Abstract
Purpose
There is increasing evidence that altered microRNA expression is associated with tumor progression and survival in cancer patients. We tested if the expression of specific microRNAs was associated with prognosis and disease progression in early stage lung adenocarcinoma.
Experimental Design
The expression of miR-21, miR-17 and miR-155 was measured by quantitative RT-PCR in tissues from 317 non small cell lung cancer (NSCLC) patients that originated from Maryland, Norway and Japan. Kaplan Meier and Cox regression analysis evaluated associations of microRNA expression with cancer-specific mortality and disease free survival.
Results
Elevated miR-21 (hazard ratio [HR] 2.06, 1.13–3.75), miR-17 (HR 2.00, 1.10–3.61), miR-155 (HR 2.37, 1.27–4.42) was associated with worse cancer-specific mortality in the Maryland cohort. These were evaluated in two additional cohorts and only miR-21 was associated with worse cancer-specific mortality in the Norwegian cohort (HR 2.78, 1.22–6.31) and worse relapse free survival in the Japanese cohort (HR 2.82, 1.57–5.07). More advanced stage tumors expressed significantly higher levels of miR-21 compared to TNM stage I tumors. TNM stage I patients were evaluated separately and high levels of miR-21 was associated with worse cancer-specific mortality (HR 2.16, 1.11–4.21) and relapse-free survival (3.40, 1.57–7.36) independent of other clinical factors.
Conclusions and Summary
This is the first study to report that increased miR-21 expression is associated with disease progression and survival in stage I lung cancer. This suggests that expression of miR-21 may contribute to lung carcinogenesis and serve as a therapeutic target or early stage prognostic biomarker for lung adenocarcinoma.
Introduction
Lung cancer is globally responsible for 1.4 million deaths annually and is the leading cause of cancer mortality (1). Once diagnosed, survival rates are low. Although the 5-year survival rate has slightly improved over the last 3 decades, that of early-stage patients is still worse than colon, gastric and breast cancer patients. Even when diagnosed early, stage I lung cancer patients have only a 60–70% five-year survival rate (2–4). Successful surgical resection remains the only curative treatment for lung cancer, highlighting the need for novel diagnostic and therapeutic strategies. Identifying factors that are associated with aggressive disease may lead to the development of novel biomarkers and identification of therapeutic targets that can help reduce the burden of this disease. MicroRNAs (miRNAs) have potential as both biomarkers and therapeutic targets for lung cancer (5–7).
MiRNAs are small noncoding RNAs that posttranscriptionally regulate the translation of target genes. They influence a variety of cellular functions, including proliferation, differentiation, and apoptosis (8). Increasing data suggests that they have a causal role in tumorigenesis (9, 10). Specific miRNAs can behave as either tumor suppressor genes or oncogenes depending on the cellular environment. MiRNA expression is systematically altered in every malignancy that has been examined (11, 12). Alteration of individual miRNAs can affect tumor growth in vivo demonstrating the potential for miRNAs to have a strong role in tumor promotion. Polymorphisms in miRNAs or miRNA binding sites may modify ones risk of developing cancer (13). The expression of miRNAs are associated with poor survival and therapeutic outcome in different cancers (5, 14–18), including lung cancer (19–24), demonstrating the potential for miRNA expression patterns to be used as prognostic biomarkers that may aid in therapeutic decisions for cancer. These associations should be validated in additional populations to assess their potential as biomarkers.
We have previously analyzed microRNA microarray data of lung cancer specimens from non small cell lung cancer (NSCLC) adenocarcinoma patients. Based on the microarray results of that study, we identified 5 microRNAs (miR-155, miR-17, miR-21, miR-145 and let-7a) whose expression were significantly altered in lung cancer and were associated with cancer-specific mortality (14). Three of these microRNAs (miR-17, miR-21 and miR-155) were potential oncogenic microRNAs showing increased expression in tumors with high expression levels being associated with poor prognosis. Based on these results, we selected miR-17, miR-21 and miR-155 to validate in three additional cohorts of NSCLC adenocarcinoma patients. Here we report the findings of the associations of the expression of miR-17, miR-21 and miR-155 with cancer-specific mortality and disease-free survival in three independent cohorts of NSCLC adenocarcinoma patients.
