Abstract
Patients with xeroderma pigmentosum (XP) have a 1,000-fold increase in ultraviolet (UV)-induced skin cancers while trichothiodystrophy (TTD) patients, despite mutations in the same genes, ERCC2 (XPD) or ERCC3 (XPB), are cancer-free. Unlike XP cells, TTD cells have a nearly normal rate of removal of UV-induced 6-4 photoproducts (6-4PP) in their DNA and low levels of the basal transcription factor, TFIIH. We examined seven XP, TTD, and XP/TTD complex patients and identified mutations in the XPD gene. We discovered large differences in nucleotide excision repair (NER) protein recruitment to sites of localized UV damage in TTD cells compared to XP or normal cells. XPC protein was rapidly localized in all cells. XPC was redistributed in TTD, and normal cells by 3 hr postirradiation, but remained localized in XP cells at 24-hr postirradiation. In XP cells recruitment of other NER proteins (XPB, XPD, XPG, XPA, and XPF) was also delayed and persisted at 24 hr (p < 0.001). In TTD cells with defects in the XPD, XPB, or GTF2H5 (TTDA) genes, in contrast, recruitment of these NER proteins was reduced compared to normals at early time points (p < 0.001) and remained low at 24 hr postirradiation. These data indicate that in XP persistence of NER proteins at sites of unrepaired DNA damage is associated with greatly increased skin cancer risk possibly by blockage of translesion DNA synthesis. In contrast, in TTD, low levels of unstable TFIIH proteins do not accumulate at sites of unrepaired photoproducts and may permit normal translesion DNA synthesis without increased skin cancer.
Keywords: DNA repair, skin cancer, ultraviolet radiation, immunofluorescence, confocal microscopy, ERCC2, XPD, XP, TTD
INTRODUCTION
Mutations in xeroderma pigmentosum group D (XPD)/excision repair cross-complementing (ERCC2) gene (MIM 126340) have been associated with different clinical disorders (MIM 278730) [Rapin et al., 2000; Lehmann, 2001; Itin et al., 2001; Bootsma et al., 2002; Friedberg et al., 2005; Kraemer et al., 2007]: XP without neurological abnormalities, XP with neurological abnormalities; trichothiodystrophy (TTD; MIM 601675) [Stefanini et al., 1986; Itin et al., 2001]; XP/TTD complex [Broughton et al., 2001]; XP/Cockayne syndrome complex (XP/CS; MIM 6105651) [Rapin et al., 2000]; and cerebro-oculo-facio-skeletal (COFS) syndrome (MIM 214150) [Graham et al., 2001]. These differences in clinical phenotypes are likely explained, at least in part, by different roles of XPD protein as a component of basal transcription factor TFIIH that operates in nucleotide excision repair (NER) as well as in transcription [Schaeffer et al., 1993, 1994; Friedberg et al., 2005]. XPD is a DNA helicase [Sung et al., 1993] with seven highly conserved helicase domains [Weber et al., 1990]. Most photosensitive TTD patients have mutations in XPD. However, several patients have been reported to carry mutations in other TFIIH components: XPB (ERCC3) [Weeda et al., 1997] and TTDA (GTF2H5) [Giglia-Mari et al., 2004].
XP is associated with sun sensitivity, sunlight-induced pigmentary changes, and a 1,000-fold increased incidence of ultraviolet (UV)-induced skin cancer [Kraemer et al., 1984, 1987, 2007; Bootsma et al., 2002]. In contrast, TTD patients have sulfur-deficient brittle hair associated with a variety of developmental abnormalities including short stature, cataracts, decreased myelin in the brain, and multiple infections. TTD patients are not reported to have XP-type skin pigmentation changes or an increased risk of cancer [Itin et al., 2001; Bootsma et al., 2002; Kraemer et al., 2007].
We examined new patients with the clinical phenotype of XP (three patients), TTD (two patients), or the XP/TTD complex (two patients) and identified defects in the XPD gene. Like earlier studies, cells from these TTD patients have reduced levels of TFIIH [Vermeulen et al., 2000, 2001; Botta et al., 2002; Dubaele et al., 2003] and nearly normal repair of UV-induced 6-4 photoproducts (6-4PP) [Broughton et al., 1990; Eveno et al., 1995; Riou et al., 1999, 2004; Nishiwaki et al., 2004]. However, we found striking differences between TTD and XP cells in the recruitment and redistribution of NER proteins to localized UV damage sites. The NER proteins accumulated at sites of unrepaired DNA damage in the XP cells and persisted for at least 24 hr. In contrast, NER proteins did not accumulate in the TTD cells probably because of instability of the TFIIH complex. These differences provide a basis for an understanding of the differences in UV-induced cancer risk in XP and TTD patients.
MATERIALS AND METHODS
Patients
After obtaining informed consent, patients with features of XP (since 1971) or TTD (since 2003) were studied at the Clinical Center, NIH, under protocols approved by the NCI Institutional Review Board. All patients had thorough skin examinations, biopsy of lesions suspicious for skin cancer, and the TTD patients had microscopic hair analysis with and without polarization [Liang et al., 2005]. Examinations included detailed ophthalmology, neurology, audiology, and other assessments as medically indicated.
Cell Strains and Culture Conditions
Normal primary dermal fibroblasts (AG13145), normal SV40-transformed dermal fibroblasts (GM00637), and primary dermal fibroblasts derived from patients—Patients XP34BE (GM16955), XP17BE (GM10428), XPTTD306BE (GM17542), XPTTD268BE (GM16877), and TTD331BE (GM15908/TTD13PV) [Giglia-Mari et al., 2004]—were obtained from the Human Genetic Mutant Cell Repository (Camden, NJ). Cell lines from Patients TTD351BE (JA1389) and TTD355BE (JA1410) were established as primary fibroblast strains from skin biopsies. Cell line XP6BE ER2-9 is an SV40-transformed fibroblast cell line of Patient XP6BE [Kraemer et al., 1975] stably transfected with wild-type XPD [Protic-Sabljic et al., 1986; Gözükara et al., 1994]. Cells from Patient TTD1VI, with mutations in the XPD gene [p.R722W and p.L461V + 716-730 del] [Sarasin et al., 1992; Takayama et al., 1996] and Patient TTD6VI’s SV40-transformed dermal fibroblasts with mutations in XPB (homozygous p.T119P) [Weeda et al., 1997; Riou et al., 1999] were a gift from Dr. A. Sarasin (Laboratory of Molecular Genetics, Villejuif, France). Cells were cultured as described previously [Oh et al., 2007].
DNA/RNA isolation, PCR Amplification, and Nucleotide Sequencing
DNA was isolated either from cells using DNAzol reagent (Invitrogen, Carlsbad, CA) or from whole blood using QIAmp DNA Blood Mini kit (Qiagen, Valencia, CA). Total cytoplasmic RNA isolated from cells using RNA queous small scale phenol-free total RNA isolation kit (Ambion, Austin, TX) was reverse-transcribed using SUPERSCRIPT first strand synthesis system (Invitrogen) with oligonucleotide (dT)12–18 primers for synthesis of the first strand cDNA for GC-rich RNA. XPD cDNA was amplified using Advantage cDNA PCR kit (BD Clontech, Mountain View, CA); PCR conditions and primers available on request (T. Ueda, J. Boyle, K.-S. Oh, S. Khan, K. Imoto, H. Inui, T. Tamura, J.J. DiGiovanna, and K.H. Kraemer, unpublished results). Sequencing reactions were as described [Oh et al., 2006]. The XPD (ERCC2) GenBank sequences used were NT_011109.15 for genomic DNA, NM_000400.15 for cDNA, and NP_000391.1 for protein. The TTDA (GTF2H5) GenBank sequences used were NT _007422.13 for genomic DNA, NM_207118.1 for cDNA (with numbering based on +1 as the A of the ATG initiation codon), and NP_997001.1 for protein.
Immunocytochemistry of NER Protein Localization Following Localized UV Irradiation
Fibroblasts were labeled with 0.8-μm (normal donors) or 2-μm (patients) latex beads (Polybead carboxylate microspheres; Polysciences, Warrington, PA) and cultured until confluent [Jaspers and Bootsma, 1982]. Cells were combined, overlaid with 5-μm Millipore filters (Millipore, Billerica, MA), irradiated with 100 J/m2 UV-C, and double-stained for immunofluorescent localization of NER proteins and photoproducts as described [Kobayashi et al., 2001; Volker et al., 2001; Imoto et al., 2002; Oh et al., 2007]. Cells were visualized with a LSM 510 confocal microscope (Carl Zeiss, Jena, Germany). For quantitation, 50 nuclei were scored as being with or without localized NER protein. Error bars indicate the standard error of the mean of more than one independent assay. Two-tailed chi-square tests were performed and p < 0.001 was considered significant.
Western Blot Analysis
Western blotting was performed as described [Oh et al., 2006] except total cell extract protein was separated on 8 to 16% gradient gels (Invitrogen) and probed with primary antibodies: anti- green fluorescent protein (GFP) monoclonal (1:2,000; BD Biosciences), anti-XPD monoclonal (1:500; a kind gift from Drs. J.-M. Egly and J. Bradsher, Institut de Genetique et de Biologie Moleculaire et Cellulaire, Strasbourg, France), or anti-β-actin monoclonal (1:4,000; Chemicon, Millipore, Billerica, MA). Horseradish peroxidase conjugated rabbit anti-mouse polyclonal immunoglobulin G (IgG) (1:10,000 for XPD; and β-actin, 1:20,000 for GFP; Novus Biochemicals, Littleton, CO) was used.
Production of Stably Transfected Positive Clones from Patient TTD1VI [XPD-GFP]
Wild-type XPD cDNA was cloned in frame to the 3′ end of the GFP gene in a pEGFP-C1 expression vector containing the selectable Neo marker gene (BD Biosciences). A total of 8×104 cells from Patient TTD1VI were cultured overnight and transfected with 1 μg XPD-GFP and 10.5 μl lipofectamine for 5 hr. Two days later cells were grown in medium supplemented with 0.6 μg/ml G418 for selection.
