Abstract
The inactivation of phosphorylated nitrate reductase (NR) by the binding of 14-3-3 proteins is one of a very few unambiguous biological functions for 14-3-3 proteins. We report here that serine and threonine residues at the +6 to +8 positions, relative to the known regulatory binding site involving serine-543, are important in the interaction with GF14ω, a recombinant plant 14-3-3. Also shown is that an increase in ionic strength with KCl or inorganic phosphate, known physical effectors of NR activity, directly disrupts the binding of protein and peptide ligands to 14-3-3 proteins. Increased ionic strength attributable to KCl caused a change in conformation of GF14ω, resulting in reduced surface hydrophobicity, as visualized with a fluorescent probe. Similarly, it is shown that the 5′ isomer of AMP was specifically able to disrupt the inactive phosphorylated NR:14-3-3 complex. Using the 5′-AMP fluorescent analog trinitrophenyl-AMP, we show that there is a probable AMP-binding site on GF14ω.
The enzyme NR (EC 1.6.6.1) plays a pivotal role in the incorporation of inorganic nitrogen into cellular constituents such as amino acids and nucleic acids. It is believed to catalyze the rate-limiting step in the reduction of nitrate to nitrite (Solomonson and Barber, 1990), and it is now apparent that this step is highly regulated in higher plants. In addition to the steady-state level of NR protein being regulated at the transcriptional and protein turnover levels (Hoff et al., 1994; Crawford, 1995), recent studies have shown posttranslational modification of NR involving protein phosphorylation to be a major regulatory mechanism (Douglas et al., 1995; Bachmann et al., 1996c; Su et al., 1996; Lillo et al., 1997).
The inactivation of NR that occurs in darkened leaves is now known to involve a two-step mechanism. First, NR is phosphorylated on a seryl residue (Ser-543 and Ser-534) in spinach (Spinacia oleracea) and Arabidopsis, respectively (Douglas et al., 1995; Bachmann et al., 1996c; Su et al., 1996), located in the hinge-1 region, which links the molybdenum cofactor and heme (Cyt b557) domains (for a review of NR structure, see Campbell and Kinghorn, 1990). This seryl phosphorylation allows the hinge-1 site to become a recognition site for a class of proteins called 14-3-3 proteins, which bind and inactivate pNR (Bachmann et al., 1996b; Moorhead et al., 1996). The 14-3-3 proteins are highly conserved among eukaryotes and typically occur in small gene families, with isoforms possibly having distinct functions (for review, see Aitken, 1996; Ferl, 1996). Their primary function appears to be as sequence-specific binding proteins involved in a variety of cellular signaling pathways. In many cases the 14-3-3-binding site involves a seryl residue that must be phosphorylated for the interaction to occur (Muslin et al., 1996). However, recent results suggest that 14-3-3 proteins can also bind to certain nonphosphorylated sequences (Petosa et al., 1998).
Previous studies have identified several metabolic and physical activators of pNR. For example, 5′-AMP, Pi, and various salts (e.g. KCl) all stimulate the rate of pNR activation in vitro in desalted crude extracts (Kaiser and Spill, 1991; Huber and Kaiser, 1996, and refs. therein). Because the 14-3-3 inhibitor protein binds directly to the regulatory phosphorylation site on pNR (Bachmann et al., 1996a), activation by these compounds could result from either direct interference with 14-3-3 binding or stimulation of endogenous protein phosphatases to dephosphorylate the requisite phosphoserine residue in NR. In the present study we have tried to address some of these issues.
We examined 14-3-3-binding interactions using two assay methods: the first was based on inactivation of enzyme activity and the second involved binding of a synthetic [32P]phosphopeptide. The assays were carried out at two pH levels, 7.5 and 6.5, in the presence or absence of divalent cations. We also compared the behavior of two different preparations of 14-3-3 proteins: purified spinach leaf 14-3-3 proteins (a mixture of isoforms) and a single recombinant isoform, Arabidopsis GF14ω (Lu et al., 1994). We show that physical (KCl and Pi) and metabolic (AMP) factors are able to disrupt the pNR:14-3-3 interaction directly, which may explain their ability to activate pNR (Huber and Kaiser, 1996, and refs. therein). These effectors were shown to interact with GF14ω directly using spectrofluorometric analysis. Of particular interest is the evidence that suggests that there is an AMP-binding site on GF14ω. New results also suggest that key residues outside of the recognized short 14-3-3-binding motif of pNR may influence binding at Ser-543.
MATERIALS AND METHODS
Plant Material
Spinach (Spinacia oleracea L. cvs Bloomsdale and Tyee) plants were grown and harvested as described previously (Huber and Huber, 1995). Experiments were carried out at least twice, usually three to five times. Typical results from a representative experiment or mean values are shown.
