Abstract
Cytochrome P450s (P450s) constitute one of the major classes of enzymes that are responsible for detoxification of exogenous molecules both in animals and plants. On the basis of its inducibility by exogenous chemicals, we recently isolated a new plant P450, CYP76B1, from Jerusalem artichoke (Helianthus tuberosus) and showed that it was capable of dealkylating a model xenobiotic compound, 7-ethoxycoumarin. In the present paper we show that CYP76B1 is more strongly induced by foreign compounds than other P450s isolated from the same plant, and metabolizes with high efficiency a wide range of xenobiotics, including alkoxycoumarins, alkoxyresorufins, and several herbicides of the class of phenylureas. CYP76B1 catalyzes the double N-dealkylation of phenylureas with turnover rates comparable to those reported for physiological substrates and produces nonphytotoxic compounds. Potential uses for CYP76B1 thus include control of herbicide tolerance and selectivity, as well as soil and groundwater bioremediation.
P450s constitute one of the largest and most extensively studied classes of enzymes. This is mainly due to their critical role in the detoxification or activation of drugs and dietary compounds in mammals (Guengerich, 1995). In plants the function of P450 hemoproteins in the biosynthesis of hormones, lipids, and secondary compounds is better documented than their role in the detoxification of exogenous chemicals (Schuler, 1996). Nevertheless, they represent a potentially significant metabolic sink for environmental contaminants (Sandermann, 1994), which can be used for controlling herbicide tolerance and selectivity (Werck-Reichhart, 1995), and could constitute a valuable potential for bioremediation (i.e. the removal of toxic and persistent organic compounds from industrial waste).
Like their animal counterparts, plant P450s are highly inducible by chemicals such as drugs or pesticides (Bolwell et al., 1994; Schuler, 1996). Some of them respond more strongly than others to chemical signals (Batard et al., 1995, 1997; Moreland et al., 1995; Potter et al., 1995). In animals the P450 isozymes having high xenobiotic-metabolizing capacity are usually more responsive to environmental chemical stimuli than those controlling the status of important endocrine regulatory factors, the expression of the latter being rather down-regulated by foreign chemicals (Waxman and Chang, 1995; Whitlock and Denison, 1995). In plants the activity of P450s metabolizing xenobiotics such as alkoxycoumarins or herbicides is usually strongly increased by treatments with exogenous chemicals (Barret, 1995; Batard et al., 1995; Frear, 1995; Persan and Schuler, 1995; Potter et al., 1995), whereas P450s with basic physiological functions, the best documented being cinnamate 4-hydroxylase, seem less responsive to such treatments (Moreland et al., 1995; Batard et al., 1997).
Because our objective was to characterize plant P450s with high xenobiotic-metabolizing capacities, we designed strategies aimed at the isolation of the forms that are the most inducible by exogenous chemicals. On the basis of Mn2+ and AP inducibility, we recently isolated two cDNAs encoding P450s from Jerusalem artichoke (Helianthus tuberosus) (Batard et al., 1998; Cabello-Hurtado et al., 1998a). One of them, CYP76B1, actively dealkylates the model xenobiotic compound 7-ethoxy-coumarin after expression in yeast (Batard et al., 1998). The other, CYP81B1, is a fatty acid in-chain hydroxylase (Cabello-Hurtado et al., 1998a). In this paper we show that CYP76B1 is the most inducible of the P450s that have been isolated from this plant, and that it metabolizes with high turnover rates a wide range of xenobiotics, including herbicides of the class of phenylureas. Phenylureas are twice dealkylated by the enzyme and thereby converted into nonphytotoxic compounds. To our knowledge, CYP76B1 is the first example of a plant P450 metabolizing with high-efficiency herbicides and structurally diverse exogenous compounds. Our data thus validate a possible strategy for the isolation of the most effective detoxifying plant P450 enzymes.
MATERIALS AND METHODS
Chemicals
AP was from Merck; benzoic acid, salicylic acid, umbelliferone, and 7-ethoxycoumarin were from Sigma. Other coumarin derivatives were gifts from Dr. J.L. Rivière (Ecole Vétérinaire, Lyon, France). Resorufin derivatives were from Pierce.