Unscheduled DNA Synthesis, Enzyme-Linked Immunoassay, and UV Sensitivity
Post-UV unscheduled DNA synthesis (UDS) using scintillation counting was performed as described [Lehmann and Stevens, 1980]. UV-induced 6-4PP and cyclobutane pyrimidine dimer (CPD) photoproducts were measured by an enzyme-linked immunoassay (ELISA) assay with TDM-2 and 64M-2 monoclonal antibodies [Mori et al., 1991; Nakagawa et al., 1998; Kobayashi et al., 2001; Imoto et al., 2002]. UV sensitivity was determined using a 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2 H-tetrazolium (MTS) assay (Promega, Madison, WI) [Imoto et al., 2002].
RESULTS
Clinical Features and Mutations in XP, TTD, and XP/TTD Patients Studied
XPD protein has dual roles in NER and transcription and is mutated in patients with different clinical diseases (Table 1; Fig. 1). At NIH we evaluated the clinical features of seven patients with XPD mutations: three had XP, two had TTD, and two had the XP/TTD complex. In addition, one patient we examined with clinical TTD had mutations in the TTDA gene (Table 1; Fig. 1D). Patient XP6BE [Robbins et al., 1974], a Caucasian woman, had characteristic XP features [Kraemer et al., 2007] of acute sun sensitivity, pigmentary abnormalities, 25 to 50 skin cancers, microcephaly, sensorineural hearing loss, and severe XP neurological symptoms (Table 1; Fig. 1A and E). She died at age 29 years. Patient XP6BE’s cells had the common XP-associated p.R683W mutation [Friedberg et al., 2005] in one allele and p.G36-R61del in the other allele resulting from a splice donor mutation [Takayama et al., 1995]. The same mutations were reported in her affected sibling, Patient XP5BE [Kobayashi et al., 2002]. Patient XP17BE [Andrews et al., 1978], a 15-year-old black male, had similar XP clinical symptoms with 25–50 skin cancers, microcephaly, sensorineural hearing loss, and intellectual impairment (intelligence quotient 50) (Table 1). Patient XP17BE’s cells had the common XP-associated p.R683W mutation and a second mutation, p.D681N, which was previously reported only in Patient XPCS2RO, a patient with the severe COFS syndrome [Graham et al., 2001]. Patient XP34BE, a 29-year-old Caucasian man, had XP features of acute sun sensitivity and mild pigmentary abnormalities but no skin cancer, hearing loss, or neurological abnormalities (Table 1). His 25-year-old affected sister, Patient XP35BE, had similar clinical features. Patient XP34BE’s cells also carry the p.R683W mutation. The second mutation was a novel splice mutation c.595–10G>A giving rise to insertion of eight nucleotides (CCCCCCAG) in the mRNA (c.625_626ins8) and a predicted truncated protein: p.I199Pfs*52 (T. Ueda, J. Boyle, K.-S. Oh, S. Khan, K. Imoto, H. Inui, T. Tamura, J.J. DiGiovanna, and K.H. Kraemer, unpublished results).
TABLE 1.
Clinical Features and Mutations in XP, XP/TTD, and TTD Patients Examined at NIH
| Diagnosis | Patient | Genetic defect |
Clinical features |
XPD (ERCC2) mutationsG
|
Mutations reported (references) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UDS (% of normal) |
Allele 1
|
Allele 2
|
||||||||||||||||
| Age/ sex |
Acute sun sensitivityA |
Abnl pigmentationA |
Skin cancers (number) |
Stature | Neuro abnlA |
Microce phalyA |
Sensori-neural hearing lossA |
Genomic | cDNA | Amino acid |
Genomic | cDNA | Amino acid |
|||||
| XP | XP6BE | XPD | DB29y/F | + | + | 25–50 | <5%-ile | + | + | + | 25–55C | g.18220C>T | c.2047C>T | p.R683W | c.105+2T>A | c.106-183del | p.G36-R61del | Takayama et al. [1995] |
| XP | XP17BE | XPD | 15y/M | + | + | 25–50 | <10%-ile | + | + | + | 33 | g.18220C>T | c.2047C>T | p.R683W | g.18061G>A | c.2041G>A | p.D681N | This paperD |
| XP | XP34BE | XPD | 29y/M | + | + | 0 | Normal | Normal | Normal | Normal | 19E | g.18220C>T | c.2047C>T | p.R683W | c.595-10G>A | c.594-595ins CCCCCCAG | p.I199Pfs*52 | This paper |
| XP/TTD | XPTTD 306BE | XPD | 11y/M | + | + | 0 | <5%-ile | + | + | + | 28 | g.18337C>T | c.2164C>T | p.R722W | g.1471C>T | c.152G>T | p.S51F | This paper |
| XP/TTD | XPTTD 268BE | XPD | 18y/M | + | + | 0 | <3%-ile | + | + | + | 8 | g.1917G>A | c.335G>A | p.R112H | g.18007T>C | c.1987T>C | p.C663R | This paperD |
| TTD | TTD351BE | XPD | 9y/M | + | Normal | 0 | <3%-ile | + | Normal | + | 115 | g.18337C>T | c.2164C>T | p.R722W | g.8944G>A | c.1133G>A | p.R378H | This paper |
| TTD | TTD355BE | XPD | 6y/M | + | Normal | 0 | <3%-ile | + | + | + | 55 | g.18363delG | c.2190delG | p.E731Rfs*14 | Unknown | Not expressed | Not expressed | This paper |
| 0 | TTDA (GTF2H5) mutationsH
|
|||||||||||||||||
| Allele 1
|
Allele 2
|
|||||||||||||||||
| TTD | TTD331BE | TTDA | 27y/M | + | Normal | 0 | Normal | + | Normal | + | 10 | g.37T>C | c.2T>C | p.M1? | Homozygous | Homozygous | Giglia-Mari et al. [2004]F | |
present.
death.
Mutation independently assessed by Dr. A. Lehmann.
XP35BE – sibling.
TTD13PV.
GenBank refeence sequences NT_011109.15 for genomic DNA, NM_000400.2 for cDNA and NP_000391.1 for protein.
GenBank reference sequences NT_007422.13 for genomic DNA, NM_207118.1 for cDNA and NP_997001.1 for protein.
FIGURE 1.
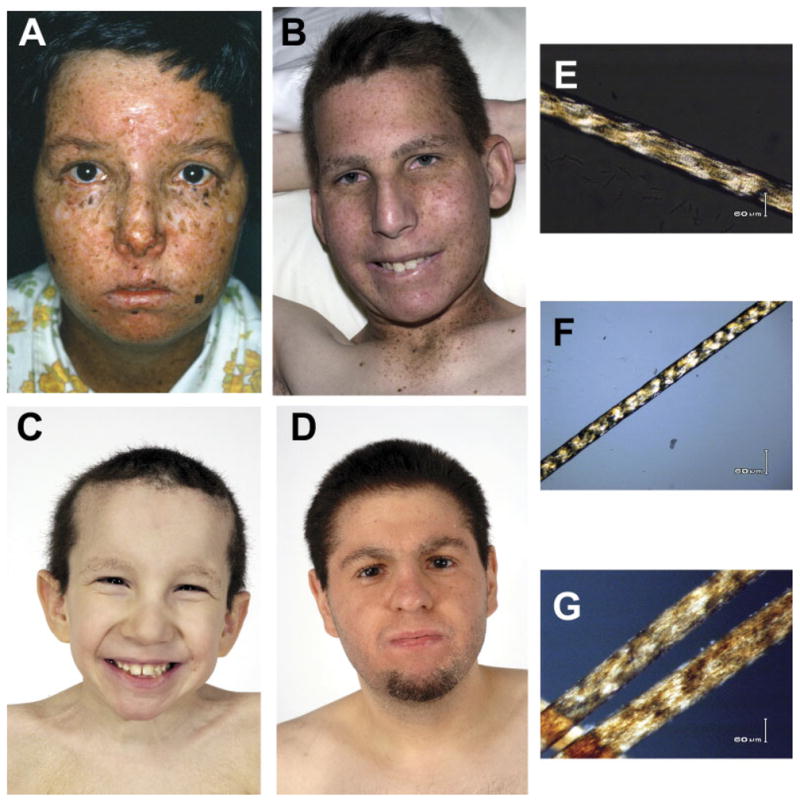
Clinical appearance of XP, XP/TTD, and TTD patients and polarizing microscopic examination of their hair. A–D: Clinical appearance of face and eyes. E–G: Microscopic examination of hair using polarized light [Liang et al., 2005]. A: Patient XP6BE, at age 18 years, had XP with neurological abnormalities [Robbins et al., 1974] and mutations in the XPD gene. Her face and neck show numerous freckle-like pigmented lesions of varying size and shape. There are scars from multiple surgical procedures. She had 25–50 basal cell and squamous cell carcinomas and two primary melanomas. The conjunctiva of her eyes show sunlight-induced inflammatory lesions. She died at the age of 29 years. B: Patient XPTTD268BE, age 18 years, has the XP/TTD complex and mutations in the XPD gene. He has the XP features of sunlight-induced freckle-like pigmented lesions of his face and neck and inflammatory lesions of the conjunctiva. His short, brittle hair and sparse eyebrows are features of TTD. E: Patient XPTTD268BE’s hair showed the TTD characteristic of alternating dark and light “tiger tail” bands with polarized light. C: Patient TTD351BE, age 9 years, has TTD and mutations in the XPD gene. He has short, brittle hair, sparse eyebrows, and ichthyosis of his upper chest. Note the absence of freckle-like pigmentation despite marked clinical sun sensitivity. F: Patient TTD351BE’s hair showed the TTD characteristic of alternating dark and light “tiger tail” bands with polarized light. D: Patient TTD331BE, age 27 years, has TTD and mutations in the TTDA gene. He had sun sensitivity without the pigmentation changes of XP.G: Patient TTD331BE’s hair showed the TTD characteristic of alternating dark and light “tiger tail” bands with polarized light.
Only two patients have previously been reported as having features of both XP and TTD [Broughton et al., 2001]. We report two additional patients with this XP/TTD complex phenotype. Patient XPTTD306BE, an 11-year-old Caucasian boy, had XP features of acute sun sensitivity and mild pigmentary abnormalities but no skin cancers (Table 1). In addition, he had the characteristic TTD features [Kraemer et al., 2007] of “tiger tail” banding of his hair on polarized microscopy [Liang et al., 2005], ichthyosis, short stature, microcephaly, and sensorineural hearing loss. Patient XPTTD306BE’s cells contain a commonly reported TTD-specific mutation, p.R722W [Friedberg et al., 2005] in the XPD gene. A mouse engineered to have this mutation was shown to have some clinical features of TTD [De Boer et al., 1998]. The second mutation in this patient, p.S51F, has not been previously reported.
