented with mannitol, GlcN or GlcNAc. Note that nagB mutants and complemented nagB suppressor mutants fail to grow (NG) on either aminosugar, while growth was restored to the nagAB double mutant and the nagB suppressor mutants, and also led to enhanced antibiotic overproduction.
When nagB mutant spores were plated at high density onto MM agar plates containing either GlcNAc or GlcN, suppressor mutants were readily obtained, with a frequency of around 1:105, which corresponds to expected single mutations or indels (insertions or deletions). This is a logical consequence of the fact that single mutations in for example nagA or any of the transporter genes should suffice to circumvent the accumulation of GlcN-6P as the likely cause for the observed toxicity. Reproducibly at least three different phenotypes were obtained for suppressor mutants allowing growth in the presence of GlcNAc restreaked on MM+GlcNAc, while suppressor mutants with restored growth in the presence of GlcN had similar phenotypes on MM+GlcN. This suggests that suppressor mutations may have arisen in different genes.
Fifteen suppressor mutants obtained on GlcNAc, designated SMA1-SMA15 (Suppressor Mutants isolated on GlcNAc) and five independent suppressor mutants obtained on GlcN-containing media, SMG1-SMG5 (Suppressor Mutant isolated on GlcN), were re-streaked on MM agar plates supplemented with mannitol, glucose, GlcN or GlcNAc. All strains developed normally on MM agar plates supplemented with mannitol (Fig. 3). However, three different phenotypes were observed for nagB suppressor mutants obtained in the presence of GlcNAc (Fig. 3 and Fig. 4). The first class of GlcNAc-selected nagB suppressor mutants displayed decreased antibiotic production but normal development (SMA1; SMA4-SMA5). The two other classes showed enhanced antibiotic production and either blocked (SMA2-SMA3; SMA6-SMA7; SMA9; SMA11-SMA15) or normal development (SMA8 and SMA10) (Fig. 3 and Fig. 4).
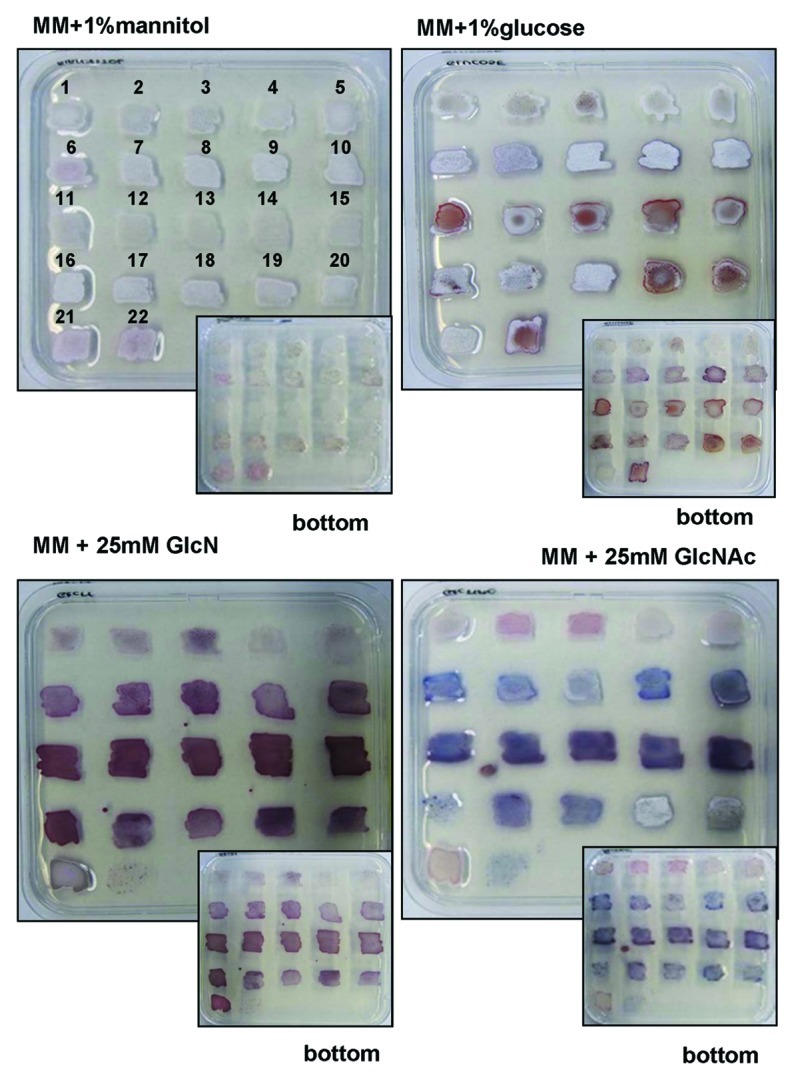
Figure 3. Analysis of antibiotic production by nagB suppressor mutants. Phenotypes of nagB suppressor mutants originally selected on GlcNAc (1–15; corresponding to SMA1-SMA15) or GlcN (16–20; corresponding to SMG1-SMG5). The parental strain M145 (21) and the nagB mutant (22) were used as controls. Strains were grown on MM agar plates supplemented with mannitol, glucose, GlcN or GlcNAc. Note that the nagB mutant failed to grow on MM supplemented with either aminosugar. Both front and reverse (inserts) of the agar plates are shown.