Protein Extraction
Frozen leaf tissue was ground into a fine powder in a chilled mortar using liquid nitrogen, followed by the addition of cold extraction buffer (2 mL g−1 fresh weight) containing 50 mm Mops-Na, pH 7.5, 10 mm MgCl2, 1 mm EDTA, 0.1% (v/v) Triton X-100, and 5 mm DTT, as described by Huber and Huber (1995), with the addition of 0.5 mm PMSF, 1 mm ε-amino-n-caproic acid, and 1 mm benzamidine-HCl.
Protein Purification
Isolation of Spinach Leaf 14-3-3 Proteins
After cell disruption, endogenous spinach leaf 14-3-3 proteins were purified by fast-protein liquid chromatography (Pharmacia2) and electroelution from native-PAGE, as described previously (Bachmann et al., 1995, 1996b).
Partial Purification, Assay, and in Vitro Phosphorylation of NR and Endogenous Kinases
Dephosphorylated NR from light-harvested leaf tissue was partially purified as described by Bachmann et al. (1996c). The only deviations made from this protocol were that NR was isolated in a 3% to 13% PEG-8000 precipitation, and the resuspended pellet was subjected to anion-exchange chromatography using a 50-mL Source 15Q media column (Pharmacia). Potential NR kinases were located using the NR6 peptide kinase assay, as described by Bachmann et al. (1996c). NRA was assayed in the presence or absence of Mg2+, as described previously (Huber et al., 1992). Partially purified NR from fast-protein liquid chromatography anion-exchange chromatography contains a co-eluting NR kinase (peak II; see McMichael et al., 1995). NR was phosphorylated in vitro with the associated kinase by incubation in a mixture containing 1 mm ATP, 4 mm MgCl2, 0.1 mm CaCl2, 2.5 mm DTT, and 1 μm microcystin-LR (Leu-Arg analog) (Calbiochem) in 50 mm Mops-Na, pH 7.5, at 25°C for 20 min. The reaction mixture was immediately desalted into 10 mm Mops-Na, pH 7.5, 2.5 mm DTT, 1 mm 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid, and 6 mm EDTA and then used for 14-3-3 protein-binding assays. The phosphorylation status was determined by enzyme inactivation (Huber and Huber, 1995), with the addition of purified spinach 14-3-3 proteins or recombinant GF14ω. The effects of pH, divalent cations, and both physical and metabolic effectors are described below. For most effectors a preincubation of 5 min on ice was allowed before assaying for NRA. Controls were included and results are presented as the percentage deviation from them. The potential dephosphorylation of pNR during incubation was determined and was not a contributing factor under these assay conditions (data not shown).
Phosphorylation and Purification of the Synthetic Peptides
The synthetic peptides designated NR2, NR6, NR24, and NR25 (see Table I) were synthesized (model 432A peptide synthesizer, Synergy, Applied Biosystems) and used for both the competition and binding assays. For competition experiments, the peptides were phosphorylated with the peak I kinase and purified using HPLC, as described previously (Bachmann et al., 1996c).
Table I.
Amino acid sequences of synthetic peptides based around the regulatory Ser-543 residue that was used in this study
| Peptide | Residue Position
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −18 | −16 | −14 | −12 | −10 | −8 | −6 | −4 | −2 | 0 | +2 | +4 | +6 | +8 | |||||||||||||||
| NR2 | R | E | R | H | L | E | I | S | D | S | G | P | T | L | K | R | T | A | S | T | P | F | ||||||
| NR6 | G | P | T | L | K | R | T | A | S | T | P | F | M | N | T | T | S | K | ||||||||||
| NR24 | G | P | T | L | K | R | T | A | S | T | P | F | M | N | A | A | A | K | ||||||||||
| NR25 | G | P | A | L | K | R | T | A | S | T | P | F | M | N | T | T | S | K | ||||||||||
All amino acids are coded using the standard single-letter codes, and positions are marked relative to the phosphorylatable Ser (Ser-543, set at position 0 in the table) of spinach NR. Substituted residues in the synthetic peptides NR24 and NR25 are shown in bold, underlined, italic type. The currently recognized 14-3-3-binding motif is underlined in each peptide.
Peptide-Binding Assay
Either purified spinach leaf 14-3-3 proteins or recombinant GF14ω (typically 500-1000 pmol), which had been dialyzed previously into 10 mm Mops-Na, pH 7.5, and 2.5 mm DTT, were mixed with synthetic [32P]phosphopeptide (typically 50–100 pmol, 80–120 cpm/pmol) and additions specified below, and then incubated on ice for 1 min. The final reaction volume was 150 μL, of which 120 μL was applied to a preequilibrated 1.5-mL Sephadex G-25 column and centrifuged at 350g for 1 min. Radiolabeled peptide bound to 14-3-3 proteins passed through with the void volume. Of the flow through, 90 μL was used for liquid-scintillation counting. Each condition tested had an internal control lacking 14-3-3 proteins. The binding values presented are corrected for 32P appearing in the flow-through fraction in the absence of 14-3-3 proteins.