Patient XPTTD268BE, a 21-year-old Caucasian male with the XP/TTD complex, had XP features of acute sun sensitivity and marked pigmentary and ocular changes (Table 1; Fig. 1B). In addition, his hair had the characteristic TTD features [Kraemer et al., 2007] of “tiger tail” banding on polarized microscopy (Fig. 1E), ichthyosis, short stature, microcephaly, neurological abnormalities, and sensorineural hearing loss. The first mutation in Patient XPTTD268BE’s cells was p.R112 H, also reported in the XP/TTD patient, Patient XP38BR [Broughton et al., 2001]. Homozygous or compound heterozygote mutations for p.R112H have been found in other TTD patients [Friedberg et al., 2005] and have been reported to be associated with an XP-like cellular phenotype [Berneburg et al., 2000]. The second mutation in Patient XPTTD268BE’s cells was c.1987T>C resulting in the novel amino acid substitution p.C663R.
Patient TTD351BE, a 10-year-old Caucasian male with TTD, had acute skin sun sensitivity and ichthyosis without the increased freckle-like pigmentation of XP (Table 1; Fig. 1C). He had characteristic TTD features of short brittle hair that showed “tiger tail” banding with polarized microscopy (Fig. 1F), short stature (<3rd percentile), multiple sinopulmonary infections, and TTD neurological abnormalities, including sensorineural hearing loss, developmental delay, and reduced myelination of the brain. Patient TTD351BE’s cells were heterozygous for the TTD-specific p.R722W mutation [Friedberg et al., 2005] and a novel mutation c.1133G>A causing p.R378H amino acid substitution. This mutation is in the region of XPD where no other mutations have been reported and is not located within any of the helicase domains.
Patient TTD355BE, a 6-year-old Caucasian male with TTD, had acute skin sun sensitivity without the increased freckle-like pigmentation of XP (Table 1). He had characteristic TTD features of short brittle hair that showed “tiger tail” banding with polarized microscopy, height <3rd percentile, cataracts requiring surgery, multiple otitis media infections, and TTD neurological abnormalities, including sensorineural hearing loss, microcephaly, developmental delay, and reduced myelination of the brain. Patient TTD355BE’s cells have the XPD mutation p.E731Rfs*14 caused by c.2190delG. The other allele was not expressed. This combination of mutations has been reported in Patient TTD1BI [Broughton et al., 1994].
Patient TTD331BE, a 27-year-old Italian-American male with TTD, had acute skin sun sensitivity without pigmentary changes of XP (Table 1; Fig. 1D). His hair showed “tiger tail” banding (Fig. 1G) diagnostic of TTD [Liang et al., 2005] but unlike most TTD patients, it was not short or brittle. He had TTD neurological abnormalities of mild sensorineural hearing loss and intellectual impairment but did not have short stature, microcephaly, or abnormal findings on brain magnetic resonance imaging (MRI). His 29-year-old brother, Patient TTD332BE, had similar clinical features. We confirmed the previously reported homozygous p.M1? mutation in the TTDA gene [Giglia-Mari et al., 2004] in Patient TTD331BE’s cells.
TFIIH Levels in XP-D and TTD Fibroblasts
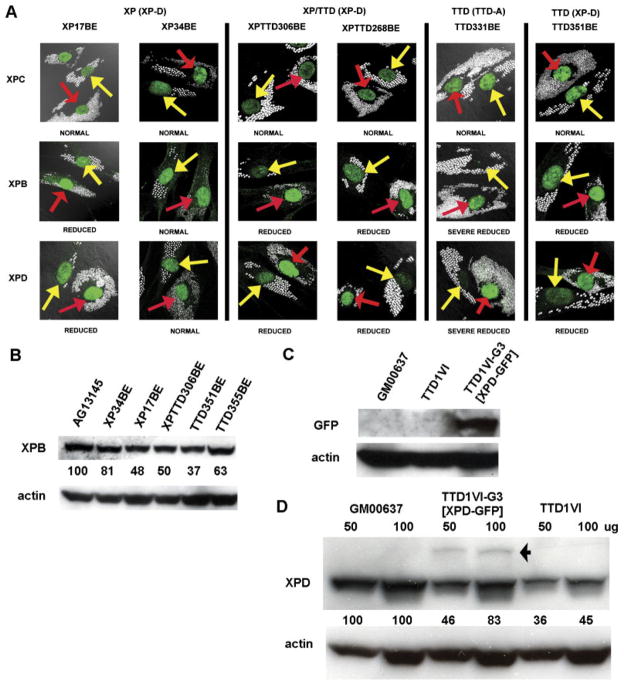
Immunofluorescence using antibodies to XPB and XPD protein in unirradiated cells served as an indicator of TFIIH levels in the various cell strains (Fig. 2A). Reduced levels of TFIIH have been reported in TTD cells of complementation groups XP-D, XP-B, and TTD-A by in vivo immunofluorescence analyses and by Western blotting [Vermeulen et al., 2000, 2001; Botta et al., 2002; Dubaele et al., 2003]. In agreement with these studies we found reduced levels of XPB and XPD protein in TTD (XP-D) (Patient TTD351BE) and TTD (TTD-A) (Patient TTD331BE) cells in vivo. Expression levels appeared normal in XP (XP-D) cells in Patient XP34BE but were reduced in Patient XP17BE. TFIIH levels were reduced in the two new XP/TTD (XP-D) cells from Patients XPTTD306BE and XPTTD268BE. In contrast, all these cells had normal levels of XPC protein.
FIGURE 2.
TFIIH levels in unirradiated cells from XP, TTD, and XP/TTD patients measured by immunofluorescence and Western blotting. A: Normal cells (AG13145) were labeled with 0.8-μm beads (red arrows) and XP (XP-D) (Patients XP17BE and XP34BE), XP/ TTD (XP-D) (Patients XPTTD306BE and XPTTD260BE), and TTD (TTD-A) (Patient TTD331BE) and TTD (XP-D) (Patient TTD351BE) cells were labeled with 2-μm beads (yellow arrows) and co-cultured. Immunofluorescence staining was performed with antibodies to XPC, XPB, and XPD proteins. The intensity of staining of the patient cells is compared to that of the normal cells. XPC levels in all patient cells are comparable to normal cells. Levels of TFIIH subunits XPB and XPD are severely reduced in TTD cells of complementation groups XP-D and TTD-A, and reduced in XP/TTD cells of complementation group XP-D. B: Western blotting measurement of XPB protein levels in normal (AG13145); XP (XP-D) (Patients XP34BE and XP17BE); XP/TTD (XP-D) (Patient XPTTD306BE); and TTD (XP-D) (Patients TTD351BE and TTD355BE) cells. The ratio of the intensity of the XPB bands to that of the β-actin bands is indicted between the rows. C: Western blotting measurement of recombinant XPD-GFP protein expression (~110 kDa) in SV40-transformed TTD1VI-G3 [XPD-GFP]+ stably transfected fibroblasts compared to normal (GM0637) and TTD (XP-D) TTD1VI SV40-transformed cells. D: Western blot showing increased levels of XPD protein in clone G3 of SV40-transformed TTD1VI [XPD-GFP]+ stably transfected fibroblasts. Lanes were loaded with 50 or 100 μg of protein as indicated. The arrow indicates the migration of GFP-XPD. The lower bands are the size of endogenous XPD protein. The ratio of the intensity of the XPD bands to that of the β-actin bands is indicated between the rows.
We used Western blot analyses to determine the steady-state levels of XPB protein as an indicator of total TFIIH levels in our XP-D cell strains (Fig. 2B). XPB levels were reduced in TTD cells (Patient TTD351BE, 37% of normal; Patient TTD355BE, 63% of normal), and XP/TTD cells (Patient XPTTD306BE, 50% of normal). Levels of XPB were slightly reduced in cells from the milder XP patient, Patient XP34BE (81% of normal). XPB levels in fibroblasts from a severely affected XP patient, Patient XP17BE, showed a greater reduction (48% of normal). This is similar to the approximately 50% reduction in the TFIIH level (p62 protein) previously reported for lymphoblastoid cells from this patient [Satoh and Hanawalt, 1997]. These results are in general agreement with the immunofluorescence assay (Fig. 2A).
Time Course of Recruitment of NER Proteins to Localized UV Damage
Patient and normal cells were labeled with differently-sized cytoplasmic beads and then cultured on the same slides. We used localized irradiation through a Millipore filter with 5-μm pores to analyze the time course of the recruitment of NER proteins to UV damage sites following 100 J/m2 UV exposure to the surface of the filter [Volker et al., 2001]. Because the filter blocks most of the UV, the cells receive a small dose of UV that permits repair and replication to continue [Imoto et al., 2002]. Cells were fixed by 0.1 hr, 0.5 hr, or 24 hr postirradiation and immunostained with pairs of antibodies to simultaneously assess the location of DNA damage and NER proteins. There were marked differences between XP, TTD, and XP/TTD cells (Figs. 3–5; Supplementary Figs. S1–S3; available online at http://www.interscience.wiley.com/jpages/1059-7794/suppmat).
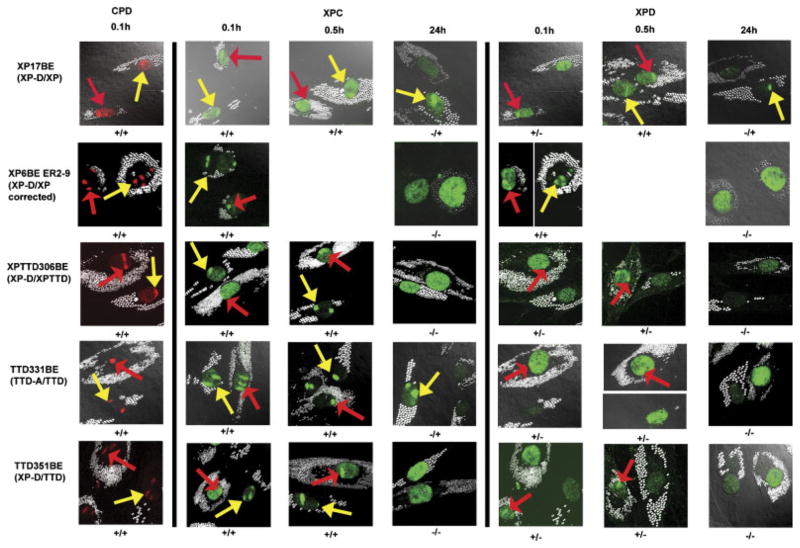
FIGURE 3.
