Abstract
Protein production through dedicated secretion systems might offer an potential alternative to the conventional cytoplasmical expression. The application of Type 1 secretion systems of Gram-negative bacteria, however, where often not successful in the past for a wide range of proteins. Recently, two studies using the E. coli maltose binding protein (MalE) and the rat intestinal fatty acid binding protein (IFABP) revealed a rational to circumvent these limitations. Here, wild-type passenger proteins were not secreted, while folding mutants with decreased folding kinetics were efficiently exported to the extracellular space. Subsequently, an one-step purification protocol yielded homogeneous and active protein. Taken together, theses two studies suggest that the introduction of slow-folding mutations into a protein sequence might be the key to use Type 1 secretion systems for the biotechnological production of proteins.
Keywords: HlyB, IFABP, Type 1 secretion, maltose, mutation, protein folding, relaxation, western blot
Introduction
Type 1 secretion systems (T1SS) of Gram-negative bacteria transport substrates directly form the cytoplasm to the exterior without the formation of any periplasmic intermediate. The substrate spectrum contains functional unrelated and different-sized proteins, i. e. the hemophore HasA (19 kDa) and the lipase LipA (65 kDa) of Serratia marcescens1,2 or the 900 kDa protein LapA that is involved in biofilm formation of Pseudomonas fluorescence.3 A transport machinery composed of two inner membrane proteins, a membrane fusion protein (MFP) and an ATP-binding-cassette (ABC) transporter, as well as an outer membrane factor (OMF) catalyzes the secretion process.4,5 The paradigm of T1SS is the hemolysin A (HlyA) secretion system of Escherichia coli (E. coli), which consists of the ABC transporter hemolysin B (HlyB), the MFP hemolysin D (HlyD) and the multi-functional TolC (OMF).6 TolC is recruited, upon interaction of the substrate (HlyA) with HlyB and HlyD, and a continuous export channel is formed bridging the cytoplasm directly to the exterior.4,7
HlyA and other T1SS substrates are suggested to be transported in an unfolded state8 and folding is triggered by binding of Ca2+ ions to so-called GG repeats in the extracellular space, directly after transport. Previously, several proteins were successfully secreted as fusion proteins with a 23 kDa C-terminal fragment of HlyA (HlyAc), for example β-lactamase or antibody fragments.9-12 However, many proteins could not be secreted limiting the exploitation of the T1SS pathway for biotechnological purposes.9 Recently, two publications investigated these limitations.8,13 Here, the maltose binding protein (MalE) and the intestinal fatty acid binding protein (IFABP) could initially not be secreted when fused to HlyAc. However, the use of slow-folding proteins with decreased folding kinetics allowed the secretion into the culture medium.8,13 Interestingly, the mutant IFABP(G121V) accumulates in E. coli as inclusion bodies, whereas secreted IFABP(G121V) was soluble and active. Taken together, both studies suggest a rational to secrete proteins of interest in good yields and in a soluble, functional state with an E. coli T1SS. However, the successful application requires proteins with slow-folding kinetics and the identification of such mutants might hamper the general applicability of the approach. In other words, the general application of this approach requires screening of protein libraries that contain random mutations generated by, for example, error-prone PCR. This should be performed with suitable activity assays that allow a simple read-out to identifiy secreted and moreover, active proteins.
Secretion of a Functional Cytoplasmic Protein
The HlyA T1SS was investigated for its applicability to secrete an eukaryotic cytoplasmic protein, the rat intestinal fatty acid binding protein (IFABP).15 IFABP is a 131 residue (15 kDa) cytoplasmical protein, which belongs to the family of fatty acid binding proteins in mammals and binds a single molecule of fatty acids.16 The binding to lipophilic drugs, for example, is used for the prediction of drug pharmacokinetic parameters in vitro.17,18 The slow-folding mutant IFBAP G121V still binds fatty acids with high affinity (KD = 120 nM ± 25 nM) and is currently produced in E. coli out of inclusion bodies.19
Both, wild-type IFABP and the slow-folding variant IFABP(G121V), were cloned inside a secretion vector in front of HlyAc. In the background of the T1SS, wild-type IFABP(wt)-HlyAc was not secreted into the medium of the E. coli culture, whereas IFABP(G121V)-HlyAc was secreted in good yields of up to 6 mg per liter cell culture (corresponding to 2 mg/OD). The secreted fusion protein could even be visualized by Coommassie brilliant blue (CBB) staining in the supernatant without concentrating the sample. Interestingly, the secreted fusion protein was soluble and stable in the culture medium and during the subsequent purification steps, which displays an advantage compared with its currently expressed form being insoluble inclusion bodies.19
Bakkes et al. demonstrated the secretion of a slow-folding mutant of MalE. The passenger MalE with two introduced slow-folding mutations was secreted and shown to be active in binding to amylose, although the major fraction did not bind to the ligand.14 The reason for this behavior, however, was not investigated. Therefore, Schwarz et al.13 investigated the folding state and the activity of secreted IFABP in more detail and compared it with published results. The folding state of secreted IFABP(G121V) was studied by intrinsic tryptophane fluorescence spectroscopy and compared with IFABP(G121V), which was purified out of cytosolic inclusion bodies and refolded.20 These studies indicated that secreted IFABP(G121V) was folded comparable to the reference protein. Since the folding state of a protein does not directly imply that the protein is active, the activity of secreted IFABP(G121V) was analyzed. The fatty acid analog 11-(dansylamino)undecanoic acid (DAUDA) was used for titration experiments to determine the binding activity of IFABP(G121V). For the validation of the experimental setup, the binding affinity of the reference protein IFABP(G121V) (non-secreted control) was examined by fluorescence spectroscopy. The determined dissociation constant (KD) of 126 nM ± 7 nM is in line with the published data (120 nM ± 25 nM).19 The KD value of secreted IFABP(G121V)-HlyAc was determined to be 195 nM ± 13 nM. This is in the same range as the KD for the non-secreted control demonstrating that secreted IFABP G121V exhibits similar binding parameters.