GF14ω Induction and Purification
The recombinant Arabidopsis 14-3-3 protein GF14ω was obtained using the Escherichia coli strain BL21 (DE3) with the overexpressing pET15b plasmid (Novagen, Madison, WI) containing the GF14ω cDNA insert (Wu et al., 1997). E. coli was grown in Luria-Bertani medium containing 50 μg/mL ampicillin. The initial inoculation was from a single-plate colony, which was grown overnight and used to seed 200 mL of Luria-Bertani medium (1:100, v/v). Cultures were incubated at 37°C with vigorous shaking until the A600 reached 0.6. Isopropyl β-d-thiogalactoside was then added to a final concentration of 1 mm to induce the T-7lac promoter and cause overexpression of GF14ω. Cultures were grown for another 2.5 to 3 h and harvested by centrifugation at 5000g for 5 min. The cell pellet was resuspended in 50 mm Tris-HCl, pH 8.0, containing 2 mm EDTA, and centrifuged again as described above. The cell paste was stored at −80°C. Frozen pellets were thawed and resuspended in binding buffer (20 mm Tris-HCl, pH 7.9, with 500 mm NaCl and 5 mm imidazole; Novagen) and sonicated at the maximum intensity (limit level 5, Micro-Tip, Sonics and Materials, Danbury, CT) for five cycles of 25 s, with intervals of 45 s of incubation in a salt-ice-water bath.
The lysate/sonicate was clarified and applied to a Ni2+-nitrilotriacetic acid-agarose-affinity column under nondenaturing conditions. The standard native protein-purification protocol was followed (Novagen). The eluted His-tagged fusion protein was concentrated using Centricon 3 concentrators (Amicon, Beverly, MA) and dialyzed against thrombin cleavage buffer containing 20 mm Tris-HCl, pH 8.4, 150 mm NaCl, and 2.5 mm CaCl2. The N-terminal His tag was then cleaved using 0.5 unit of biotinylated thrombin per milligram of recombinant protein at 20°C for 4 h. Thrombin was removed by affinity capture using streptavidin agarose, following the manufacturer's instructions (Novagen). The cleaved protein was then purified by fast-protein liquid chromatography using a 1-mL Source 15Q column (Pharmacia). The major protein peak was collected and concentrated, as described above, and then divided into aliquots and stored at −80°C until use.
Fluorescence Spectroscopy
All fluorescence measurements were made using a spectrofluorophotometer (model RF-5301 PC, Shimadzu, Columbia, MD). Metal-free GF14ω protein was prepared by extensive dialysis against 10 mm Mops-Na, pH 7.5, containing 2.5 mm DTT, 5 mm EDTA, and 5 mm EGTA. The protein was then dialyzed against the same buffer without EDTA or EGTA. A stock solution of 100 μm bis-ANS (Molecular Probes, Eugene, OR) was prepared and diluted to a final concentration of 1 μm during measurements. A stock solution of 400 μm TNP-AMP (Molecular Probes) was prepared and diluted to 60 μm (unless stated otherwise) during measurements. The final concentrations of GF14ω used for fluorescence measurements are given below, as are the excitation and emission wavelengths used for each chromogenic probe.
Protein Estimation
Protein concentration was determined by the dye-binding Bio-Rad microassay using BSA as a standard (Bradford, 1976).
RESULTS AND DISCUSSION
The Ser-543-Binding Motif
Regions of NR that interact with the 14-3-3 inhibitor protein can be identified by determining whether the corresponding phosphorylated peptides disrupt the pNR:14-3-3 complex and thereby increase NRA. During the course of the studies to characterize the known 14-3-3-binding site within NR, we used two control phosphopeptides, pNR2 and pNR6. Both are based on Ser-543 (see Table I) and thus were able to compete with pNR for 14-3-3 binding. However, these two peptides differed considerably in their ability to disrupt the pNR:14-3-3 complex. The presence of 40 μm pNR6 was enough to provide half-maximal reactivation of pNR, which is consistent with the earlier report by Bachmann et al. (1996a). The pNR2 peptide also competed with pNR for binding with 14-3-3 proteins, but at 40 μm it resulted in only about 12% reactivation of pNR (data not shown). One possible explanation for the observation with pNR2 is that residues outside of the well-recognized motif (Muslin et al., 1996) may have a role in the binding of NR to the 14-3-3 protein. Therefore, we synthesized two additional synthetic peptides, NR24 and NR25 (see Table I); NR24 had Ala substituted for the Thr and Ser residues at the +6 to +8 positions, and NR25 had a single Thr residue at the −6 position replaced with an Ala residue.