Recruitment of XPC and XPD proteins to localized DNA damage in XP, XP/TTD, and TTD cells at 0.1 hr, 0.5 hr, and 24 hr after UV irradiation. Normal cells (AG13145) were labeled with 0.8-μm latex beads (red arrows) and cells (from Patients XP17BE, XP6BE ER2-9, XPTTD306BE, TTD331BE, and TTD351BE) were labeled with 2-μm latex beads (yellow arrows). UV irradiation at 100 J/m2 was delivered through a 5-μm filter. Cells were fixed by 0.1 hr, 0.5 hr, or 24 hr postirradiation and immunostained with pairs of antibodies to simultaneously assess the location of DNA damage and NER proteins. The arrows indicate sites of localized immunostaining: normal (red arrows), patient (yellow arrows). Where necessary, images were captured at two exposure times to show any localization with low levels of protein (images separated by white line). Symbols below each image indicate localization (+) or nonlocalization (−) of NER proteins or photoproducts in normal cells/patient cells.
FIGURE 5.
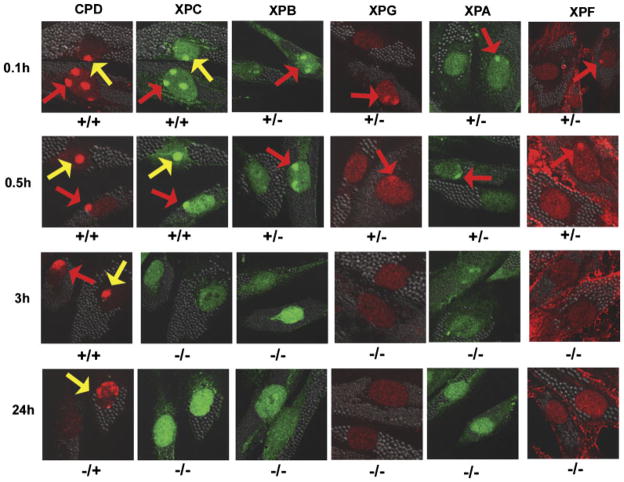
Recruitment of XPC, XPB, XPG, XPA, and XPF proteins to sites of localized UV damage in cells from Patient TTD6VI (XP-B) at 0.1hr, 0.5 hr, 3 hr, and 24 hr after UV irradiation. Normal AG13145 cells were labeled with 0.8-μm latex beads (red arrows) and cells from Patient TTD6VI (XP-B) were labeled with 2-μm latex beads (yellow arrows). Cells were fixed by 0.1hr, 0.5 hr, 3 hr, or 24 hr postirradiation and immunostained with pairs of antibodies to simultaneously assess the location of DNA damage and NER proteins. The arrows indicate sites of localized immunostaining: normal (red arrows), patient (yellow arrows). Symbols below each image indicate localization (+) or nonlocalization (−) of NER proteins or photoproducts in normal cells/patient cells. Experimental details are as in Fig.3.
CPD formation and XPC protein recruitment
DNA damage was rapidly observed in all cell strains following exposure to UV irradiation, as demonstrated by CPD foci at 0.1 hr (Fig. 3). By 0.5 hr post-UV-irradiation, CPD were still present in the nuclei of normal and patient cell strains (data not shown). By 24 hr CPD foci were reduced in normal cells and had a lesser reduction in patient cells, reflecting reduced CPD repair (data not shown). XPC protein, which is involved in the initial recognition of these bulky lesions [Friedberg et al., 2005], was recruited to these damage sites within 0.1 hr of irradiation in all cell strains tested, with about 60 to 75% of the cells having localized XPC (Figs. 3 and 4A). By 0.5 hr postirradiation, XPC protein continued to be localized in about 60 to 90% of the nuclei in all of the cell strains (Figs. 3 and 4A). By 3 hr postirradiation, XPC was redistributed in normal and TTD cells but remained localized in the XP (XP-D) cells (data not shown). By 24 hr, about 13% of normal and 2 to 28% of TTD cells showed localized XPC. In contrast, more than 60% of the XP (XP-D) cells from Patient XP17BE showed persistent XPC localization (p<0.001). Of interest, like the XP cells, there was persistent localization of XPC in most cells from Patient XPTTD268BE (XP-D) cells at 24 hr (estimates from limited counts). In contrast, like TTD cells, approximately 30% of cells from Patient XPTTD306BE showed XPC localization at 24 hr (estimates from limited counts).
FIGURE 4.
Frequency of XPC, XPD, and XPG NER protein localization in XP, XP/TTD, and TTD cells at 0.1 hr, 0.5 hr, and 24 hr after UV irradiation. A total of 50 nuclei were scored as being with (positive) or without (negative) DNA damage or NER protein localization at the indicated time points postirradiation. A: XPC localization. B: XPD localization. C: XPG localization. Where shown, error bars indicate the SEM of at least two independent counts.*p<0.001 significant difference between patient cell count and normal cell count at the equivalent time postirradiation. Bar numbers: 1, Patient AG13145; 2, Patient XP34BE; 3, Patient XP17BE; 4, Patient XPTTD306BE; 5, Patient TTD331BE; 6, Patient TTD351BE; 7, Patient TTD355BE; 8, Patient TTD1VI; and 9, Patient TTD1VI [XPD-GPF]+.
Localization of XPD protein
XPD helicase is recruited to DNA damage and carries out localized DNA unwinding in the damaged region [Friedberg et al., 2005]. In normal fibroblasts XPD was localized to damage sites within 0.1 hr of irradiation. However, at this time XPD localization was only at low frequency in XP (XPD), XP/TTD (XP-D), or TTD (XP-D or TTD-A) cells (p<0.001; Figs. 3 and 4B). In normal cells, localization of XPD was still apparent at 0.5 hr, indicating that repair was ongoing at this time (Figs. 3 and 4B). Interestingly, by 0.5 hr post-UV, XPD localized in XP (XP-D) cells and the frequency of XPD-positive nuclei in cells from Patient XP17BE (74%) and Patient XP34BE (50%) was not significantly different from that in normal cells (63%) (Fig. 4B). In contrast, XPD continued to be localized in only a small proportion of TTD cells (Patient TTD351BE [XP-D], 9%; Patient TTD355BE [XP-D], 0%; Patient TTD1VI [XP-D], 8%; and Patient TTD331BE [TTD-A], 0%) and XP/TTD cells (Patient XPTTD306BE [XP-D], 6%) (p<0.001 compared to normal cells; Figs. 3 and 4B). Observations at 3 hr and 8 hr confirmed the persistent localization of XPD in XP cells and low to absent levels of localization in TTD (XP-D and TTD-A) and XP/TTD (XP-D) cells (Supplementary Fig. S3). By 24 hr, the frequency had declined to less than 10% of normal cells showing localized XPD staining, indicating redistribution of the XPD protein and the completion of repair. In marked contrast, the frequency of Patient XP17BE cells showing localized XPD remained significantly elevated (61%; p<0.001), indicating the persistence of XPD protein at the damage site. The low frequency of XP/TTD or TTD cells with localized XPD continued through to 24 hr postirradiation, such that localization values were similar to those in normal cells at 24 hr, which had completed repair (Fig. 4B).
Localization of XPG, XPB, XPA, and XPF proteins
XPG protein recruitment followed a similar pattern as XPD (Fig. 4C; Supplementary Fig. S1; data not shown). Thus there was rapid recruitment of XPG in normal cells by 0.1 hr but not in XP, TTD, or XP/TTD cells. The XP (XP-D) cells showed recruitment of XPG by 0.5 hr that persisted as long as 24 hr. In contrast, there was a significantly lower frequency (p<0.001) of TTD or XP/TTD cells showing XPG recruitment at 0.5 hr compared to normal cells. However, by 24 hr, the redistribution of XPG protein in normal cells resulted in a frequency of XPG positive cells that was similar to the persistently low frequency in TTD or XP/TTD cells.
Post-UV localization of NER proteins XPB, XPA, and XPF was assessed in cells from Patients XP17BE (XP-D), TTD331BE (TTDA), and TTD351BE (XP-D). These NER proteins followed a similar course as XPD and XPG proteins (Supplementary Figs. S1, S2A, S2B, and S2C). Thus at 0.1 hr post-UV all three patient cell strains had significantly (p<0.001) reduced frequency of XPB, XPA, and XPF compared to normal cells. However, the recruitment of these proteins in the TTD cells remained low even after 24 hr postirradiation, whereas XPB, XPA, and XPF became localized in XP (XP-D) cells by 0.5 hr and persisted to 24 hr. In both XP/TTD cell strains, from Patients XPTTD306BE and XPTTD268BE, localization of XPG was minimal throughout the postirradiation time course (and localization of XPB and XPD was comparable between cells from both patients), similar to that in TTD cells. However, XPA and XPF localization was also minimal in cells from Patient XPTTD306BE (Supplementary Fig. S1). This is similar to the localization in TTD cells (Supplementary Fig. S2B and S2C). In contrast, the localization of XPA and XPF in Patient XPTTD268BE’s cells was delayed (Supplementary Fig. S1). This is similar to the localization in XP (XP-D) cells, which occurred at normal levels by 0.5 hr (Supplementary Fig. S2B and S2C).
We also studied cells from Patient TTD6 VI, a TTD patient with a mutation in the XPB gene [Weeda et al., 1997; Riou et al., 1999; Oh et al., 2006]. These TTD (XP-B) cells showed results similar to that of the TTD (XP-D) cells (Fig. 5). There was normal localization of XPC at 0.1 h and 0.5 h post-UV and redistribution by 3 h. The NER proteins XPB, XPA, XPG and XPF were not localized at any post-UV time point (0.1 h, 0.5 h, 3 h, 8 h or 24 h) in the cells from Patient TTD6 VI (Fig. 5; data not shown). In contrast, like the XP (XP-D) cells, XP (XP-B) cells showed delayed localization of these proteins and persistence at sites of unrepaired DNA damage at 24 h [Oh et al., 2007].