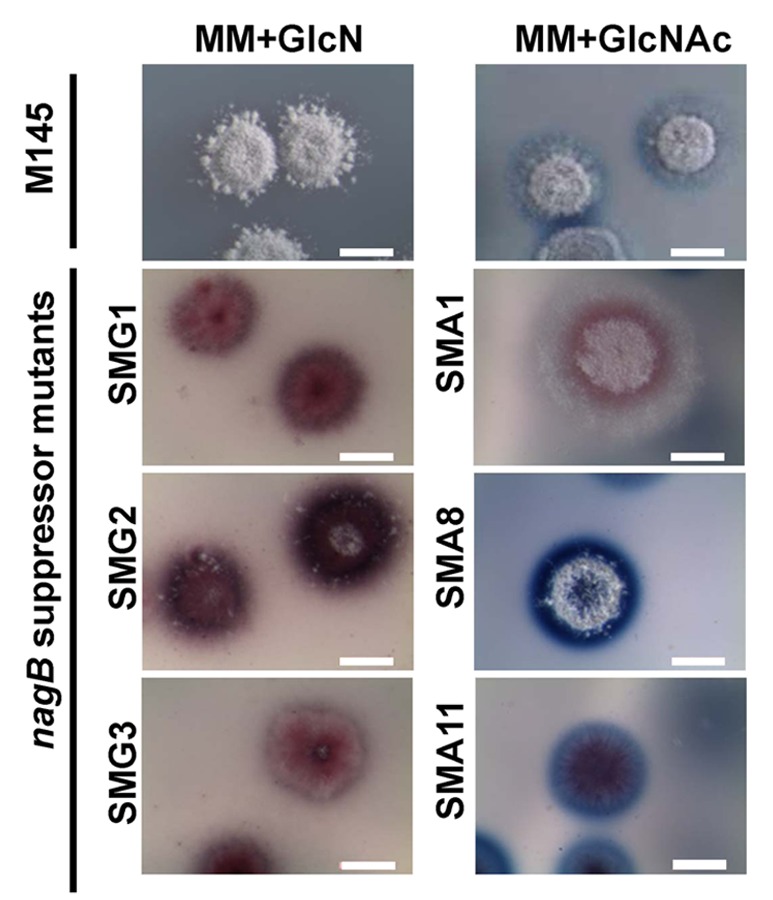
Figure 4. Phenotypic diversity of nagB suppressor mutants. Stereomicrographs illustrating the phenotypes of S. coelicolor M145 and representatives of three different classes of SMA mutants (SMA1, SMA8, SMA11) and of SMG mutants (SMG1-SMG3) grown on MM agar plates supplemented with GlcN (M145 and SMG mutants; left) or GlcNAc (M145 and SMA mutants; right). Colonies were grown for 5 days. Note that the majority of nagB suppressor mutants overproduce antibiotics.
What happens when GlcN-obtained suppressor mutants are grown in the presence of GlcNAc or vice versa, when GlcNAc-obtained mutants are grown on GlcN-containing media? All suppressor mutants except one, were able to grow on both aminosugars. However, GlcN-obtained nagB suppressor SMG1 is of particular interest, as it fails to grow on GlcNAc. This suggests that the second-site mutation specifically relieved the toxicity of GlcN and not of GlcNAc. Therefore, elucidation of this mutation would be of great interest for our understanding of GlcN(Ac) metabolism. A likely candidate is a mutation in the GlcN transport system, which so far has not been elucidated. All other SMG strains overproduced antibiotics, and for SMG2 and SMG3 development was blocked (Fig. 3). All GlcNAc-selected nagB suppressor mutants showed impaired development when grown on MM with GlcN, while some also displayed reduced antibiotic production (SMA1-SMA10) as compared with the wild-type strain (Fig. 3 and Fig. 4). Like for SMA suppressors, SMG mutants grown on MM agar plates supplemented with GlcNAc displayed three distinct phenotypes.