Methods
Patients and Tissue samples
Primary lung tumors and adjacent noncancerous tissues were procured from patients undergoing surgical resections. All tissues were snap-frozen immediately after surgery and stored at −80°C until use. Detailed background information for each tissue donor, including age, gender, clinical staging, and smoking status was collected. Tumor histopathology was classified according to the World Health Organization Classification of Tumor system. For this study, only patients with the diagnosis of non-small lung cancer and with adenocarcinoma histology were used. All others were excluded prior to beginning the study.
The Maryland cohort consisted of 89 patients recruited from hospitals in the Metropolitan Baltimore area between 1987 and 2009. In this cohort, 13 cases were overlapping with our previous study (14). For the Norwegian cohort, 37 patients were recruited from Haukeland University Hospital, Bergen, Norway from 1988 to 2003. These patients were recruited consecutively at the time of surgery, whenever practically feasible. Patients from the Japanese cohort (n=191) were selected from a larger lung cancer population of 393 stage I–II patients recruited from National Cancer Center Hospital, Tokyo, Japan from 1998 to 2008. The selection criteria for the Japanese cohort required all cases to have undergone potentially curative radical surgery without any evidence of tumor cells present in the resection margins or mediastinal lymph node involvement. These patients did not receive neoadjuvant therapy and they could not have been diagnosed with cancer in the 5 years before lung cancer diagnosis. To maximize the statistical efficiency to identify factors associated with cancer relapse, all stage I patients with cancer relapse (n=30) and stage II (n=52) patients for which RNA was available were selected. Incidence density sampling (25) selected the 109 non-relapsed stage I patients from the larger cohort. There was no selection based on cancer-specific mortality for the Maryland or Norway cohorts and all cases with lung adenocarcinoma were included. The differences in selection criteria of the Japanese cohort, at least in part, contributes to the improved prognosis of this cohort compared to the American and Norwegian cohorts.
Survival time for the Maryland cohort was determined with a combination of searching the National Death Index (www.cdc.gov/nchs/ndi.htm) through 12/31/2006 and searching the Social Security death Index through 8/17/2010 for those still alive after according to the National Death Index. The Norwegian cohort was followed through 12/31/2007 and the Japanese cohort was followed through 6/1/2010. Written informed consent was obtained from all participants. This study was approved by the Institutional Review Board (IRB) for the National Institutes of Health, Regional Committees for Medical and Health Research Ethics in Norway and for National Cancer Center at Japan.
RNA isolation and miRNA measurement
Total RNA was extracted from grossly dissected, snap-frozen tissue samples using TRIZOL (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. abundance and integrity of 18S and 28S ribosomal bands were assessed via the Bioanalyser 2100 system (Agilent Technologies, Santa Clara, CA) and this was used as quality control for RNA samples. RNA was isolated in the country of origin and was shipped to the Laboratory of Human Carcinogenesis (USA) for data collection while blinded to clinical outcomes. Quantitative reverse transcriptase-PCR (qRTPCR) of mature microRNA was performed using Taqman MicroRNA assays (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions with the 7900 HT Fast Real-Time PCR System (Applied Biosystems). All assays were performed in triplicate. All Taqman probes were purchased from Applied Biosystems; hsa-miR-17 (Assay ID 002308), hsa-miR-21 (ID 000397), hsa-miR-155 (ID 002623) with RNU66 (ID 001002) as a normalization control.
Statistical analysis
Relative expression quantitation of microRNA was calculated with RQ manager 1.2 (Applied Biosystems). Patients were dichotomized into high or low expression groups based on the median expression value for each microRNA. Univariate and multivariate Cox regression was used to evaluate the associations between clinical covariates and cancer-specific mortality (Maryland and Norwegian cohorts) and relapse-free survival (Japanese cohort) in Stata 9.2 (StataCorp LP, College station, TX). For all analyses, age was treated as a categorical variable as greater than or less than 65. Smoking status was evaluated with two variables. First, smoking was dichotomized as never smokers (0 pack years) and ever smokers (> 0 pack years). Second, smoking was dichotomized as > 20 pack years and < 20 pack years. TNM staging was categorized as stage I versus stage II/III. For final multivariable Cox regression models, all variables were included that were moderately associated (p < 0.10) with cancer-specific mortality or relapse-free survival in at least one cohort in any of the analyses. Expression graphs were used to analyze differences in microRNA expression between cancer tissues and adjacent noncancerous tissues for all qRT-PCR data using Graphpad Prism v5.0 (Graphpad Software Inc, San Diego, CA.) Kaplan-Meier analysis was performed using Graphpad Prism v5.0.