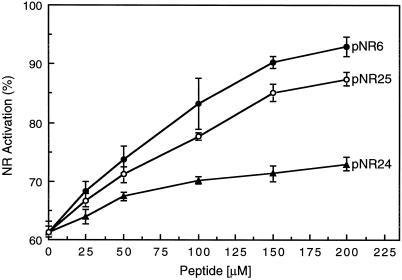
Figure 1 shows that the ability of pNR24 to disrupt the pNR:14-3-3 complex was significantly reduced relative to that of pNR6 or pNR25. This suggests that Ser and/or Thr residues C terminal to Ser-543 could play some role in the binding of the target ligand to the 14-3-3 protein. Alternatively, these residues may enhance the peptides' secondary structure, making a more favorable “competitor” to disrupt the pNR:14-3-3 complex. A pileup of some recognized 14-3-3-binding proteins (Table II) indicated that there are often Thr and Ser residues at the +6 to +9 positions relative to the phosphorylated Ser residue. With the exception of one protein (Bcr), all of the 14-3-3-binding proteins shown in Table II have at least one Ser/Thr residue within 10 residues C terminal to the phosphorylated Ser residue. The notion that residues outside of the recognized, short binding motifs (Yaffe et al., 1997) may play a role in 14-3-3 interactions is novel, and may be a part of the answer to the elusive question of 14-3-3 specificity and/or selectivity.
Figure 1.
Disruption of the pNR:14-3-3 inactive complex by three phosphorylated synthetic peptides, pNR6, pNR24, and pNR25. Partially purified NR was phosphorylated and immediately desalted, as described in Methods. The mixtures of pNR, GF14ω (5 μm), and increasing concentrations of the indicated phosphopeptide were preincubated at 25°C for 5 min before assaying for NRA. Activities are expressed as a percentage of the control, which contained no phosphopeptides or GF14ω, pH 7.5, plus 5 mm Mg2+. Values are means of three determinants ± se. See Table I for peptide sequences.
Table II.
Alignment of sites in various proteins involved in the interaction with 14-3-3 proteins
| Protein | Phospho-Ser | Residue
Position
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −2 | 0 | +2 | +4 | +6 | +8 | +10 | ||||||||||
| Raf-1 | Ser-259 | R | S | T | S | T | P | N | V | H | M | V | S | T | T | L |
| Ser-621 | R | S | A | S | E | P | S | L | H | R | A | A | H | T | E | |
| β-Raf-1 | Ser-364 | R | S | S | S | A | P | N | V | H | I | N | T | I | E | P |
| Ser-728 | R | S | A | S | E | P | S | L | N | R | A | G | F | Q | T | |
| PKCβ | Ser-241 | R | R | L | S | V | E | I | W | D | W | D | L | T | S | R |
| Bcr | Ser-95 | A | S | A | S | R | P | Q | P | A | P | A | D | G | A | D |
| Ser-371 | R | S | P | S | Q | N | S | Q | Q | S | F | D | S | S | P | |
| cdc25a | Ser-106 | R | I | H | S | L | P | Q | K | L | L | G | C | S | P | A |
| Ser-191 | R | D | S | S | E | P | G | N | F | I | P | L | F | T | P | |
| cdc25b | Ser-216 | R | P | S | S | A | P | D | L | M | C | L | S | P | D | R |
| Tyr hyd | Ser-349 | R | H | A | S | S | P | M | H | S | P | E | P | D | C | C |
| Trp hyd | Ser-260 | R | H | S | S | D | P | F | Y | T | P | E | P | D | T | C |
| NR | Ser-543 | R | T | A | S | T | P | F | M | N | T | T | S | K | M | Y |
All amino acids are coded using the standard single-letter codes. The abbreviated protein names are: Raf-1 kinase, Raf-1; β Raf-1 kinase, β-Raf-1; protein kinase C β, PKCβ; Tyr hydroxylase, Tyr hyd; and Trp hydroxylase, Trp hyd. The numbering of a residue position is relative to the phosphorylatable Ser at position 0. Ser and Thr residues that are present C terminal to the phosphorylatable Ser are shown in boldface, underlined type. Note that with one exception, all target proteins contain at least one Ser/Thr residue C terminal to the recognized binding motif (for review, see Aitken, 1996).
Recently, Petosa et al. (1998) used x-ray diffraction to determine specific site interactions between 14-3-3ω and either a phosphopeptide or a novel unphosphorylated peptide ligand. They showed that the two peptides bound to the 14-3-3ω amphipathic groove differently based on their sequence differences. However, the peptides, especially the one based on a Raf-1 kinase-binding motif (which closely resembles the NR-binding motif), did not extend far enough to include the outlying Ser/Thr residues that we believe may influence binding. Thus, it was not determined if these residues bind directly with 14-3-3ω or have a role in determining the secondary structure of the ligand.