Synthesis of [14C]ferulic acid, isoscopoletin, and scopoletin was described by Cabello-Hurtado et al. (1998b). [3-14C]trans-Cinnamic acid was from Isotopchim (Ganagobie, France), [1-14C]lauric acid was from Commissariat à l'Energie Atomique (Gif-sur-Yvette, France), and [1-14C]-capric acid and [phenyl-U-14C]2,4-D were from Sigma. [7-14C]Benzoic acid, [1-14C]myristic acid, [1-14C]palmitic acid, [1-14C]stearic acid, [1-14C]oleic acid, and [1-14C]linoleic acid were from DuPont. [14C]Geraniol and [14C]-S-naringenin were kindly provided by Dr. D. Hallahan (IACR, Rothamsted, UK) and Dr. G. Kochs (Institut für Biologie II, Freiburg, Germany), respectively. [Dichlorophenyl-U-14C]diclofop was kindly provided by Hoechst (Frankfurt, Germany), [triazine-2-14C]chlorsulfuron by DuPont, and [phenyl-U-14C]bentazon by BASF (Ludwigshafen, Germany). [PhenylU-14C]chlortoluron, [phenyl-U-14C]isoproturon, [triazine-U-14C]simazine, and reference metabolites were generous gifts from Novartis (Basel, Swizerland).
Plant Material
Jerusalem artichoke (Helianthus tuberosus L. var. Blanc commun) tubers were grown in an open field, harvested in November, and stored in plastic bags at 4°C in the dark. For aging experiments, tubers were sliced (1.5 mm thick), washed, and aged for 48 h in aerated (4 L min−1) distilled water containing 35 mm DMSO, 20 mm AP, 8 mm sodium PB, 25 mm MnCl2, 1.7 mm Flav, 100 μm NA, or 260 μm B(a)P. The MnCl2 solution was adjusted to pH 7.0. Water-insoluble compounds (Flav, NA, and B(a)P) were added to the aging medium dissolved in DMSO (0.25% of the total volume).
RNA Isolation and RNA-Blot Hybridization
Isolation of total RNA and RNA-blot hybridization was performed as described previously (Batard et al., 1998). The hybridization signal was recorded and quantified with a phosphor imager (model BAS1000, Fuji, Japan), using hybridization at 55°C to a 300-bp Capsicum annuum 25S rRNA probe for standardization. The same membrane was successively hybridized with the CYP81B1 (46% GC), CYP73A1 (47% GC), CYP76B1 (43% GC), CYP81B1 (46% GC), and 25S rRNA probes (45 × 106 cpm in each case), respectively. It was stripped by boiling in 0.1% SSC between successive hybridizations. Slot-blot analysis was performed in the same conditions using 10 μg of total RNA in each slot.
Expression in Yeast
The BamHI and EcoRI sites were introduced by PCR just upstream of the ATG and downstream of the stop codon of the CYP76B1 coding sequence using the primers 5′-ATATATGGATCCATGGATTTTCTTATAATAGTGAGTAC (sense) and 5′-TATATAGAATTCATGCTAGTTCAATGGTATTGGAACAACAC (reverse). The PCR mixture was preheated for 2 min at 92°C before addition of 1 unit of Pfu DNA polymerase (Stratagene). After 3 min of additional heating at 92°C, 30 cycles of amplification were carried out as follows: 1 min of denaturation at 92°C, 1 min of annealing at 52°C, and 2 min of extension at 72°C. The reaction was completed by 10 min of extension at 72°C. After BamHI/EcoRI digestion, the 1470-bp coding sequence was inserted into the vector pYeDP60 (Pompon et al., 1996). Transformation of the yeast Saccharomyces cerevisiae W(R), WAT11 and WAT21, and yeast growth were performed according to the method of Pompon et al. (1996). When indicated, 0.5 mm δ-aminolevulinic acid methyl ester (Sigma) was added to the induction medium, or cells were stored at 4°C for 24 h before preparation of the microsomes. Yeast microsomes were prepared as described in Batard et al. (1998).
Production of Antibodies and Western-Blot Analysis
CYP76B1 4-His-tagged at the C terminus was generated by PCR modification of its coding sequence using the reverse primer 5′-TATATAGAATTCATGCTAATGATGATGATGGTTCAATGGTATTGGAACAACAC with sense primer and PCR conditions as above. The modified protein was expressed in yeast using the same procedure as for the wild type. Microsomes were solubilized in 2% Triton X-114 followed by phase partitioning as described previously (Gabriac et al., 1991). The protein was purified on a Ni2+-loaded chelating column (HiTrap, Pharmacia Biotech) using the procedure recommended by the manufacturer, with elution in sodium phosphate 50 mm, pH 7.4, containing 0.5 m NaCl and 1 m imidazole. Polyclonal antibodies were raised in rabbits by successive injections of one time with 16 μg and five times with 8 μg of purified protein emulsified in Freund's complete and incomplete adjuvants, respectively, and used for western-blot analysis as in Werck-Reichhart et al. (1993).