Correction of the defect in localization of NER proteins by transfection of XPD
To demonstrate specificity of the XPD cellular abnormality, SV40-transformed fibroblasts from Patient TTD1VI were transfected with wild-type XPD cloned in-frame with GFP. Western blotting using antibodies against GFP (Fig. 2C) confirmed the presence of the construct in a stable clone, TTD1VI-G3 [XPD-GFP]+. This clone had increased post-UV survival (Supplementary Fig. S4A). Blotting with XPD antibodies demonstrated increased levels of XPD in TTD1VI [XPD-GFP]+ compared to untransfected cells from Patient TTD1VI (Fig. 2D). Two bands are present. The upper band (arrow, Fig. 2D) is at the location of GFP-XPD and the lower band is the location of XPD protein. Thus the presence of wild-type GFP-XPD protein increased the steady state level of endogenous (mutated) XPD (from 45% of normal to 83%) probably by stabilizing the TFIIH complex. Similar increased TFIIH stability following XPD-GFP transfection of TTD cells was previously reported [Botta et al., 2002].
To determine the specificity of the abnormality of localization of NER proteins, we studied XP (XP-D) and TTD (XP-D) cell lines, which were stably corrected by addition of the wild-type XPD gene. The failure to recruit NER proteins was corrected in XP6BE ER2-9 (Fig. 3; data not shown) and TTD1VI [XPD-GFP]+ cell lines, which were stably transfected with wild-type XPD (Fig. 4A–C). For example, in TTD1VI [XPD-GFP]+ cells, 39% of the nuclei had localized XPD within 0.1 hr of irradiation, compared to 47% in normal cells (p=not significant), and 8% in uncorrected nuclei in cells from Patient TTD1VI (p<0.001) (Fig. 4B).
UV Damage Repair
The rate of DNA repair, as measured by the post-UV UDS rate, was 8 to 55% of normal in all the patients’ cells studied except for Patient TTD351BE, which was 115% of normal (Table 1). There have been no other reports of such high UDS in photosensitive TTD (XP-D) cells. Post-UV cell survival among the TTD cells appeared to correlate with UDS (Supplementary Fig. S4A). Thus Patient TTD351BE’s cells had nearly normal post-UV cell survival, Patient TTD355BE’s cells (55% UDS) had intermediate survival, and Patient TTD331BE’s cells (10% UDS) had the greatest sensitivity
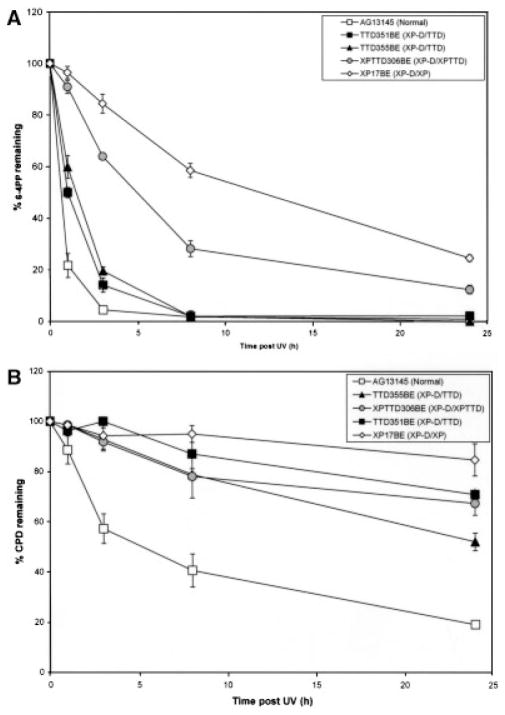
UDS at early times after UV is primarily a reflection of repair of 6-4PP. Using an ELISA assay we compared CPD and 6-4PP repair in normal cells (AG13145) with that of other reported normal cell strains [Nishiwaki et al., 2004; Riou et al., 2004]. Results were comparable, with almost 100% 6-4PP removed by 24 hr post-UV (Fig. 6A). We also observed 19% CPD remaining at 24 hr in AG13145 (Fig. 5B). This was comparable to other normal cells: 25% in 198 VI [Riou et al., 2004] and 30% in MSU-1 [Nishiwaki et al., 2004].
FIGURE 6.
Repair of UV-induced 6-4PP and CPD in XP, TTD, and XP/TTD cells. Fibroblasts from normal (AG13146) or XP (XP-D) (Patient XP17BE); TTD (XP-D) (Patients TTD351BE and TTD355BE); and XP/TTD (XP-D) (Patient XPTTD306BE) cells were irradiated with 10 J/m2 UV and harvested at various times postirradiation. The percentage of the initial number of photoproducts was determined using an ELISA with antibodies specific to: (A) 6-4PP or (B) CPD. Error bars indicate the SEM of at least two independent experiments.
6-4PP and CPD repair was greatly reduced in Patient XP17BE’s cells, with 25% 6-4PP, and 85% CPD remaining at 24 hr (Fig. 6A and B). These cells also showed the greatest post-UV cell sensitivity (Supplementary Fig. S4B). In the XP/TTD cell strain from Patient XPTTD306BE, 6-4PP repair was intermediate between XP and TTD cells, with 12% 6-4PP remaining at 24 hr. Cells from Patients XP17BE and XPTTD306BE had the lowest levels of UDS (33% and 28%, respectively) (Table 1).
In agreement with previous reports [Broughton et al., 1990; Eveno et al., 1995; Riou et al., 1999, 2004; Nishiwaki et al., 2004], we found that the TTD cells had reduced CPD repair and nearly normal repair of 6-4PP. In cells from Patient TTD351BE, 6-4PP repair was initially delayed, with 14% 6-4PP remaining at 3 hr in Patient TTD351BE’s cells compared to 4% in normal AG13145 cells. CPD repair was greatly reduced, with 71% CPD remaining at 24 hr. Patient TTD355BE’s cells had an intermediate 6-4PP repair activity at earlier time points (20% remaining at 3 hr) (Fig. 6A). CPD repair was also reduced in this cell strain (52% CPD remaining at 24 hr) (Fig. 6B). UDS and post-UV cell survival in Patient TTD355BE’s fibroblasts was intermediate (Table 1; Supplementary Fig. S4). Remarkably, the other cells (from Patient TTD1BI) reported with the p.E731Rfs*14 frameshift mutation in one allele of the XPD gene and the other allele not expressed also had 55% UDS [Broughton et al., 1994]. The cells from Patient TTD6VI (XP-B) were reported to have nearly normal post-UV 6-4PP repair and reduced CPD repair (73% CPD remaining at 24 hr) [Riou et al., 1999].
DISCUSSION
Clinical Features of Patients With Mutations in the XPD Gene
The different clinical phenotypes (XP, TTD, or XP/TTD complex) of patients are associated with specific mutations in the XPD gene. Some of these mutations were reported to be null [Taylor et al., 1997] but others suggested that clinical symptoms could reflect the combined effects of each allele [Dubaele et al., 2003]. We report XP patients heterozygous for the common p.R683W mutation with both severe (Patients XP17BE and XP6BE) and mild phenotypes (Patient XP34BE) (Table 1), suggesting a contribution of the second allele or other modifying genes [Broughton et al., 2001] (T. Ueda, J. Boyle, K.-S. Oh, S. Khan, K. Imoto, H. Inui, T. Tamura, J.J. DiGiovanna, and K.H. Kraemer, unpublished results). Similarly, the four known patients with the XP/TTD complex (Patients XPTTD306BE and XPTTD268BE from this report, and Patients XP38BR and XP189MA from Broughton et al. [2001]) were compound heterozygotes. Three had a mutation that was reported in TTD patients plus a second novel mutation which might contribute to the XP phenotype.
The TTD Paradox
While TTD patients have mutations in the same genes as XP patients, they do not have the increased freckle-like pigmentation or an increased frequency of sunlight induced skin cancers [Itin et al., 2001; Bootsma et al., 2002; Kraemer et al., 2007]. Further, the level of the basal transcription factor, TFIIH, which might be expected to protect against skin cancer because of its role in NER, is lower in TTD (XP-D) cells than in XP (XP-D) cells (Fig. 2A; data not shown) [Botta et al., 2002; Dubaele et al., 2003; Nishiwaki et al., 2004]. A proposed explanation is that a NER defect gives rise to XP, whereas a transcription defect gives rise to TTD [Bootsma and Hoeijmakers, 1993; Vermeulen et al., 1994; Friedberg et al., 2005]. Thus the cancer-free phenotype of TTD would be unrelated to its repair defect [Berneburg et al., 2000; Nishiwaki et al., 2004] and the defect in transcription would prevent skin abnormalities from being expressed. Certain mutations in XPD have been shown to reduce its repair activity while preserving some transcription activity [Winkler et al., 2000; Dubaele et al., 2003]. Some TTD cells have altered temperature sensitivity of transcription [Vermeulen et al., 2001]. Other reports have attempted to explain these differences on the basis of differential repair of 6-4PP and CPD [Eveno et al., 1995; Riou et al., 1999, 2004], or on levels of post-UV expression of p53 [Dumaz et al., 1997]. There is substantial heterogeneity in the level of NER in TTD cells [Lehmann et al., 1988; Nishiwaki et al., 2004]. However, the UV-induced plasmid mutation frequency was similarly elevated in XP and TTD cells [Marionnet et al., 1995]. A defective immune response was proposed to contribute to UV induced carcinogenesis in XP-D cells in one study [Ahrens et al., 1997] but was discounted in a later study [Berneburg et al., 2000].