pNR Activators Disrupt the pNR:14-3-3 Complex
The effects of previously identified physical and metabolic activators of pNR (for review, see Huber et al., 1996) were examined using the two assay methods to assess the interaction between pNR and 14-3-3 proteins. Table III shows that 5 mm 5′-AMP was able to partially prevent the inhibition of pNR by a mixture of 14-3-3 proteins, or GF14ω, suggesting that 5′-AMP interfered with the binding of the 14-3-3 inhibitor protein. The effect was relatively specific, in that the 3′ and 2′ isomers of AMP did not significantly reduce inhibition of pNR, nor did 5′-AMS. Thus, the effect required a phosphate group at the 5′ position. Not only were the effects evident with both preparations of 14-3-3 proteins, but also when assays were done at pH 6.5. The observation that 5′-AMP was effective at pH 6.5 in the absence of divalent cations is significant, because it suggests that the effect at pH 7.5 (with Mg2+) cannot be explained by chelation of the divalent cation that is strictly required for the pNR:14-3-3 protein interaction at pH 7.5 (Huber et al., 1996). At pH 6.5, the interaction between pNR and the 14-3-3 protein is known to be less dependent on divalent cations (Kaiser and Huber, 1994; Huber and Kaiser, 1996). It is also clear that 5′-AMP interacts directly with pNR, because activity in the absence of 14-3-3 proteins was increased slightly by 5′-AMP, but not by its other isomers or 5′-AMS. However, there is also a site of interaction of 5′-AMP on the 14-3-3 protein; these results are presented below.
Table III.
Metabolites and physical activators reduce the association of 14-3-3 proteins with pNR
| Addition | Inhibition of NRA
|
Inhibition by 14-3-3 | ||||
|---|---|---|---|---|---|---|
| −GF14ω | +GF14ω | GF14ω | −14-3-3 | +14-3-3 | ||
| % of control | % | |||||
| pH 7.5, +Mg2+ | ||||||
| None | 100 | 54 | 46 | 100 | 73 | 27 |
| 5 mm 5′-AMP | 126 | 107 | 15 | 134 | 125 | 8 |
| 5 mm 3′-AMP | 109 | 67 | 39 | 115 | 92 | 20 |
| 5 mm 2′-AMP | 105 | 61 | 42 | 111 | 88 | 21 |
| 5 mm 5′-AMS | 101 | 60 | 41 | 93 | 71 | 24 |
| 10 mm KCl | 98 | 59 | 40 | 103 | 79 | 8 |
| 50 mm KCl | 111 | 83 | 25 | 117 | 99 | 15 |
| 100 mm KCl | 118 | 103 | 13 | 121 | 114 | 6 |
| 10 mm Pi | 109 | 77 | 30 | 106 | 85 | 20 |
| pH 6.5, −metal ion | ||||||
| None | 100 | 69 | 31 | 100 | 79 | 21 |
| 5 mm 5′-AMP | 120 | 102 | 15 | 123 | 112 | 9 |
| 5 mm 3′-AMP | 109 | 82 | 25 | 105 | 85 | 19 |
| 5 mm 2′-AMP | 106 | 76 | 28 | 108 | 89 | 18 |
| 5 mm 5′-AMS | 102 | 74 | 27 | 107 | 87 | 19 |
| 10 mm KCl | 104 | 78 | 25 | 101 | 85 | 16 |
| 50 mm KCl | 107 | 85 | 21 | 105 | 96 | 9 |
| 100 mm KCl | 127 | 113 | 11 | 131 | 127 | 3 |
| 10 mm Pi | 115 | 102 | 11 | 109 | 91 | 17 |
pNR was incubated with and without endogenous spinach 14-3-3 proteins (mixture of isoforms, 20 μm) or recombinant GF14ω (5 μm), with additions as indicated, on ice for 5 min. NRA was then determined in the same conditions as the preincubation. Results are expressed as a percentage of the control activity and all values are means of two determinations.