Enzyme Assays
7-Alkoxycoumarins and 7-alkoxyresorufins O-dealkylation were measured by fluorometry (Werck-Reichhart et al., 1990). The conversion of other molecules was assayed by TLC or HPLC analysis of polar metabolites formed from radiolabeled compounds (Cabello-Hurtado et al., 1998a, 1998b). NADPH-Cyt c reductase was assayed according to the method of Benveniste et al. (1986). Kinetic data were fitted using the nonlinear regression program DNRPEASY (Duggleby, 1984).
Analytical Methods
Liquid chromatography-MS was performed by coupling a HPLC system (model 140A, Perkin Elmer) on a VG BioQ triple quadrupole. The separation on a reverse-phase column (300–5 C18 2 × 125 mm, Macherey-Nagel, Duren, Germany) was flow spliced via a stainless steel tee with approximately 1/15 of column effluent directed toward the mass spectrometer, 14/15 directed toward the UV detector (model 496, Waters). The mass spectra were obtained by scanning from m/z 300−1800 in 6 s with a cone voltage of 50 V.
Spectrophotometric measurements of P450 content and substrate binding were performed as in Gabriac et al. (1991). Protein was quantified using the Bio-Rad protein assay.
RESULTS
CYP76B1 Is More Strongly Induced by Xenobiotics than Other P450s
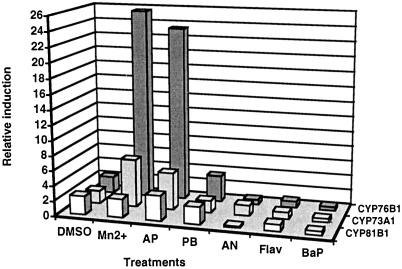
Isolation of the CYP76B1 cDNA, based on PCR amplification of a conserved P450 region from a library of AP-treated Jerusalem artichoke tuber tissues, and subsequent selection of AP-induced P450 cDNAs, has been described previously (Batard et al., 1998). Two other P450 cDNAs were also isolated from Jerusalem artichoke: cinnamate 4-hydroxylase CYP73A1 (Teutsch et al., 1993) and fatty acid in-chain hydroxylase CYP81B1 (Cabello-Hurtado et al., 1998a). Increased accumulation of all three of the P450 mRNAs was detected following treatment of tuber tissues with xenobiotics. However, successive hybridizations of CYP76B1, CYP73A1, and CYP81B1 probes with the same RNA gel blot prepared with tuber tissues treated with different chemicals (DMSO, MnCl2, AP, PB, Flav, NA, and B(a)P) indicated that by far the highest increase in the steady-state level of P450 transcripts was achieved in the case of CYP76B1 (Fig. 1). Similar results were obtained by parallel slot-blot hybridization analysis of the same set of total RNA. Tuber tissues treated with chemicals, in particular Mn2+ and AP, accumulated 2- to 6-fold more transcripts coding for CYP76B1 than for CYP73A1 or CYP81B1.
Figure 1.
Relative increase in the steady-state level of CYP76B1, CYP81B1, and CYP73A1 transcripts induced by xenobiotics. RNA was prepared from Jerusalem artichoke tuber tissues, sliced, and aged in water or in solutions of different chemicals. The same RNA-blot membrane (20 μg of total RNA in each lane) was successively hybridized with CYP81B1, CYP73A1, and CYP76B1 32P-radiolabeled probes. Hybridization signals were quantified with a phosphor imager. Induction is calculated by comparison with the signal recorded with RNA from tissues aged in water. Induction obtained with Flav, NA, and B(a)P, which were dissolved in DMSO, was compared with that obtained for tissues treated with DMSO alone.