Unfolding Domains – Useful Tools For Secretion?
The major bottleneck of this secretion technology is its dependency on slow-folding proteins. As an alternative route to directed evolution appoaches (see below), a destabilizing domain was analyzed to transfer instability to the fusion proteins. The destabilizing domain ddFKBP, an engineered version of the human FKBP12, was used, which was already applied successfully in eukaryotes to destabilize fused proteins,21-24 however, no studies were performed in bacteria so far. Schwarz et al.13 investigated, whether the ddFKBP destabilizes fusion proteins in E. coli and if this domain allows the secretion of proteins via the T1SS without the introduction of slow-folding mutations. The ddFKBP was fused N-terminally to HlyAc, IFABP(wt)-HlyAc and IFABP(G121V)-HlyAc, respectively. All protein constructs were expressed as indicated by CBB staining and western blot analysis of whole cell extracts. Obviously, ddFKBP did not increase secretion levels of the fusion constructs. These results indicate that the intrinsic folding state of HlyAc, IFABP(wt) and IFABP(G121V) is not changed drastically and that the ddFKBP does not facilitate the secretion of a protein of interest with the T1SS. This raises the question whether the ddFKBP transmits instability to fused proteins or induces their degradation in bacteria?
Engineering of Proteins with Slow Folding Characteristics
The successful secretion of proteins via the HlyA T1SS depends on the availability of slow-folding mutants. Besides wild-type proteins with slow-folding kinetics, many proteins fold too quickly inside cells, which hampers the general applicability of T1SS. Up to now, the a priori prediction and alteration of folding properties is impossible with biochemical, computational or bioinformatical approaches. Nevertheless, directed evolution methods are available to generate proteins with desired characteristics, i. e. altered folding characteristics.25,26 Basically, this can be performed within three relatively easy experimental steps (see Figure 1).
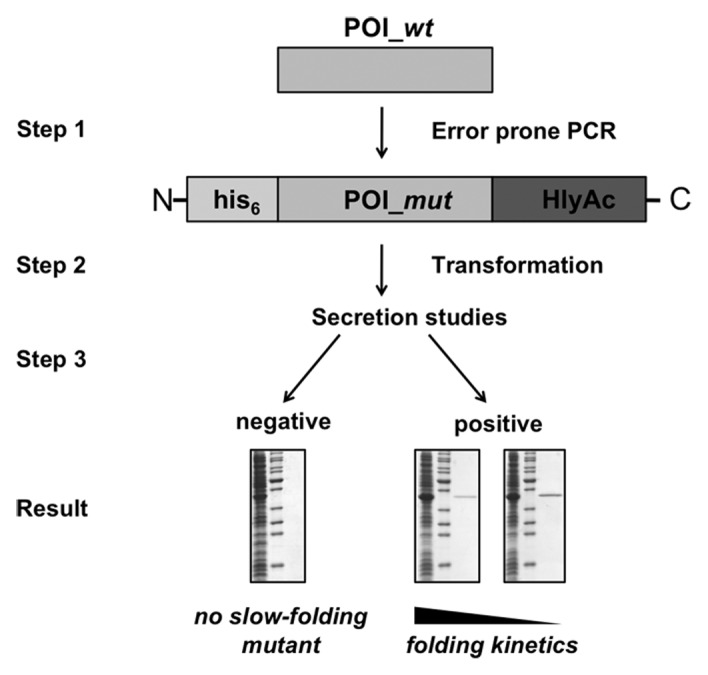
Figure 1.Identification of slow-folding mutants employing an E. coli T1SS. Scheme of the three-step protocol for the generation of slow-folding mutants of a protein of interest. Step 1: Random mutation of a POI_wt sequence and insertion of the resulting POI_mut in front of HlyAc. Step 2: Transformation of E. coli cells with the generated plasmid library in combination with the plasmid encoding the T1SS components. Step 3: Small-scale secretion studies to identify clones that secrete fusion proteins. The plasmids of secretion-positive clones are subsequently sequenced to identify the corresponding slow-folding mutations. If available, agar plate assays might be used to test the activity of these secreted proteins (see text).