Results
We examined the expression of miR-21, miR-17 and miR-155 in three independent cohorts of NSCLC adenocarcinoma patients. These cohorts consist of 89 cases recruited from Maryland, 37 cases recruited from Norway, and 191 cases recruited from Japan (Table 1). The 5-year survival rate was 45% for the Maryland cohort, 37% for the Norway cohort, and 85% for the Japan cohort. These variations were largely due to differences in TNM staging and selection criteria. In addition to the obvious racial, geographic, and cultural differences among these cohorts, smoking history also differed.
Table 1.
Characteristics of study populations of patients with non small cell lung adenocarcinoma
| Maryland Cohort (n=89) | Norway Cohort (n=37) | Japan Cohort (n=191) | p-value1 | |
|---|---|---|---|---|
| Age-years | p<0.0005 | |||
| Mean (SD) | 63.6 (10.5) | 64.4 (11.7) | 59.6 (7.7) | |
| Range | 32–90 | 37–82 | 30–76 | |
| Gender-no. (%) | p=0.742 | |||
| Male | 46 (52) | 20 (54) | 92 (48) | |
| Female | 43 (48) | 17 (46) | 99 (52) | |
| Race-no. (%) | p<0.0005 | |||
| Caucasian | 58 (65) | 37 (100) | 0 | |
| African-American | 31 (35) | 0 | 0 | |
| Asian | 0 | 0 | 191 (100) | |
| TNM Stage-no. (%) | p<0.0005 | |||
| I | 57 (64) | 21 (57) | 142 (74) | |
| II | 22 (25) | 5 (14) | 49 (26) | |
| III | 10 (11) | 11 (29) | 0 (0) | |
| Smoking (pack year)-no. (%) | p<0.0005 | |||
| never | 7 (8) | 3 (8) | 95 (50) | |
| <20 pack years | 12 (13) | 12 (32) | 30 (16) | |
| ≥20 pack years | 66 (74) | 19 (52) | 66 (34) | |
| unknown | 4 (5) | 3 (8) | 0 (0) |
p-value from one-way ANOVA or Fisher’s exact test, where appropriate
Cox regression analysis evaluated the association between the expression of miR-21, miR-17 and miR-155 and prognosis. Cancer-specific mortality was used as an endpoint for the Maryland and Norway cohorts while relapse-free was used for the Japan cohort based on data availability. High tumor expression (based on median) of miR-21 (hazard ratio [HR] = 2.06, 95% confidence interval [CI] = 1.13–3.75, p = 0.018), miR-17 (HR = 2.00, 95% CI = 1.10–3.61, p = 0.022) and miR-155 (HR = 2.37, 95% CI = 1.27–4.42, p = 0.006) were significantly associated with cancer-specific mortality in the Maryland cohort (Table 2). MiR-21, miR-17 and miR-155 were then evaluated in the Norway and Japan cohorts while blinded to all clinical characteristics. High miR-21 was associated with cancer specific-mortality (HR = 2.78, 95% CI = 1.22–6.31, p = 0.014) in the Norway cohort and relapse-free survival (HR = 2.82, 95% CI = 1.57–5.07, p < 0.0005) in the Japan cohort (Table 2). These associations were significant in each cohort providing strong evidence that miR-21 expression may be a useful prognostic biomarker for lung adenocarcinoma. The expression of miR-17 and miR-155 were not significantly associated with prognosis in either the Norway or Japan cohort.
Table 2.