NER Protein Recruitment and Redistribution is Dependent on XPD, XPB, and TTDA
NER proteins are sequentially recruited to sites of DNA damage in a dynamic process [Volker et al., 2001; Friedberg et al., 2005; Giglia-Mari et al., 2006; Essers et al., 2006]. Our data indicate that the specific XP or TTD mutations in XPD, XPB, or TTDA influence recruitment and redistribution of other NER proteins (Fig. 7). In normal cells the NER proteins are rapidly recruited to localized UV damage at the earliest time studied (0.1 hr) (Figs. 3,4, 5, and 7; Supplementary Fig. S2) in agreement with studies with labeled NER proteins [Giglia-Mari et al., 2006]. These proteins persist for 0.5 hr and are substantially redistributed by 3 hr in normal cells (Figs. 3, 4, 5, and 7; Supplementary Fig. S3). In XP (XP-D) cells, with the exception of XPC, all studied NER proteins (XPD, XPG, XPB, XPA, and XPF) were delayed in their recruitment to localized sites of DNA damage (Figs. 2, 3, and 7; Supplementary Figs. S1 and S2). This pattern was similar in XP (XP-D) cells from patients with (Patients XP17BE, XP29BE, and XP32BE) or without (Patients XP34BE and XPLABE) neurological symptoms (data not shown for all cells). Furthermore, there was a persistence of localization of all NER proteins including XPC up to 24 hr post-UV-irradiation. This similar pattern of NER protein recruitment in cells from XP-D patients with a range of XP phenotypes indicates a fundamental NER defect specific to XP that is not related to the presence or absence of neurological defects. There was similar delayed localization and persistent accumulation of these NER proteins in cells from XP patients with mutations in the XPB gene [Oh et al., 2007].
FIGURE 7.
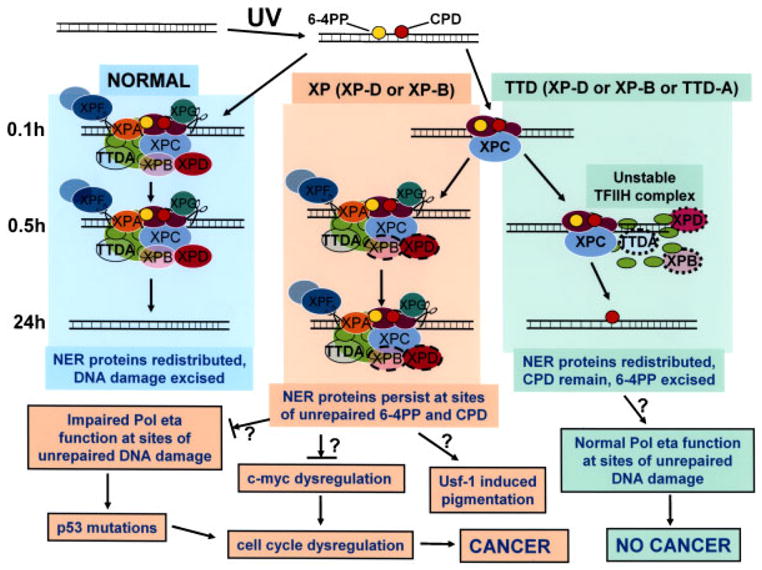
Schematic diagram of repair of UV-induced DNA damage (6-4PP and CPD) in normal, XP (XP-D or XP-B), and TTD (XP-D, XP-B, or TTD-A) cells. DNA is damaged by UV producing 6-4PP (yellow circles) and CPD (red circles). In normal cells (left column) by 0.1hr NER proteins are localized at the site of DNA damage. These proteins remain localized at 0.5 hr but by 24 hr the NER proteins are redistributed and the DNA damage has been excised in the normal cells. In contrast, in XP (XP-D or XP-B) cells (center column) by 0.1hr only XPC is visualized at the site of localized DNA damage. By 0.5 hr the NER proteins, including mutated XPD (dark pink oval with dashed border) and mutated XPB (light pink oval with dashed border), are recruited to the site of localized DNA damage. These proteins persist at 24 hr at sites of unrepaired 6-4PP and CPD. In TTD (XP-D, or XP-B or TTD-A) cells (right column) by 0.1hr only XPC is visualized at the site of localized DNA damage as in the XP cells. By 0.5 hr the NER proteins includingmutated XPD (dark pink oval with dotted border), mutated XPB (light pink oval with dotted border), and mutated TTDA (blue/green oval with dotted border) are recruited to the site of localized DNA damage in only a small proportion of the cells because of the instability of the TFIIH complex. At 24 hr the NER proteins have redistributed, CPD remain and 6-4PP are repaired. The NER proteins include DDB1-DDB2 (XPE) (purple ovals); XPC (light blue oval), which is complexed to HR23b and centrin 2; components of TFIIH: XPD (dark pink oval), XPB (light pink oval), TTDA (blue/green oval), and seven other proteins (light green ovals); and endonucleases XPF (dark gray oval) complexed to ERCC1 (light gray oval) and XPG (dark green oval). In the XP (XP-D or XP-B) cells the persistent NER proteins at the site of unrepaired 6-4PP and CPD may block progression of the translesion polymerase, pol eta, thereby creating a partial mimic of XP variant cells. This impaired pol eta function could result in increased mutagenesis of genes such as p53, which lead to cell cycle dysregulation that my eventually lead to cancer. In addition, mutations in TFIIH components have been shown to result in c-myc dysregulation with resulting cell-cycle dysregulation. The increased pigmentation in XP patients may be related to stimulation of tyrosinase activity via the UV-responsive transcription factor, Usf-1. In contrast, in TTD (XP-D, XP-D, or TTD-A) cells the unstable TFIIH complex is sufficient to repair 6-4PP but NER proteins do not persist at the site of unrepaired CPD. The translesion polymerase, pol eta can function normally to bypass the unrepaired DNA damage and thus does not induce an elevated frequency of cancer. (See discussion for more details.)
In a cell free assay, there is a tight coordination between the “comings and goings” of NER factors that are involved in repair of DNA lesions [Riedl et al., 2003]. Impairment of this coordination by “coming and staying” would be expected to severely impede the normal functioning of this region of DNA. This persistence of NER proteins at sites of unrepaired 6-4PP and CPD may result in disruption of a number of cellular pathways leading to the clinical features of XP (Fig. 7). In normal cells, a translesion DNA polymerase, pol eta, serves to bypass persistent DNA damage [Lehmann et al., 2007]. Patients with the variant form of XP have a defect in pol eta [Broughton et al., 2002; Kannouche and Stary, 2003]. Like other XP patients, the XP variant patients have clinical XP with increased pigmentation and a high frequency of skin cancers [Bootsma et al., 2002; Kraemer et al., 2007]. We hypothesize that the persistent NER proteins at the site of unrepaired DNA damage serves as a block to access of pol eta. Following UV exposure pol eta was reported to relocalize into intranuclear foci in a greater frequency of cells and at a lower UV dose level in NER deficient XP-A cells than in normal cells [Kannouche et al., 2001]. A similar block to RNA polymerase at DNA lesions is thought to serve as a signal for transcription-coupled DNA repair [Friedberg et al., 2005]. This block to pol eta would create the equivalent of an XP variant in cells with defective NER. As in XP variant patients, another (presently unidentified) error-prone polymerase would bypass the lesion, permitting replication at the expense of increased mutagenesis. As a consequence mutations would accumulate in genes such as p53 [Kraemer, 1997]. Alterations in p53 lead to cell-cycle dysregulation, and UV-specific mutations of p53 are often found in cancers of XP patients [Sarasin and Giglia-Mari, 2002]. In addition, failure to release TFIIH may impair its shuttling between NER and transcription [Riedl et al., 2003]. The XP cells we tested have the common p.R683W mutation in the XPD gene, which has been shown to rescue lethality in a yeast assay [Taylor et al., 1997]. XPD cells with the p.R683W mutation have impaired TFIIH [Dubaele et al., 2003]. Defective interaction of the TFIIH/FUSE/FBR/FIR system in c-myc expression can result in dysregulation of the cell cycle, thereby linking transcription abnormalities to cancer induction [Liu et al., 2001, 2006; Weber et al., 2005]. In addition, persistence of NER proteins may lead to impaired chromatin and nucleosome assembly by hindering the CAF-1 localization, which normally leads to incorporation of new H3.1 histones at sites of repaired UV damage. This process occurs at a late stage in the damage response, after repair of UV lesions and its failure may contribute to genetic instability [Green and Almouzni, 2003; Polo et al., 2006]. The pigmentation in XP patients may be stimulated through increased tyrosinase activity induced by the UV responsive transcription factor Usf-1 [Galibert et al., 2001; Corre et al., 2004].
Unstable TFIIH in TTD
The XPD protein, a component of the basal transcription factor, TFIIH, is essential for life [Friedberg et al., 2005] and thus all cells must have a minimal level of functional XPD protein [Lehmann, 2001]. TTD cells have low levels of TFIIH proteins and after UV damage, these NER proteins do not accumulate to the same extent as in the XP cells (Figs. 3, 4, and 7; Supplementary Figs. S1–S3). This may be explained on the basis of instability of the TFIH complex because of ineffective assembly or rapid redistribution. XPD mutations leading to XP or to TTD have been shown to have different effects on the stability of TFIIH [Winkler et al., 2000; Dubaele et al., 2003; Riedl et al., 2003]. The mutations in the XPD protein in TTD cells serve as a signal for lower levels of TFIIH [Winkler et al., 2000; Dubaele et al., 2003; Riedl et al., 2003]. Addition of wild-type XPD protein to TTD cells serves to increase the level of the endogenous TFIIH proteins (Fig. 2C) [Botta et al., 2002], probably by increasing the stability of the TFIIH complex. However, the lower levels of TFIIH proteins in the TTD cells (Fig. 2A) are sufficient to repair most 6-4PP (Figs. 5A and 7) [Eveno et al., 1995; Riou et al., 2004]. Similarly, only a few percent of normal XPC mRNA and protein is required for nearly normal removal of 6-4PP in cells from Patient XP72TMA with a mild form of XP [Khan et al., 2004, 2006]. Like the XP cells, the TTD cells have persistent DNA damage in the form of CPD lesions (Fig. 6B). Normal repair of 6-4PP and reduced repair of CPD is also observed in cells from patients with Cockayne syndrome [Barrett et al., 1991; Parris and Kraemer, 1993], which, like TTD, has defective DNA repair but no increase in cancer. The TTD cells differ from the XP cells in that by 24 hr the NER proteins are redistributed (Figs. 3, 4, 5, and 7; Supplementary Figs. S1, S2, and S3). We hypothesize that this absence of NER protein accumulation permits normal functioning of the translesion polymerase, pol eta, in bypassing of the unrepaired DNA lesions. Thus the TTD patients do not develop cancer (Fig. 7). These observations do not assess transcription-coupled repair of 6-4PP, which can be measured at a reduced rate in TTD cells [Riou et al., 2004] and might also contribute to the cancer-free phenotype of TTD patients.