Step 1 represents the random mutagenesis of the wild-type protein of interest (POI_wt). Various methods are available to mutate the gene, which are summarized in, for example Wong et al.27 The mutagenesis products are inserted inside the secretion plasmid in front of the secretion signal (HlyAc) DNA sequence, This results in a library of mutated POI (POI_mut) fusion proteins. E. coli cells are transformed with the plasmid library together with the plasmid encoding the T1SS complex (Step 2) and obtained colonies are analyzed in secretion experiments (Step 3). Since the secretion signal is localized at the C-terminal end of the fusion protein, only full-length fusion proteins will be secreted; fusion proteins with artificial stop codons introduced via the error prone PCR reaction are not secreted and thereby automatically eliminated from the screen. Mutations that do not alter the folding kinetics will not be secreted either.
However, proteins with slow-folding kinetics can be detected, by SDS-PAGE analysis of the culture supernatants. Staining of the gels or immunochemical detection visualizes the secreted proteins and allows the densometrical quantification of secreted proteins. A great advantage is that the amount of secreted protein is inversely correlated with the folding rate of the protein. In other words, the protein secreted most efficiently will have the slowest folding kinetics. Furthermore, supernatant samples can be used immediately for enzyme-linked activity or immunosorbent assays (ELISA). If such an activity of the POI can be visualized, the secretion system enables high-throughput screening of a vast pool of mutants. For example, the lipolytic activity of secreted lipases can be visualized on agar plates, which contain the appropiate substrate of the lipase.28 In summary, the technology described in Schwarz et al.13 represents a valuable and novel tool, usuable for various experimental setups, for example, the generation of folding mutants allowing secretion and subsequently the screening of secreted proteins with improved characteristics which are interesting within the biotechnological area.
Acknowledgments
We are indebted to Diana Clausnitzer, Protein Production Facility, of the Heinrich Heine University, for valuable discussion during the cloning of the constructs. We thank the Ministry of Innovation, Science and Research of the German Federal State North Rhine-Westphalia (NRW) and the Heinrich Heine University Düsseldorf (scholarship from the CLIB Graduate Cluster Industrial Biotechnology to CKWS and scholarship from the Biostruct NRW Graduate School to MHHL).
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/20712
References
- 1.Meier R, Drepper T, Svensson V, Jaeger KE, Baumann U. A calcium-gated lid and a large beta-roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens. J Biol Chem. 2007;282:31477–83. doi: 10.1074/jbc.M704942200. [DOI] [PubMed] [Google Scholar]
- 2.Létoffé S, Ghigo JM, Wandersman C. Secretion of the Serratia marcescens HasA protein by an ABC transporter. J Bacteriol. 1994;176:5372–7. doi: 10.1128/jb.176.17.5372-5377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–18. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 4.Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E.coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–96. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holland IB, Schmitt L, Young J. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review) Mol Membr Biol. 2005;22:29–39. doi: 10.1080/09687860500042013. [review] [DOI] [PubMed] [Google Scholar]
- 6.Jarchau T, Chakraborty T, Garcia F, Goebel W. Selection for transport competence of C-terminal polypeptides derived from Escherichia coli hemolysin: the shortest peptide capable of autonomous HlyB/HlyD-dependent secretion comprises the C-terminal 62 amino acids of HlyA. Mol Gen Genet. 1994;245:53–60. doi: 10.1007/BF00279750. [DOI] [PubMed] [Google Scholar]
- 7.Benabdelhak H, Kiontke S, Horn C, Ernst R, Blight MA, Holland IB, et al. A specific interaction between the NBD of the ABC-transporter HlyB and a C-terminal fragment of its transport substrate haemolysin A. J Mol Biol. 2003;327:1169–79. doi: 10.1016/S0022-2836(03)00204-3. [DOI] [PubMed] [Google Scholar]