Univariate Cox Regression analysis of miRNAs expression levels and survival in cancerous tissue.
| microRNA (high vs low)a | Maryland cohort (n=89) (Cancer-Specific Mortality)
|
Norway cohort (n=37) (Cancer-Specific Mortality)
|
Japan cohort (n=191) (Relapse Free Survival)
|
|||
|---|---|---|---|---|---|---|
| HR (95% Cl) | P value | HR (95% Cl) | P value | HR (95% Cl) | P value | |
| miR-17 | 2.00 (1.10–3.61) | 0.022 | 1.23 (0.56–2.70) | 0.603 | 1.37 (0.80–2.37) | 0.245 |
| miR-21 | 2.06 (1.13–3.75) | 0.018 | 2.78 (1.22–6.31) | 0.014 | 2.82 (1.57–5.07) | <0.0005 |
| miR-155 | 2.37 (1.27–4.42) | 0.006 | 1.60 (0.73–3.52) | 0.245 | 1.33 (0.77–2.29) | 0.309 |
Abbreviations: HR, hazard ratio; CI, confidence interval
High expression in cancerous tissue for all miRNAs was defined based on the median. MicroRNA expression was measured with qRT-PCR.
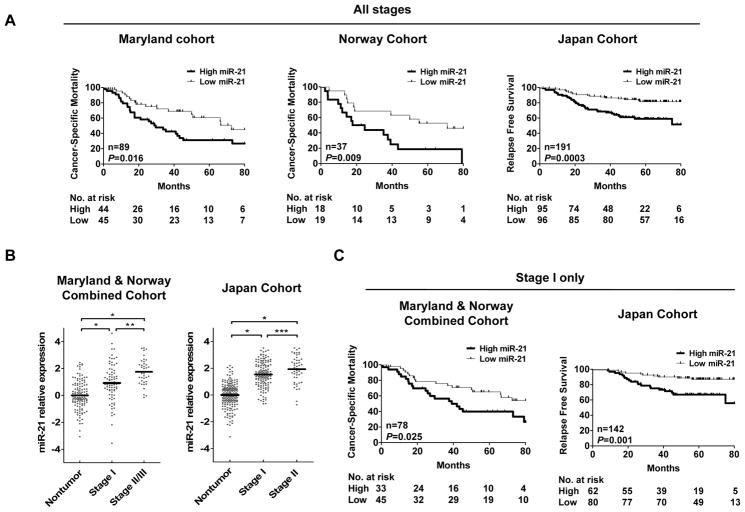
Kaplan-Meier analysis further demonstrated that patients with elevated miR-21 expression had significantly worse cancer-specific mortality in the Maryland (p = 0.016) and Norway cohort (p = 0.009) and worse relapse-free survival in the Japan cohort (p = 0.0003) (Figure 1A). We also analyzed the expression of miR-21 comparing tumors with adjacent noncancerous tissue. MiR-21 expression was consistently elevated in tumors from the Maryland cohort (2.11-fold higher, p < 0.001, Wilcoxon matched pairs test), Norway cohort (2.78-fold higher, p < 0.001) and Japan cohort (3.11-fold, p < 0.001) consistent with an oncogenic role for this miRNA.
Figure 1.
The expression of miR-21 is associated with survival in the Maryland, Norway and Japan cohorts. (A) Kaplan-Meier survival analysis of all stage cases in the Maryland, Norway and Japan cohort stratified by median miR-21 tumor expression. (B) Expression differences of miR-21 between tumor and nontumor in the Maryland and Norway combined cohort and Japan cohort. Dot plots represent miR-21 relative threshold cycle values from qRT-PCR and expression levels in tumor were normalized to nontumor. Relative threshold cycle values greater than 0 in tumor indicate higher expression than that of mean in nontumor log2 scale. Horizontal bars indicate mean expression value. *P < 0.0001, **P = 0.001, ***P = 0.009. Wilcoxon matched-pairs test. (C) Kaplan-Meier survival analysis of TNM stage I cases in the Maryland and Norway combined cohort and Japan cohort stratified by median miR-21 tumor expression.
The Maryland and Norway cohorts were similar in 5-year survival rates, TNM staging, gender and age at diagnosis. To increase the statistical power for all further analyses, the Maryland and Norway cohorts were combined. MiR-21 expression was significantly elevated in more advanced-stage tumors consistent with a role for miR-21 in the progression of cancer (Figure 1B). Multivariate Cox regression demonstrated that miR-21 expression was associated with poor prognosis in the Maryland/Norway and Japan cohorts independent of staging (Table 3) supporting that miR-21 may be a useful prognostic biomarker for lung cancer.
Table 3.