NER protein localization in XP/TTD complex cells
In XP/TTD complex cells immunocytochemistry and Western blotting for XPD and XPB proteins demonstrated that, like TTD cells, the TFIIH levels were substantially reduced (Fig. 2A). These patients have features of both XP and TTD (Fig. 1; Table 1). In Patient XPTTD306BE, XPA and XPF localization was at a very reduced level throughout the time course as in TTD cells, perhaps attributable to the p.R722W TTD-specific mutation. However, in the cells from Patient XPTTD268BE, recruitment of XPA and XPF was delayed but present by 0.5 hr and persisted to 24 hr, as seen in XP (XP-D) cells (Supplementary Fig. S1). These cells had a p.R112H mutation that has been associated with a cellular phenotype which is indistinguishable from that of XP (XP-D) cells with a p.R683W mutation [Berneburg et al., 2000]. An XP/TTD complex patient (Patient XP38BR) with this p.R112H mutation had more severe XP-like skin involvement including skin cancer [Broughton et al., 2001]. Patient XPTTD268BE had more XP type pigmentary changes (Fig. 1B) than the other XP/TTD complex patient, Patient XPTTD306BE, perhaps reflecting a role of persistent NER proteins as sites of unrepaired DNA damage in stimulating pigmentation (Fig. 7).
Supplementary Material
Acknowledgments
Grant sponsor: Intramural Research Program of the National Institutes of Health; Center for Cancer Research, National Cancer Institute.
We thank Dr. Alan R. Lehmann for generous assistance and advice and Ms. Heather Fawcett for assistance in the UDS assay. Dr. Christine Liang and medical students Andrea Morris and Tara Rao took the photomicrographs of the hairs from TTD patients while working in our laboratory. We thank Drs. David Levens, Larry Loeb, and Roger Woodgate for helpful discussions.
Footnotes
The Supplementary Material referred to in this article can be accessed at http://www.interscience.wiley.com/jpages/1059-7794/suppmat.
References
- Ahrens C, Grewe M, Berneburg M, Grether-Beck S, Quilliet X, Mezzina M, Sarasin A, Lehmann AR, Arlett CF, Krutmann J. Photocarcinogenesis and inhibition of intercellular adhesion molecule 1 expression in cells of DNA-repair-defective individuals. Proc Natl Acad Sci USA. 1997;94:6837–6841. doi: 10.1073/pnas.94.13.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews AD, Barrett SF, Robbins JH. Xeroderma pigmentosum neurological abnormalities correlate with colony-forming ability after ultraviolet radiation. Proc Natl Acad Sci USA. 1978;75:1984–1988. doi: 10.1073/pnas.75.4.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SF, Robbins JH, Tarone RE, Kraemer KH. Evidence for defective repair of cyclobutane pyrimidine dimers with normal repair of other DNA photoproducts in a transcriptionally active gene transfected into Cockayne syndrome cells. Mutat Res. 1991;255:281–291. doi: 10.1016/0921-8777(91)90032-k. [DOI] [PubMed] [Google Scholar]
- Berneburg M, Clingen PH, Harcourt SA, Lowe JE, Taylor EM, Green MH, Krutmann J, Arlett CF, Lehmann AR. The cancer-free phenotype in trichothiodystrophy is unrelated to its repair defect. Cancer Res. 2000;60:431–438. [PubMed] [Google Scholar]
- Bootsma D, Hoeijmakers JH. DNA repair. Engagement with transcription. Nature. 1993;363:114–115. doi: 10.1038/363114a0. [DOI] [PubMed] [Google Scholar]
- Bootsma D, Kraemer KH, Cleaver JE, Hoeijmakers JHJ. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In: Vogelstein B, Kinzler KW, editors. The Genetic Basis of Human Cancer. 2. New York: McGraw- Hill; 2002. pp. 211–237. [Google Scholar]
- Botta E, Nardo T, Lehmann AR, Egly JM, Pedrini AM, Stefanini M. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum Mol Genet. 2002;11:2919–2928. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- Broughton BC, Lehmann AR, Harcourt SA, Arlett CF, Sarasin A, Kleijer WJ, Beemer FA, Nairn R, Mitchell DL. Relationship between pyrimidine dimers, 6-4 photoproducts, repair synthesis and cell survival: studies using cells from patients with trichothiodystrophy. Mutat Res. 1990;235:33–40. doi: 10.1016/0921-8777(90)90020-6. [DOI] [PubMed] [Google Scholar]
- Broughton BC, Steingrimsdottir H, Weber CA, Lehmann AR. Mutations in the xeroderma pigmentosum group D DNA repair/ transcription gene in patients with trichothiodystrophy. Nat Genet. 1994;7:189–194. doi: 10.1038/ng0694-189. [DOI] [PubMed] [Google Scholar]
- Broughton BC, Berneburg M, Fawcett H, Taylor EM, Arlett CF, Nardo T, Stefanini M, Menefee E, Price VH, Queille S, Sarasin A, Bohnert E, Krutmann J, Davidson R, Kraemer KH, Lehmann AR. Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene. Hum Mol Genet. 2001;10:2539–2547. doi: 10.1093/hmg/10.22.2539. [DOI] [PubMed] [Google Scholar]
- Broughton BC, Cordonnier A, Kleijer WJ, Jaspers NG, Fawcett H, Raams A, Garritsen VH, Stary A, Avril MF, Boudsocq F, Masutani C, Hanaoka F, Fuchs RP, Sarasin A, Lehmann AR. Molecular analysis of mutations in DNA polymerase eta in xeroderma pigmentosum-variant patients. Proc Natl Acad Sci USA. 2002;99:815–820. doi: 10.1073/pnas.022473899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre S, Primot A, Sviderskaya E, Bennett DC, Vaulont S, Goding CR, Galibert MD. UV-induced expression of key component of the tanning process, the POMC and MC1R genes, is dependent on the p-38-activated upstream stimulating factor-1 (USF-1) J Biol Chem. 2004;279:51226–51233. doi: 10.1074/jbc.M409768200. [DOI] [PubMed] [Google Scholar]
- De Boer J, De Wit J, Van Steeg H, Berg RJ, Morreau H, Visser P, Lehmann AR, Duran M, Hoeijmakers JH, Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- Dubaele S, Proietti dS, Bienstock RJ, Keriel A, Stefanini M, Van Houten B, Egly JM. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- Dumaz N, Duthu A, Ehrhart JC, Drougard C, Appella E, Anderson CW, May P, Sarasin A, Daya-Grosjean L. Prolonged p53 protein accumulation in trichothiodystrophy fibroblasts dependent on unrepaired pyrimidine dimers on the transcribed strands of cellular genes. Mol Carcinog. 1997;20:340–347. [PubMed] [Google Scholar]
- Essers J, Vermeulen W, Houtsmuller AB. DNA damage repair: anytime, anywhere? Curr Opin Cell Biol. 2006;18:240–246. doi: 10.1016/j.ceb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Eveno E, Bourre F, Quilliet X, Chevallier-Lagente O, Roza L, Eker AP, Kleijer WJ, Nikaido O, Stefanini M, Hoeijmakers JH. Different removal of ultraviolet photoproducts in genetically related xeroderma pigmentosum and trichothiodystrophy diseases. Cancer Res. 1995;55:4325–4332. [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. Washington, DC: ASM Press; 2006. p. 1164. [Google Scholar]
- Galibert MD, Carreira S, Goding CR. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UVinduced tyrosinase expression. EMBO J. 2001;20:5022–5031. doi: 10.1093/emboj/20.17.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NG, Raams A, Argentini M, Van der Spek PJ, Botta E, Stefanini M, Egly JM, Aebersold R, Hoeijmakers JH, Vermeulen W. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Miquel C, Theil AF, Mari PO, Hoogstraten D, Ng JM, Dinant C, Hoeijmakers JH, Vermeulen W. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol. 2006;4:e156. doi: 10.1371/journal.pbio.0040156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gözükara EM, Parris CN, Weber CA, Salazar EP, Seidman MM, Watkins JF, Prakash L, Kraemer KH. The human DNA repair gene, ERCC2 (XPD), corrects ultraviolet hypersensitivity and ultraviolet hypermutability of a shuttle vector replicated in xeroderma pigmentosum group D cells. Cancer Res. 1994;54:3837–3844. [PubMed] [Google Scholar]
- Graham JM, Jr, Anyane-Yeboa K, Raams A, Appeldoorn E, Kleijer WJ, Garritsen VH, Busch D, Edersheim TG, Jaspers NG. Cerebrooculo- facio-skeletal syndrome with a nucleotide excision-repair defect and a mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am J Hum Genet. 2001;69:291–300. doi: 10.1086/321295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto K, Kobayashi N, Katsumi S, Nishiwaki Y, Iwamoto TA, Yamamoto A, Yamashina Y, Shirai T, Miyagawa S, Dohi Y, Sugiura S, Mori T. The total amount of DNA damage determines ultraviolet-radiationinduced cytotoxicity after uniform or localized irradiation of human cells. J Invest Dermatol. 2002;119:1177–1182. doi: 10.1046/j.1523-1747.2002.19514.x. [DOI] [PubMed] [Google Scholar]
- Itin PH, Sarasin A, Pittelkow MR. Trichothiodystrophy: update on the sulfur-deficient brittle hair syndromes. J Am Acad Dermatol. 2001;44:891–920. doi: 10.1067/mjd.2001.114294. [DOI] [PubMed] [Google Scholar]
- Jaspers NG, Bootsma D. Genetic heterogeneity in ataxiatelangiectasia studied by cell fusion. Proc Natl Acad Sci USA. 1982;79:2641–2644. doi: 10.1073/pnas.79.8.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Stary A. Xeroderma pigmentosum variant and errorprone DNA polymerases. Biochimie. 2003;85:1123–1132. doi: 10.1016/j.biochi.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Khan SG, Metin A, Gozukara E, Inui H, Shahlavi T, Muniz-Medina V, Baker CC, Ueda T, Aiken JR, Schneider TD, Kraemer KH. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum Mol Genet. 2004;13:343–352. doi: 10.1093/hmg/ddh026. [DOI] [PubMed] [Google Scholar]
- Khan SG, Boyle J, Imoto K, Oh KS, Armstrong N, Tamura D, DiGiovanna JJ, Kraemer KH. Differential expression of XPC protein and localization at UV damage is associated with severe or mild clinical phenotypes in xeroderma pigmentosum group C patients. J Invest Dermatol. 2006;126:79. (abstract) [Google Scholar]