- 8.Bakkes PJ, Jenewein S, Smits SH, Holland IB, Schmitt L. The rate of folding dictates substrate secretion by the Escherichia coli hemolysin type 1 secretion system. J Biol Chem. 2010;285:40573–80. doi: 10.1074/jbc.M110.173658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blight MA, Holland IB. Heterologous protein secretion and the versatile Escherichia coli haemolysin translocator. Trends Biotechnol. 1994;12:450–5. doi: 10.1016/0167-7799(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 10.Fernández LA, Sola I, Enjuanes L, de Lorenzo V. Specific secretion of active single-chain Fv antibodies into the supernatants of Escherichia coli cultures by use of the hemolysin system. Appl Environ Microbiol. 2000;66:5024–9. doi: 10.1128/AEM.66.11.5024-5029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chervaux C, Sauvonnet N, Le Clainche A, Kenny B, Hung AL, Broome-Smith JK, et al. Secretion of active beta-lactamase to the medium mediated by the Escherichia coli haemolysin transport pathway. Mol Gen Genet. 1995;249:237–45. doi: 10.1007/BF00290371. [DOI] [PubMed] [Google Scholar]
- 12.Mackman N, Baker K, Gray L, Haigh R, Nicaud JM, Holland IB. Release of a chimeric protein into the medium from Escherichia coli using the C-terminal secretion signal of haemolysin. EMBO J. 1987;6:2835–41. doi: 10.1002/j.1460-2075.1987.tb02580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz CK, Landsberg CD, Lenders MH, Smits SH, Schmitt L. Using an E. coli Type 1 secretion system to secrete the mammalian, intracellular protein IFABP in its active form. J Biotechnol. 2012;159:155–61. doi: 10.1016/j.jbiotec.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Bakkes PJ, Jenewein S, Smits SH, Holland IB, Schmitt L. The rate of folding dictates substrate secretion by the Escherichia coli hemolysin type 1 secretion system. J Biol Chem. 2010;285:40573–80. doi: 10.1074/jbc.M110.173658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sweetser DA, Heuckeroth RO, Gordon JI. The metabolic significance of mammalian fatty-acid-binding proteins: abundant proteins in search of a function. Annu Rev Nutr. 1987;7:337–59. doi: 10.1146/annurev.nu.07.070187.002005. [DOI] [PubMed] [Google Scholar]
- 16.Veerkamp JH, Peeters RA, Maatman RG. Structural and functional features of different types of cytoplasmic fatty acid-binding proteins. Biochim Biophys Acta. 1991;1081:1–24. doi: 10.1016/0005-2760(91)90244-c. [DOI] [PubMed] [Google Scholar]
- 17.Chuang S, Velkov T, Horne J, Porter CJ, Scanlon MJ. Characterization of the drug binding specificity of rat liver fatty acid binding protein. J Med Chem. 2008;51:3755–64. doi: 10.1021/jm701192w. [DOI] [PubMed] [Google Scholar]
- 18.Rowland A, Knights KM, Mackenzie PI, Miners JO. Characterization of the binding of drugs to human intestinal fatty acid binding protein (IFABP): potential role of IFABP as an alternative to albumin for in vitro-in vivo extrapolation of drug kinetic parameters. Drug Metab Dispos. 2009;37:1395–403. doi: 10.1124/dmd.109.027656. [DOI] [PubMed] [Google Scholar]
- 19.Kim K, Frieden C. Turn scanning by site-directed mutagenesis: application to the protein folding problem using the intestinal fatty acid binding protein. Protein Sci. 1998;7:1821–8. doi: 10.1002/pro.5560070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Frieden C. Observation of sequential steps in the folding of intestinal fatty acid binding protein using a slow folding mutant and 19F NMR. Proc Natl Acad Sci U S A. 2007;104:11993–8. doi: 10.1073/pnas.0705253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods. 2007;4:1007–9. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 23.Herm-Götz A, Agop-Nersesian C, Münter S, Grimley JS, Wandless TJ, Frischknecht F, et al. Rapid control of protein level in the apicomplexan Toxoplasma gondii. Nat Methods. 2007;4:1003–5. doi: 10.1038/nmeth1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park EC, Finley D, Szostak JW. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci U S A. 1992;89:1249–52. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–9. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 26.Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–91. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 27.Wong TS, Roccatano D, Zacharias M, Schwaneberg U. A statistical analysis of random mutagenesis methods used for directed protein evolution. J Mol Biol. 2006;355:858–71. doi: 10.1016/j.jmb.2005.10.082. [DOI] [PubMed] [Google Scholar]
- 28.Eom GT, Song JK, Ahn JH, Seo YS, Rhee JS. Enhancement of the efficiency of secretion of heterologous lipase in Escherichia coli by directed evolution of the ABC transporter system. Appl Environ Microbiol. 2005;71:3468–74. doi: 10.1128/AEM.71.7.3468-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farinas ET, Bulter T, Arnold FH. Directed enzyme evolution. Curr Opin Biotechnol. 2001;12:545–51. doi: 10.1016/S0958-1669(01)00261-0. [DOI] [PubMed] [Google Scholar]