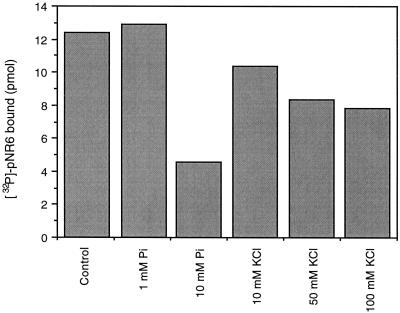
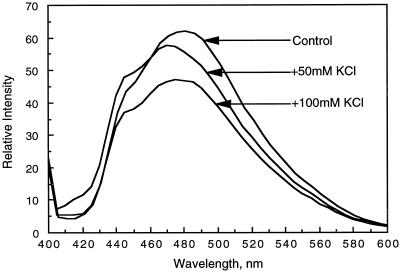
Also shown in Table III are the effects of KCl and Pi on the inhibition of NRA by 14-3-3 proteins. Addition of KCl or Pi reduced inhibition of pNR caused by 14-3-3 proteins. This was true for both the mixture of 14-3-3 proteins and recombinant GF14ω in the presence and absence of Mg2+ and in the two pH environments. Direct evidence that both physical effectors examined had an effect on 14-3-3 association with pNR is shown in Figure 2. Of the two activators, Pi at 10 mm (ionic strength = 0.09) inhibited binding of [32P]NR6 to GF14ω by about 63% relative to the control. KCl (100 mm) at a similar ionic strength (0.10) also inhibited peptide binding but to a lesser extent, suggesting that the Pi effect cannot be explained entirely by increased ionic strength. However, ionic strength does appear to have a direct effect on 14-3-3 proteins. To study this further we used the fluorescent probe bis-ANS (Takashi et al., 1977). We were able to show that an increase in ionic strength directly affects the surface hydrophobicity of GF14ω. The addition of up to 100 mm KCl caused a sequential reduction in the hydrophobic surface area (Fig. 3), as monitored by a reduction in bis-ANS fluorescence. A similar effect was also seen with Pi, and the reduced bis-ANS fluorescence was attributed to an increase in ionic strength only (data not shown). Thus, increased ionic strength caused a conformational change in the 14-3-3 protein, which may directly affect its ability to interact with binding ligands such as native pNR or the synthetic peptide pNR6. This could explain the increase in NRA reported in Table III.
Figure 2.
The physical effectors Pi and KCl inhibit the binding of pNR6 to GF14ω at pH 6.5. GF14ω (500 pmol) was incubated with 60 pmol of [32P]pNR6 (60 cpm/pmol) plus additions as shown. Representative results are shown.
Figure 3.
Ionic strength affects the surface hydrophobicity of 14-3-3 proteins. Emission spectra of bis-ANS, a fluorescent probe, to monitor changes in GF14ω surface hydrophobicity. A final concentration of 5 μm GF14ω in 100 mm Mops, pH 7.5, 10 mm Mg2+, with the addition of 1 μm bis-ANS, was used as the control. To this mixture 50 or 100 mm KCl was added, and the bis-ANS emission spectrum was recorded. The excitation wavelength was 385 nm, and the emission spectra were recorded from 400 to 600 nm.
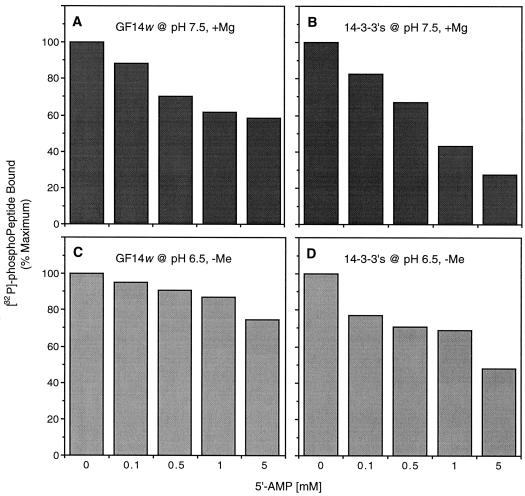
Possible 5′-AMP-Binding Site on 14-3-3 Proteins
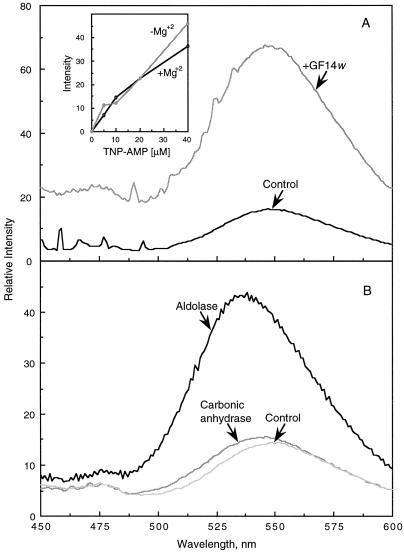
Figure 4 shows that 5′-AMP was also able to interfere with the association of pNR6 and 14-3-3 proteins. This was evident with both 14-3-3 protein sources at either pH 7.5 or 6.5 and in the presence or absence of divalent cations. The most pronounced effect was seen with the 14-3-3 protein mixture at pH 7.5 (in the presence of 5 mm Mg2+). These results suggest that 5′-AMP was directly interacting with either the phosphopeptide or the 14-3-3 protein, and prompted us to determine if a direct effect of 5′-AMP on GF14ω could be established. We tested this possibility using TNP-AMP, which fluoresces when bound to proteins (Bishop et al., 1986). As shown in Figure 5A, TNP-AMP was able to bind to the recombinant GF14ω, as demonstrated by a characteristic increase in fluorescence at approximately 543 nm and the associated blue shift (Bishop et al., 1986). As expected, the binding and fluorescence were independent of Mg2+ (Fig. 5A, inset). The binding of TNP-AMP was confirmed to be specific using other proteins such as aldolase, which is known to have an AMP-binding site (Kasprzak and Kochman, 1981), and carbonic anhydrase, which has no known AMP-binding capacity (Fig. 5B). As expected, only aldolase was able to significantly enhance TNP-AMP fluorescence. These data strongly suggest that GF14ω has an AMP-binding site.