Univariate and Multivariate Cox Regression analysis of miR-21 expression levels and other clinical covariates in three cohorts on all cases.
| Univariate Analysis
| ||||
|---|---|---|---|---|
| Combined Maryland/Norway cohort (n=126) (Cancer-Specific Mortality)
|
Japan cohort (n=191) (Relapse Free Survival)
|
|||
| HR(95% Cl) | P value | HR(95% Cl) | P value | |
| miR-21 (High vs Low)a | 2.31 (1.42–3.76) | 0.001 | 2.82 (1.57–5.07) | <0.0005 |
| Stage (II, III vs I)b | 2.35 (1.46–3.80) | <0.0005 | 3.17 (1.82–5.52) | <0.0005 |
| Sex (M vs F) | 1.09 (0.68–1.73) | 0.734 | 1.29 (0.75–2.32) | 0.351 |
| Age (≥65 vs <65) | 1.42 (0.89–2.28) | 0.142 | 1.58 (0.91–2.77) | 0.106 |
| Ever smoke (ever vs never)c | 0.94 (0.48–1.83) | 0.847 | 1.34 (0.78–2.32) | 0.287 |
| Packyears (≥20 vs <20) | 1.53 (0.85–2.78) | 0.155 | 1.60 (0.92–2.79) | 0.098 |
|
| ||||
|
Multivariate Analysis, final modeld
| ||||
|
Combined Maryland/Norway cohort (n=126) (Cancer-Specific Mortality)
|
Japan cohort (n=191) (Relapse Free Survival)
|
|||
| HR(95% Cl) | P value | HR(95% Cl) | P value | |
|
| ||||
| miR-21 (High vs Low)a | 2.25 (1.32–3.82) | 0.003 | 2.66 (1.47–4.83) | 0.001 |
| Stage (II, III vs I)b | 2.37 (1.42–3.95) | 0.001 | 2.78 (1.57–4.94) | <0.0005 |
| Age (≥65 vs <65) | 1.32 (0.80–2.19) | 0.270 | 1.88 (1.06–3.32) | 0.030 |
| Packyears (≥20 vs <20) | 2.00 (1.07–3.72) | 0.028 | 1.50 (0.85–2.65) | 0.160 |
Abbreviations: HR, hazard ratio; CI, confidence interval.
High expression in cancerous tissue for all miR-21 was defined based on the median.
There are no TNM stage III patients in the Japan cohort therefore this comparison is Stage II vs. I for that cohort.
Never smoker defined as 0 pack years.
Final models included all variables that were moderately associated with survival (p<0.10) in at least one cohort in the univariate analysis.
Metastases are not clinically detectable in TNM stage I lung cancer patients, yet many patients have undetectable micro-metastases that will progress. Prognostic biomarkers for TNM stage I patients may help identify those at high risk of disease progression and provide an opportunity to intervene at an earlier time. Validated prognostic biomarkers may also influence future revisions of tumor staging system.
We evaluated the potential of miR-21 expression as an early stage prognostic biomarker using a stratified analysis on TNM stage I tumors. Based on Kaplan Meier analysis, high miR-21 expression was associated with poor cancer-specific mortality for stage I cases in the Maryland/Norway cohort (P = 0.025, Kaplan-Meier log-rank test) and with worse relapse-free survival in the Japan cohort (P = 0.001) (Figure 1C). These associations were significant and independent of all clinical covariates in multivariate models (Table 4) demonstrating the potential for miR-21 as an early stage prognostic biomarker for lung cancer. High expression of miR-21 was also associated with cancer-specific mortality in the 48 TNM stage II/III patients from the Maryland/Norway cohort (HR = 3.35, 95% CI = 1.43–7.82, p = 0.005) but not disease-free survival in the 49 stage II patients from the Japan cohort.
Table 4.