Interestingly, we also observed a strong effect of the inactivation of nagB and the subsequent suppressor mutants on antibiotic production on media not supplemented with GlcNAc or GlcN (Fig. 3). The nagB mutant and most of the suppressor mutants obtained from either GlcNAc or GlcN showed enhanced antibiotic production and strong delay in aerial mycelium formation on MM agar plates supplemented with glucose. Glucose is transported to the cell via the GlcP permease and enters glycolysis after subsequent phosphorylation by glucose kinase (Glk), respectively29 (Fig. 1). Glucose-6P can enter glycolysis or be converted in two steps into GlcN-6P, mediated via glucose-6P isomerase and GlmS (Fig. 1). Accumulation of part of the glucose as GlcN-6P may explain the effect of glucose on antibiotic production in the mutants.
Genetic Analysis of the nagB Suppressor Mutants Reveals Unsuspected Involvement of NagA in GlcN Utilization
Likely candidates for the suppressor mutations obtained on GlcNAc were the GlcNAc transporter genes nagE2 and nagF or the metabolic genes nagA or nagK. These genes and nagE1, a direct homolog and neighboring gene of nagE2, were amplified by PCR and subjected to DNA sequencing. In total 10 individual suppressor mutants obtained on GlcN and 10 obtained on GlcNAc were analyzed. While GlcNAc suppressors did not reveal mutations in any of the sequenced genes or their promoter regions, one of nagB suppressor mutants selected on GlcN (SMG4) contained an out-of-frame deletion of 416 bp in nagA (nt positions 261- 676). In a control experiment, this suppressor mutant was complemented with plasmid pHJL401 harboring the nagKA operon under the control of its own promoter. pHJL401 is a low-copy number vector which is very suitable for complementation experiments due to its stability and low copy number.30 Indeed, this transformant had regained sensitivity to both GlcN or GlcNAc, indicating that the deletion in nagA was the sole cause of the suppressor phenotype (Fig. 2). Full complementation of SMG4 was also achieved with pHJL401 harboring only nagA under the control of the nagKA promoter (not shown).
In line with the idea that the accumulation of GlcN-6P was the cause of the toxicity, nagAB double mutants (which do not accumulate GlcN-6P for the lack of NagA activity) grew well regardless of whether they were challenged with GlcN or GlcNAc. Moreover, the mutants displayed enhanced antibiotic production (Fig. 2). To establish if other suppressor mutants had also sustained a mutation or indel in nagA, 19 additional suppressor mutants obtained on GlcNAc and 15 obtained on GlcN were selected and complemented with a wild-type copy of the nagA gene. Of the transformants, 10 GlcN-selected and three GlcNAc-selected nagB suppressor mutants lost their ability to grow in the presence of either aminosugar, suggesting they may also have been mutated in nagA.
The high frequency of second-site nagA mutations relieving GlcN toxicity to nagB mutants strongly suggests that NagA is somehow involved in the conversion of internalized GlcN into the toxic compound GlcN-6P, or a derivative thereof, thus revealing a novel and unexpected intersection between the GlcN and GlcNAc utilization pathways in S. coelicolor. Up to date there are no reports presenting the involvement of the enzyme NagA in GlcN metabolism, nor is there any evidence of conversion of GlcN to the NagA substrate GlcNAc-6P (see KEGG database31). This presents one example of the high relevance of the suppressor mutants, and closer analysis of GlcN and GlcNAc metabolism is currently in progress.
Since only three out of the 19 GlcNAc-selected nagB suppressor mutants were due to a second-site mutation in nagA, the 16 remaining mutants most likely sustained a mutation in one or more other genomic loci that could also relieve aminosugar toxicity of nagB mutants. We are currently analyzing the genomes of several of the nagB suppressors. Identification of these mutations and the corresponding genes will shed new light on aminosugar metabolism in streptomycetes. These molecules play an important role in central metabolism and cell-wall biosynthesis and accumulation of GlcN(Ac)-derived metabolites serves as a trigger of antibiotic production. Better understanding of aminosugar metabolism in streptomycetes may provide alternative tools for the improvement of antibiotic productivity as well as the possible activation and discovery of new antibacterial compounds.