Figure 4.
Concentration-dependent disruption of the pNR6:14-3-3 association by 5′-AMP in the presence or absence of 5 mm Mg2+. Either a spinach 14-3-3 protein mixture (1100 pmol) or recombinant GF14ω (900 pmol) was incubated with [32P]pNR6 (100 pmol; 100 cpm/pmol), as described in Methods. Results are expressed as a percentage of the maximum binding in the controls, which were 58 pmol (A), 18 pmol (B), 28 pmol (C), and 13 pmol (D), respectively.
Figure 5.
The fluorescence emission spectra of TNP-AMP in the presence of GF14ω, aldolase, and carbonic anhydrase. A, Intrinsic fluorescence of 40 μm TNP-AMP in 100 mm Mops, pH 7.5, 10 mm Mg2+ (Control), followed by the addition of 150 μg of GF14ω. The inset shows the titration of 0 to 40 μm TNP-AMP in the presence or absence of 10 mm Mg2+. B, Intrinsic fluorescence of TNP-AMP in the absence of protein (Control), and two spectra after the addition of 150 μg of aldolase or carbonic anhydrase. For all assays, the excitation wavelength was 410 nm and the emission spectra was recorded from 450 to 600 nm, with maximal intensity at approximately 543 nm. Before recording the emission spectra a preincubation of 5 min at 22°C after the addition of TNP-AMP was allowed.
CONCLUDING REMARKS
In this study we made several major findings with regard to NR regulation. In general, many of the characteristics of the NR regulatory system can now be understood to reflect features of the 14-3-3 proteins. We show that the interaction of 14-3-3 proteins with the Ser-543-binding site may involve residues outside of the currently recognized motif. In particular, Ser and Thr residues at the +6 to +8 position relative to the phosphorylated Ser residue in the recognized 14-3-3 protein-binding motif may also have an unrecognized importance in ligand binding. To our knowledge, the role of these outlying residues has not been investigated with any other protein that interacts with 14-3-3 proteins. We also show that ligand binding most likely involves electrostatic forces, because it can be disrupted with KCl and Pi. In addition, Pi and 5′-AMP are able to disrupt the interaction in a manner that cannot be entirely explained by an increase in ionic strength. This led us to one of the most exciting and unexpected findings of the present study, evidence for a putative 5′-AMP-binding site on 14-3-3 proteins. The effect seen with 5′-AMP may be of physiological significance, because elevated levels of 5′-AMP are thought to occur under some stress conditions, such as anoxia (for review, see Huber and Kaiser, 1996). In addition to a direct effect on the 14-3-3 protein, both Pi and 5′-AMP may also interact directly with NR, since both compounds stimulated NRA in the absence of 14-3-3 proteins. It is likely that interaction at this additional site may also contribute to the disruption of 14-3-3 protein interactions with native pNR.
ACKNOWLEDGMENT
The authors thank Dr. R. Ferl's laboratory for the generous donation of the GF14ω 14-3-3 clone and specifically Dr. P. Sehnke for advice on expression.
Abbreviations:
- AMS
5′-adenosine monosulfate
- bis-ANS
4,4′-bis(1-anilinonaphthalene 8-sulfonate)
- NR
nitrate reductase
- NRA
nitrate reductase activity
- pNR
phosphorylated nitrate reductase
- pNRX
phosphorylated synthetic peptide X, X is any number
- TNP-AMP
trinitrophenyl-AMP
Footnotes
This work was a cooperative investigation of the U.S. Department of Agriculture-Agricultural Research Service and the North Carolina Agricultural Research Service (Raleigh, NC), and was supported by a grant from the U.S. Department of Agriculture-National Research Initiative (grant no. 93-37305-9231 to J.L.H. and S.C.H.).
Mention of a trademark or proprietary product does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture or the North Carolina Agricultural Service, nor does it imply its approval to the exclusion of other products that might also be suitable.