Univariate and Multivariate Cox Regression analysis of miR-21 expression levels and other clinical covariates in TNM stage I cases.
| Univariate Analysis
| ||||
|---|---|---|---|---|
| Combined Maryland/Norway cohort (n=78) (Cancer-Specific Mortality)
|
Japan cohort (n=142) (Relapse Free Survival)
|
|||
| HR (95% Cl) | P value | HR (95% Cl) | P value | |
| miR-21 (High vs Low)a | 2.06 (1.08–3.91) | 0.028 | 3.24 (1.51–6.95) | 0.003 |
| Sex (M vs F) | 1.12 (0.59–2.14) | 0.715 | 1.30 (0.75–2.23) | 0.351 |
| Age (≥65 vs <65) | 1.02 (0.54–1.96) | 0.933 | 0.89 (0.42–1.84) | 0.751 |
| Ever smoke (ever vs never)b | 1.66 (0.51–5.41) | 0.398 | 0.86 (0.41–1.79) | 0.686 |
| Packyears (≥20 vs <20) | 1.61 (0.65–3.95) | 0.301 | 1.43 (0.67–3.05) | 0.361 |
|
| ||||
|
Multivariate Analysis, final modelc
| ||||
|
Combined Maryland/Norway cohort (n=78) (Cancer-Specific Mortality)
|
Japan cohort (n=142) (Relapse Free Survival)
|
|||
| HR (95% Cl) | P value | HR (95% Cl) | P value | |
|
| ||||
| miR-21 (High vs Low)a | 2.16 (1.11–4.21) | 0.023 | 3.40 (1.57–7.36) | 0.002 |
| Age (≥65 vs <65) | 0.90 (0.46–1.76) | 0.766 | 0.97 (0.44–2.11) | 0.931 |
| Packyears (≥20 vs <20) | 1.64 (0.65–4.11) | 0.291 | 1.62 (0.75–3.52) | 0.217 |
Abbreviations: HR, hazard ratio; CI, confidence interval.
High expression in cancerous tissue for all miR-21 was defined based on the median.
Never smoker defined as 0 pack years.
Final models included all variables that were used in the Supplemental Table 2 multivariate models.
Discussion
Altered expression of miRNAs has been found in every malignancy examined and appears to play an important role in tumorigenesis. There is increasing evidence that miRNAs have utility as both biomarkers and therapeutic targets for cancer. Here, we report that elevated miR-21 expression is associated with worse prognosis in NSCLC patients.
We evaluated the expression of miRNAs from snap-frozen lung tissue from 317 NSCLC patients originating from three independent cohorts. To our knowledge, this is the largest study to date using flash-frozen tissue in NSCLC and the only study to use three independent cohorts. The association of increased miR-21 expression with worse lung cancer prognosis was significant in each of the three cohorts from different geographical regions of the world. This suggests that our findings are representative of the majority of lung adenocarcinoma patients. A limitation of the study is that we were only studying NSCLC with adenocarcinoma histology; therefore our findings may not be relevant to small cell lung cancer or NSCLC with other histologies.
We assayed the expression of miR-21 from snap-frozen tissue while blinded to clinical outcomes and found that miR-21 was associated with NSCLC prognosis. This association is consistent with our original microRNA array findings using frozen tissue (14), but these results are in apparent contrast to another study that we recently published evaluating miRNA expression in formalin-fixed paraffin-embedded (FFPE) tumor specimens (26). In that study we evaluated miR-21 expression, in addition to other miRNAs, in FFPE lung cancer tissue from 639 patients enrolled in the International Adjuvant Lung Cancer Trial (IALT), of which 218 patients were diagnosed with NSCLC with adenocarcinoma histology. We did not find that increased miR-21 was significantly associated with worse prognosis in those tissues. This is likely due to the fact that the quality of RNA extracted from FFPE tissue is not as high of a quality as RNA extracted from frozen tissue. Therefore, qRTPCR measurements on RNA extracted from FFPE tissue may be less reliable than qRTPCR measurements on RNA extracted from snap-frozen tissue. Three additional reports support this assumption. When using snap frozen tissue, Markou and colleagues found that overexpression of miR-21 was associated with poor survival in 48 NSCLC patients (19). Gao and colleagues also used snap-frozen tissue as a source for RNA extraction and found an association of increased miR-21 expression and poor survival in 47 NSCLC patients (27). But when FFPE tissue was used, Duncavage and colleagues did not find that miR-21 was associated with worse patient outcomes in 46 NSCLC patients (28). The fact that we observed similar associations with high miR-21 expression and poor NSCLC prognosis in three independent cohorts when using snap-frozen tissues provides confidence that our observations in this study are accurate and representative.