Acknowledgments
This work was supported by a VICI grant from the Dutch Technology Foundation (STW) to G.P.vanW.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/21371
References
- 1.Chater KF, Losick R. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives. In: Shapiro JA, Dworkin M, eds. Bacteria as multicellular organisms. New York: Oxford University Press, 1997:149-82. [Google Scholar]
- 2.Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 3.Hopwood DA. Streptomyces in nature and medicine: the antibiotic makers. New York: Oxford University Press, 2007. [Google Scholar]
- 4.Sánchez S, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, et al. Carbon source regulation of antibiotic production. J Antibiot (Tokyo) 2010;63:442–59. doi: 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- 5.van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep. 2011;28:1311–33. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 6.Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci U S A. 2003;100(Suppl 2):14555–61. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8:557–63. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Martín JF, Liras P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol. 2010;13:263–73. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 9.van Wezel GP, McKenzie NL, Nodwell JR. Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 2009;458:117–41. doi: 10.1016/S0076-6879(09)04805-8. [DOI] [PubMed] [Google Scholar]
- 10.Butler MJ, Bruheim P, Jovetic S, Marinelli F, Postma PW, Bibb MJ. Engineering of primary carbon metabolism for improved antibiotic production in Streptomyces lividans. Appl Environ Microbiol. 2002;68:4731–9. doi: 10.1128/AEM.68.10.4731-4739.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Smanski MJ, Shen B. Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl Microbiol Biotechnol. 2010;86:19–25. doi: 10.1007/s00253-009-2428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nothaft H, Rigali S, Boomsma B, Świątek M, McDowall KJ, van Wezel GP, et al. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol Microbiol. 2010;75:1133–44. doi: 10.1111/j.1365-2958.2009.07020.x. [DOI] [PubMed] [Google Scholar]
- 13.van Wezel GP, Krabben P, Traag BA, Keijser BJ, Kerste R, Vijgenboom E, et al. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. Appl Environ Microbiol. 2006;72:5283–8. doi: 10.1128/AEM.00808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–5. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Müller M, et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol. 2006;61:1237–51. doi: 10.1111/j.1365-2958.2006.05319.x. [DOI] [PubMed] [Google Scholar]
- 16.D’Alia D, Eggle D, Nieselt K, Hu WS, Breitling R, Takano E. Deletion of the signalling molecule synthase ScbA has pleiotropic effects on secondary metabolite biosynthesis, morphological differentiation and primary metabolism in Streptomyces coelicolor A3(2) Microb Biotechnol. 2011;4:239–51. doi: 10.1111/j.1751-7915.2010.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsiao NH, Söding J, Linke D, Lange C, Hertweck C, Wohlleben W, et al. ScbA from Streptomyces coelicolor A3(2) has homology to fatty acid synthases and is able to synthesize gamma-butyrolactones. Microbiology. 2007;153:1394–404. doi: 10.1099/mic.0.2006/004432-0. [DOI] [PubMed] [Google Scholar]
- 18.Brückner R, Titgemeyer F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol Lett. 2002;209:141–8. doi: 10.1016/S0378-1097(02)00559-1. [DOI] [PubMed] [Google Scholar]
- 19.Nothaft H, Dresel D, Willimek A, Mahr K, Niederweis M, Titgemeyer F. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J Bacteriol. 2003;185:7019–23. doi: 10.1128/JB.185.23.7019-7023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nothaft H, Dresel D, Willimek A, Mahr K, Niederweis M, Titgemeyer F. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J Bacteriol. 2003;185:7019–23. doi: 10.1128/JB.185.23.7019-7023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Świątek MA, Tenconi E, Rigali S, van Wezel GP. Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in control of development and antibiotic production. J Bacteriol. 2012;194:1136–44. doi: 10.1128/JB.06370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plumbridge J, Vimr E. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J Bacteriol. 1999;181:47–54. doi: 10.1128/jb.181.1.47-54.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogler AP, Lengeler JW. Analysis of the nag regulon from Escherichia coli K12 and Klebsiella pneumoniae and of its regulation. Mol Gen Genet. 1989;219:97–105. doi: 10.1007/BF00261163. [DOI] [PubMed] [Google Scholar]
- 24.Teplyakov A, Obmolova G, Badet-Denisot MA, Badet B. The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase. Protein Sci. 1999;8:596–602. doi: 10.1110/ps.8.3.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C, Rodionov DA, Li X, Laikova ON, Gelfand MS, Zagnitko OP, et al. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J Biol Chem. 2006;281:29872–85. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- 26.Bernheim NJ, Dobrogosz WJ. Amino sugar sensitivity in Escherichia coli mutants unable to grow on N-acetylglucosamine. J Bacteriol. 1970;101:384–91. doi: 10.1128/jb.101.2.384-391.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plumbridge J. An alternative route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in Escherichia coli. J Bacteriol. 2009;191:5641–7. doi: 10.1128/JB.00448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White RJ. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968;106:847–58. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Wezel GP, Mahr K, König M, Traag BA, Pimentel-Schmitt EF, Willimek A, et al. GlcP constitutes the major glucose uptake system of Streptomyces coelicolor A3(2) Mol Microbiol. 2005;55:624–36. doi: 10.1111/j.1365-2958.2004.04413.x. [DOI] [PubMed] [Google Scholar]
- 30.van Wezel GP, White J, Hoogvliet G, Bibb MJ. Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Mol Microbiol Biotechnol. 2000;2:551–6. [PubMed] [Google Scholar]
- 31.Kyoto Encyclopedia of Genes and Genomes (KEGG) Database [Internet]. 1995 - .Release 63.0. Kyoto (Japan): Kanehisa Laboratories. [updated 2012 July 1; cited 2012 July 5]. Available from: http://www.genome.jp/kegg-bin/show_pathway?org_name=sco&mapno=00520.