LITERATURE CITED
- Aitken A. 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 14-3-3 proteins associated with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduced dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 1996a;398:26–30. doi: 10.1016/s0014-5793(96)01188-x. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Liao P-C, Gage DA, Huber SC. The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett. 1996b;387:127–131. doi: 10.1016/0014-5793(96)00478-4. [DOI] [PubMed] [Google Scholar]
- Bachmann M, McMicheal RW, Jr, Huber JL, Kaiser WM, Huber SC. Partial purification and characterization of a calcium-dependent protein kinase and an inhibitor protein required for inactivation of spinach leaf nitrate reductase. Plant Physiol. 1995;108:1083–1091. doi: 10.1104/pp.108.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M, Shiraishi N, Campbell WH, Yoo B-C, Harmon AC, Huber SC. Identification of Ser 543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell. 1996c;8:505–517. doi: 10.1105/tpc.8.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JE, Nakamoto RK, Inesi G. Modulation of the binding characteristics of a fluorescent nucleotide derivative to the sarcoplasmic reticulum adenosinetriphosphatase. Biochemistry. 1986;25:696–703. doi: 10.1021/bi00351a029. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Campbell WH, Kinghorn JR. Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem Sci. 1990;15:315–319. doi: 10.1016/0968-0004(90)90021-3. [DOI] [PubMed] [Google Scholar]
- Crawford NM. Nitrate: nutrient and signal for plant growth. Plant Cell. 1995;7:859–868. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Morrice N, MacKintosh C. Identification of a regulatory phosphorylation site in the hinge 1 region of nitrate reductase from spinach (Spinacia oleracea) leaves. FEBS Lett. 1995;377:113–117. doi: 10.1016/0014-5793(95)01300-8. [DOI] [PubMed] [Google Scholar]
- Ferl RJ. 14-3-3 protein and signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:49–73. doi: 10.1146/annurev.arplant.47.1.49. [DOI] [PubMed] [Google Scholar]
- Hoff T, Truong H-N, Caboche M. The use of mutants and transgenic plants to study nitrate assimilation. Plant Cell Environ. 1994;17:489–506. [Google Scholar]
- Huber JL, Huber SC, Campbell WH, Redinbaugh MG. Reversible light/dark modulation of spinach leaf nitrate reductase activity involves protein phosphorylation. Arch Biochem Biophys. 1992;296:58–65. doi: 10.1016/0003-9861(92)90544-7. [DOI] [PubMed] [Google Scholar]
- Huber SC, Bachmann M, Huber JL. Post-translational regulation of nitrate reductase activity: a role for Ca2+ and 14-3-3 proteins. Trends Plant Sci. 1996;1:432–438. [Google Scholar]
- Huber SC, Huber JL. Metabolic activators of spinach leaf nitrate reductase: effects on enzymatic activity and dephosphorylation by endogenous phosphatases. Planta. 1995;196:180–189. [Google Scholar]
- Huber SC, Kaiser WM. 5-Aminoimidazole-4-carboxamide riboside activates nitrate reductase in darkened spinach and pea leaves. Physiol Plant. 1996;98:833–837. [Google Scholar]
- Kaiser WM, Huber SC. Modulation of nitrate reductase in vivo and in vitro: effects of phosphoprotein phosphatase inhibitors, free Mg2+ and 5′-AMP. Planta. 1994;193:358–364. [Google Scholar]
- Kaiser WM, Spill D. Rapid modulation of spinach leaf nitrate reductase by photosynthesis. II. In vitro modulation by ATP and AMP. Plant Physiol. 1991;96:368–375. doi: 10.1104/pp.96.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzak AA, Kochman M. Characterisation of nucleotide-binding site of rabbit liver fructose-1,6-bisphosphate aldolase. J Biol Chem. 1981;256:6127–6133. [PubMed] [Google Scholar]
- Lillo C, Kazazaic S, Ruoff P, Meyer C. Characterization of nitrate reductase from light- and dark-exposed leaves. Plant Physiol. 1997;114:1377–1383. doi: 10.1104/pp.114.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Sehnke PC, Ferl RJ. Phosphorylation and calcium binding properties of an Arabidopsis GF14 brain protein homolog. Plant Cell. 1994;6:501–510. doi: 10.1105/tpc.6.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael RW, Bachmann M, Huber SC. Spinach leaf sucrose-phosphate synthase and nitrate reductase are phosphorylated/inactivated by multiple protein kinases in vitro. Plant Physiol. 1995;108:1077–1082. doi: 10.1104/pp.108.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, Mackintosh C. Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr Biol. 1996;6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Petosa C, Masters SC, Bankston LA, Pohl J, Wang B, Fu H, Liddington RC. 14-3-3ω binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J Biol Chem. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- Solomonson LP, Barber MJ. Assimilatory nitrate reductase: functional properties and regulation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:225–253. [Google Scholar]
- Su W, Huber SC, Crawford NM. Identification in vitro of a post-translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell. 1996;8:519–527. doi: 10.1105/tpc.8.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashi R, Tonomura Y, Morales MF. 4′,4′-Bis(1-anilinonaphthalene 8-sulfonate) (bis-ANS): a new probe of the active site of myosin. Proc Natl Acad Sci USA. 1977;74:2334–2338. doi: 10.1073/pnas.74.6.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Lu G, Sehnke P, Ferl RJ. The heterologous interaction among plant 14-3-3 proteins and identification of regions that are important for dimerization. Arch Biochem Biophys. 1997;339:2–8. doi: 10.1006/abbi.1996.9841. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]