Biomarkers that are capable of identifying patients at high risk of dying of stage I lung cancer may provide physicians with tools to help choose appropriate therapeutic strategies. The current clinical guidelines recommend surgical resection for TNM stage I lung cancer and additional therapy is not recommended unless the disease progresses (29). Here we find evidence that miR-21 has the potential to identify high risk early stage patients in three cohorts. To our knowledge, this is the first report that demonstrates an association of miR-21 with prognosis in TNM stage I patients. Stage I patients with elevated miR-21 expression are at high risk of disease progression, therefore these patients may be suitable for earlier or more aggressive intervention than current recommendations indicate. Further studies are warranted and required to address this important area of research.
MiR-21 is an oncogenic microRNA with a suggested role in multiple cancer types. Overexpression of miR-21 has been found in at least 18 malignancies indicating that altered expression of miR-21 may be a common mechanism in carcinogenesis (11, 30). The cause of miR-21 overexpression in cancer is still being elucidated. The altered activity of at least four cancer related regulatory factors is thought to contribute to the altered expression of miR-21, including signal transducer and activator of transcription 3 (STAT3), activator protein 1 (AP-1), transforming growth factor β (TGFβ), and epidermal growth factor receptor (EGFR). STAT3 is a transcription factor that is downstream of IL-6 (interleukin 6) and can influence cell transformation. The miR-21 promoter has STAT3 binding sites and IL-6 can induce the expression of miR-21 in a STAT3 dependent manner indicating that altered inflammatory states may be affecting miR-21 expression in the context of cancer (31). AP-1 can also increase the expression of miR-21. AP-1 can regulate cell proliferation, apoptosis and cell migration and has been implicated in a number of cancers, therefore it may be contributing to the altered expression of miR-21. TGFβ has been shown to increase miR-21 expression through the Smad signaling pathways and may be involved in the increased expression of miR-21 in cancer. EGFR is an important cell surface receptor whose activity has important implications in cancer. The function of EGFR has also been linked to miR-21 expression, raising the possibility that altered function of EGFR may contribute to cancer in part by deregulating miR-21 expression (7, 32).
The finding that increased expression of miR-21 is associated with worse prognosis is similar to findings in other cancers. Increased levels of miR-21 are associated with worse prognosis and/or therapeutic outcome in multiple cancer types including colon cancer (15), breast cancer (33), chronic lymphocytic leukemia (34), pancreatic cancer (35), tongue cancer (36), astrocytomas (37) and head and neck cancers (38). We previously reported that increased miR-21 expression was associated with poor cancer-specific mortality in early, stage II colon cancer (15), similar to the finding that miR-21 is associated with prognosis in early, stage I NSCLC. Taken together, miR-21 expression has the potential to be a prognostic biomarker in multiple cancer types, including early stage disease where it may help guide therapeutic decisions.
Mechanistic studies have demonstrated the oncogenic potential of miR-21 (39) suggesting that targeting miR-21 may provide some therapeutic benefit. Overexpression of miR-21acts as an anti-apoptotic factor and promotes cell proliferation. Inhibition of miR-21 inhibits cell growth in vitro and inhibits tumor growth in xenograft mouse models. MiR-21 expression can affect cell migration, cell invasion and tumor metastasis. MiR-21 can target several tumor suppressor genes including programmed cell death 4 (PDCD4) (40, 41), phosphatase and tensin homolog (PTEN)(42), reversion-inducing-cysteine-rich protein with kazal motifs (RECK) (43), TIMP3 (43), maspin (44), nuclear factor 1 B-type (NFIB) (45), tropomyosin 1 (TPM1) (46), sprouty 2 (SPRY2) (47) and Ras homolog gene family, member B (RHOB) (48). Overexpression of miR-21 can cause a malignant transformation in vivo and loss of miR-21 expression results in tumor regression in these models (49). MiR-21 expression is also required for K-Ras-dependent lung tumor formation in mice, demonstrating necessary role of this microRNA in lung tumorigeneses (50). Inhibiting miR-21 can also sensitize cancer cells to chemotherapy (7, 50), further demonstrating the potential for targeting miR-21, alone or in combination with other therapies, as a cancer treatment. Therefore, miR-21 may be both a useful biomarker and a therapeutic target for multiple cancer types.