- Kobayashi N, Katsumi S, Imoto K, Nakagawa A, Miyagawa S, Furumura M, Mori T. Quantitation and visualization of ultraviolet-induced DNA damage using specific antibodies: application to pigment cell biology. Pigment Cell Res. 2001;14:94–102. doi: 10.1034/j.1600-0749.2001.140204.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Uchiyama M, Fukuro S, Tanaka K. Mutations in the XPD gene in xeroderma pigmentosum group D cell strains: confirmation of genotype-phenotype correlation. Am J Med Genet. 2002;110:248–252. doi: 10.1002/ajmg.10465. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Coon HG, Petinga RA, Barrett SF, Rahe AE, Robbins JH. Genetic heterogeneity in xeroderma pigmentosum: complementation groups and their relationship to DNA repair rates. Proc Natl Acad Sci USA. 1975;72:59–63. doi: 10.1073/pnas.72.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Scotto J. DNA repair protects against cutaneous and internal neoplasia: evidence from xeroderma pigmentosum. Carcinogenesis. 1984;5:511–514. doi: 10.1093/carcin/5.4.511. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- Kraemer KH. Sunlight and skin cancer: another link revealed. Proc Natl Acad Sci USA. 1997;94:11–14. doi: 10.1073/pnas.94.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Stevens S. A rapid procedure for measurement of DNA repair in human fibroblasts and for complementation analysis of xeroderma pigmentosum cells. Mutat Res. 1980;69:177–190. doi: 10.1016/0027-5107(80)90187-6. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Arlett CF, Broughton BC, Harcourt SA, Steingrimsdottir H, Stefanini M, Malcolm A, Taylor R, Natarajan AT, Green S. Trichothiodystrophy, a human DNA repair disorder with heterogeneity in the cellular response to ultraviolet light. Cancer Res. 1988;48:6090–6096. [PubMed] [Google Scholar]
- Lehmann AR. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Liang C, Kraemer KH, Morris A, Schiffmann R, Price VH, Menefee E, DiGiovanna JJ. Characterization of tiger tail banding and hair shaft abnormalities in trichothiodystrophy. J Am Acad Dermatol. 2005;52:224–232. doi: 10.1016/j.jaad.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC, Reinberg D, Levens D. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–363. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- Liu J, Kouzine F, Nie Z, Chung HJ, Elisha-Feil Z, Weber A, Zhao K, Levens D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006;25:2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marionnet C, Benoit A, Benhamou S, Sarasin A, Stary A. Characteristics of UV-induced mutation spectra in human XP-D/ERCC2 gene-mutated xeroderma pigmentosum and trichothiodystrophy cells. J Mol Biol. 1995;252:550–562. doi: 10.1006/jmbi.1995.0519. [DOI] [PubMed] [Google Scholar]
- Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Kobayashi N, Muramatsu T, Yamashina Y, Shirai T, Hashimoto MW, Ikenaga M, Mori T. Three-dimensional visualization of ultraviolet-induced DNA damage and its repair in human cell nuclei. J Invest Dermatol. 1998;110:143–148. doi: 10.1046/j.1523-1747.1998.00100.x. [DOI] [PubMed] [Google Scholar]
- Nishiwaki Y, Kobayashi N, Imoto K, Iwamoto TA, Yamamoto A, Katsumi S, Shirai T, Sugiura S, Nakamura Y, Sarasin A, Miyagawa S, Mori T. Trichothiodystrophy fibroblasts are deficient in the repair of ultraviolet-induced cyclobutane pyrimidine dimers and (6-4)photoproducts. J Invest Dermatol. 2004;122:526–532. doi: 10.1046/j.0022-202X.2004.22226.x. [DOI] [PubMed] [Google Scholar]
- Oh KS, Khan SG, Jaspers NG, Raams A, Ueda T, Lehmann A, Friedmann PS, Emmert S, Gratchev A, Lachlan K, Lucassan A, Baker CC, Kraemer KH. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- Oh KS, Imoto K, Boyle J, Khan SG, Kraemer KH. Influence of XPB helicase on recruitment and redistribution of nucleotide excision repair proteins at sites of UV-induced DNA damage. DNA Repair (Amst) 2007;6:1359–1370. doi: 10.1016/j.dnarep.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parris CN, Kraemer KH. Ultraviolet-induced mutations in Cockayne syndrome cells are primarily caused by cyclobutane dimer photoproducts while repair of other photoproducts is normal. Proc Natl Acad Sci USA. 1993;90:7260–7264. doi: 10.1073/pnas.90.15.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Protic-Sabljic M, Seetharam S, Seidman MM, Kraemer KH. An SV40-transformed xeroderma pigmentosum group D cell line: establishment, ultraviolet sensitivity, transfection efficiency and plasmid mutation induction. Mutat Res. 1986;166:287–294. doi: 10.1016/0167-8817(86)90028-3. [DOI] [PubMed] [Google Scholar]
- Rapin I, Lindenbaum Y, Dickson DW, Kraemer KH, Robbins JH. Cockayne syndrome and xeroderma pigmentosum. Neurology. 2000;55:1442–1449. doi: 10.1212/wnl.55.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou L, Zeng L, Chevallier-Lagente O, Stary A, Nikaido O, Taieb A, Weeda G, Mezzina M, Sarasin A. The relative expression of mutated XPB genes results in xeroderma pigmentosum/Cockayne’s syndrome or trichothiodystrophy cellular phenotypes. Hum Mol Genet. 1999;8:1125–1133. doi: 10.1093/hmg/8.6.1125. [DOI] [PubMed] [Google Scholar]
- Riou L, Eveno E, van Hoffen A, van Zeeland AA, Sarasin A, Mullenders LH. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 2004;64:889–894. doi: 10.1158/0008-5472.can-03-2070. [DOI] [PubMed] [Google Scholar]
- Robbins JH, Kraemer KH, Lutzner MA, Festoff BW, Coon HG. Xeroderma pigmentosum. An inherited disease with sun sensitivity, multiple cutaneous neoplasms, and abnormal DNA repair. Ann Intern Med. 1974;80:221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- Sarasin A, Blanchet-Bardon C, Renault G, Lehmann A, Arlett C, Dumez Y. Prenatal diagnosis in a subset of trichothiodystrophy patients defective in DNA repair. Br J Dermatol. 1992;127:485–491. doi: 10.1111/j.1365-2133.1992.tb14845.x. [DOI] [PubMed] [Google Scholar]
- Sarasin A, Giglia-Mari G. p53 gene mutations in human skin cancers. Exp Dermatol. 2002;11(Suppl 1):44–47. doi: 10.1034/j.1600-0625.11.s.1.11.x. [DOI] [PubMed] [Google Scholar]
- Satoh MS, Hanawalt PC. Competent transcription initiation by RNA polymerase II in cell-free extracts from xeroderma pigmentosum groups B and D in an optimized RNA transcription assay. Biochim Biophys Acta Gene Struct Expression. 1997;1354:241–251. doi: 10.1016/s0167-4781(97)00102-4. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers JH, Chambon P, Egly JM. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers JHJ, Egly JM. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanini M, Lagomarsini P, Arlett CF, Marinoni S, Borrone C, Crovato F, Trevisan G, Cordone G, Nuzzo F. Xeroderma pigmentosum (complementation group D) mutation is present in patients affected by trichothiodystrophy with photosensitivity. Hum Genet. 1986;74:107–112. doi: 10.1007/BF00282072. [DOI] [PubMed] [Google Scholar]
- Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature. 1993;365:852–855. doi: 10.1038/365852a0. [DOI] [PubMed] [Google Scholar]
- Takayama K, Salazar EP, Lehmann A, Stefanini M, Thompson LH, Weber CA. Defects in the DNA repair and transcription gene ERCC2 in the cancer-prone disorder xeroderma pigmentosum group D. Cancer Res. 1995;55:5656–5663. [PubMed] [Google Scholar]
- Takayama K, Salazar EP, Broughton BC, Lehmann AR, Sarasin A, Thompson LH, Weber CA. Defects in the DNA repair and transcription gene ERCC2(XPD) in trichothiodystrophy. Am J Hum Genet. 1996;58:263–270. [PMC free article] [PubMed] [Google Scholar]
- Taylor EM, Broughton BC, Botta E, Stefanini M, Sarasin A, Jaspers NG, Fawcett H, Harcourt SA, Arlett CF, Lehmann AR. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc Natl Acad Sci USA. 1997;94:8658–8663. doi: 10.1073/pnas.94.16.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen W, Van Vuuren AJ, Chipoulet M, Schaeffer L, Appeldoorn E, Weeda G, Jaspers NG, Priestley A, Arlett CF, Lehmann AR. Three unusual repair deficiencies associated with transcription factor BTF2(TFIIH): evidence for the existence of a transcription syndrome. Cold Spring Harb Symp Quant Biol. 1994;59:317–329. doi: 10.1101/sqb.1994.059.01.036. [DOI] [PubMed] [Google Scholar]
- Vermeulen W, Bergmann E, Auriol J, Rademakers S, Frit P, Appeldoorn E, Hoeijmakers JH, Egly JM. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat Genet. 2000;26:307–313. doi: 10.1038/81603. [DOI] [PubMed] [Google Scholar]
- Vermeulen W, Rademakers S, Jaspers NG, Appeldoorn E, Raams A, Klein B, Kleijer WJ, Hansen LK, Hoeijmakers JH. A temperaturesensitive disorder in basal transcription and DNA repair in humans. Nat Genet. 2001;27:299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- Weber CA, Salazar EP, Stewart SA, Thompson LH. ERCC2: cDNA cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3. EMBO J. 1990;9:1437–1447. doi: 10.1002/j.1460-2075.1990.tb08260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Liu J, Collins I, Levens D. TFIIH operates through an expanded proximal promoter to fine-tune c-myc expression. Mol Cell Biol. 2005;25:147–161. doi: 10.1128/MCB.25.1.147-161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G, Eveno E, Donker I, Vermeulen W, Chevallier-Lagente O, Taieb A, Stary A, Hoeijmakers JH, Mezzina M, Sarasin A. A mutation in the XPB/ERCC3 DNA repair transcription gene, associated with trichothiodystrophy. Am J Hum Genet. 1997;60:320–329. [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT, Weeda G. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.