In conclusion, we found strong evidence that high miR-21 expression is associated with poor lung cancer survival outcomes, including early stage patients. This was found in three separate cohorts and is consistent with our previous report (14) and two additional reports (19, 27). These data suggest that increased miR-21 may in part be responsible for the aggressive nature and poor survival outcomes of NSCLC adenocarcinoma patients. These results are likely representative of the majority of NSCLC adenocarcinomas. Therefore, miR-21 expression has potential as a diagnostic and prognostic biomarker for lung adenocarcinoma, including stage I lung cancer.
Translational Relevance.
We report that elevated miR-21 expression is independently associated with worse prognosis in three independent NSCLC adenocarcinoma cohorts from different regions of the world. This suggests that our findings are representative of the majority of NSCLC adenocarcinoma patients. These associations were significant when evaluating TNM stage I tumors alone. Biomarkers that are capable of identifying patients at high risk of dying of stage I lung cancer may provide physicians with key tools to help diagnose lung cancer and choose appropriate therapeutic strategies. Therefore miR-21 expression has potential utility as an early stage prognostic biomarker for NSCLC adenocarcinoma patients. We also find increased levels of miR-21 expression in more advanced tumors, consistent with a possible role for miR-21 in the progression of NSCLC. These findings suggest that miR-21 may also be a useful therapeutic target for NSCLC patients.
Acknowledgments
This research was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Defense Congressionally Directed Medical Research Program Grant PR093793, the Norwegian Cancer Society, and a Grant-in-Aid from the Ministry of Health, Labor and Welfare for the 3rd-term Comprehensive 10-year Strategy for Cancer Control, Japan.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010 doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 3.van Rens MT, de la Riviere AB, Elbers HR, van Den Bosch JM. Prognostic assessment of 2,361 patients who underwent pulmonary resection for non-small cell lung cancer, stage I, II, and IIIA. Chest. 2000;117:374–9. doi: 10.1378/chest.117.2.374. [DOI] [PubMed] [Google Scholar]
- 4.Nesbitt JC, Putnam JB, Jr, Walsh GL, Roth JA, Mountain CF. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg. 1995;60:466–72. doi: 10.1016/0003-4975(95)00169-l. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 6.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27 (Suppl 2):S52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 11.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 17.Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
- 20.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 21.Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–9. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 22.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Landi MT, Zhao Y, Rotunno M, Koshiol J, Liu H, Bergen AW, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 16:430–41. doi: 10.1158/1078-0432.CCR-09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 25.Lubin JH, Gail MH. Biased selection of controls for case-control analyses of cohort studies. Biometrics. 1984;40:63–75. [PubMed] [Google Scholar]
- 26.Voortman J, Goto A, Mendiboure J, Sohn JJ, Schetter AJ, Saito M, et al. MicroRNA Expression and Clinical Outcomes in Patients Treated with Adjuvant Chemotherapy after Complete Resection of Non-Small Cell Lung Carcinoma. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao W, Yu Y, Cao H, Shen H, Li X, Pan S, et al. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother. 2010;64:399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 28.Duncavage E, Goodgame B, Sezhiyan A, Govindan R, Pfeifer J. Use of MicroRNA Expression Levels to Predict Outcomes in Resected Stage I Non-small Cell Lung Cancer. J Thorac Oncol. 2010;5:1755–63. doi: 10.1097/JTO.0b013e3181f3909d. [DOI] [PubMed] [Google Scholar]
- 29.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–9. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 30.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–25. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 31.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–55. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 33.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–52. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–6. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Huang H, Sun L, Yang M, Pan C, Chen W, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 37.Zhi F, Chen X, Wang S, Xia X, Shi Y, Guan W, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur J Cancer. 2010;46:1640–9. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Avissar M, McClean MD, Kelsey KT, Marsit CJ. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. 2009;30:2059–63. doi: 10.1093/carcin/bgp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 41.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–9. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 45.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 Targets the Tumor Suppressor Gene Tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 47.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, et al. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–82. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Connolly EC, Van Doorslaer K, Rogler LE, Rogler CE. Overexpression of miR-21 promotes an in vitro metastatic phenotype by targeting the tumor suppressor RHOB. Mol Cancer Res. 2010;8:691–700. doi: 10.1158/1541-7786.MCR-09-0465. [DOI] [PubMed] [Google Scholar]
- 49.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 50.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]