Optimized Expression of CYP76B1 in Yeast
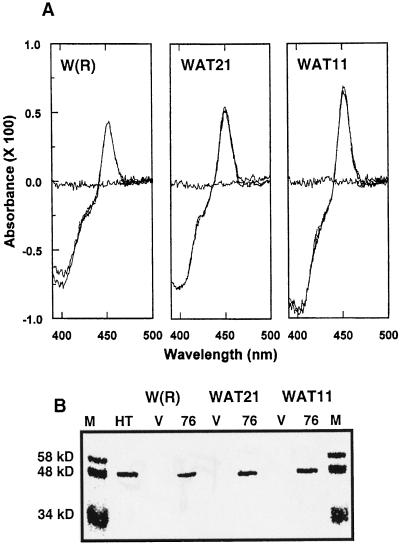
The CYP76B1 coding sequence was expressed in three engineered yeast strains, W(R), WAT11, and WAT21, which also expressed yeast or Arabidopsis P450 reductases ATR1 and ATR2, respectively, under the transcriptional control of the same Gal-inducible promoter GAL10-CYC1 (Urban et al., 1997). The levels of expression of CYP76B1 were compared in the different yeast strains after 12, 16, 24, and 36 h of induction with Gal, or continuously grown in the presence of Gal. Highest expression was obtained using the WAT11 strain continuously grown in the presence of Gal, reaching 157 pmol mg−1 microsomal protein (0.86% of microsomal protein). Contrary to CYP81B1 (Cabello-Hurtado et al., 1998a), CYP76B1 was expressed at a significant level in the presence of yeast reductase, but its expression was consistently 15% to 40% lower in WAT21 or W(R) than in WAT11 (Fig. 2). Expression did not increase when δ-aminolevulinic acid was added to the growth medium, and significantly decreased when cells were grown at 25°C or stored for 24 h at 4°C before extraction.
Figure 2.
Comparison of the expression of CYP76B1 in yeast strains coexpressing yeast [W(R)], Arabidopsis ATR1 (WAT11), or ATR2 (WAT21) reductases. A, CO/reduced versus reduced difference spectra measured in microsomes of the three yeast strains after 16 h of induction with Gal. Microsomal protein concentration in the cuvettes was 900 μg mL−1. No P450 was detected in microsomes prepared from the same yeast strains transformed with a void plasmid. B, Western-blot analysis of the same microsomes. M, Mr markers; HT, microsomes from Jerusalem artichoke tuber treated with AP; V, microsomes from yeast transformed with a void plasmid; 76, microsomes from yeasts transformed with CYP76B1. Each lane was loaded with 20 μg of microsomal protein.
CYP76B1 Metabolizes Several Classes of Exogenous Molecules
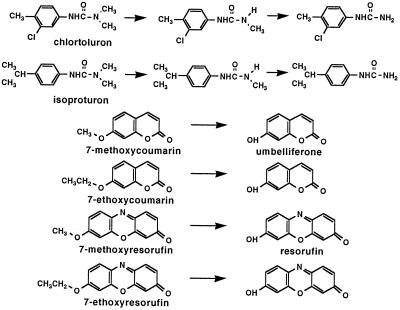
CYP76B1 was previously reported to dealkylate 7-ethoxycoumarin (Batard et al., 1998). After optimized expression in yeast, its metabolic capacity was more thoroughly investigated. Using transformed WAT11 yeast microsomes, we first assayed for possible metabolism of a number of endogenous molecules known or suspected to be substrates of P450 oxygenases from Jerusalem artichoke tuber tissues. No metabolites were formed from phenolics (cinnamate, benzoate, ferulate, naringenin, scopoletin, and isoscopoletin), isoprenoids (geraniol, ABA, and obtusifoliol), or fatty acids (capric, lauric, myristic, palmitic, oleic, linoleic, and linolenic acids). We then tested different classes of foreign compounds, including molecules previously shown to be metabolized by the plant-tuber microsomes (Higashi et al., 1981; Fonné, 1985; Fonné-Pfister et al., 1988; Werck-Reichhart et al., 1990; Batard et al., 1995): B(a)P and the drug AP, fluorescent alkoxycoumarins (7-methoxycoumarin, 7-propoxycoumarin, and 7-butoxycoumarin) and alkoxyphenoxazones (7-methoxy-resorufin, 7-ethoxyresorufin, 7-pentoxyresorufin, and 7-benzyloxyresorufin), and representative members of different classes of herbicides (2,4-D, diclofop, chlorsulfuron, bentazon, dicamba, chlortoluron, and isoproturon). In addition to 7-ethoxycoumarin, five xenobiotics were actively converted into more polar metabolites (Table I). No metabolism was detected in the absence of NADPH or in control microsomes prepared from yeast transformed with a void plasmid.
Table I.
CYP76B1-dependent metabolism of xenobiotics
| Strain | NADPH | Activity
|
|||||
|---|---|---|---|---|---|---|---|
| MCOD | ECOD | MROD | EROD | CTUDM | IPUDM | ||
| pkat mg−1 protein | |||||||
| Control | + | n.d.a | n.d. | n.d. | n.d. | n.d. | n.d. |
| W(R) | + | 10.9 ± 0.3 | 8.4 ± 1.5 | 12.6 ± 1.6 | 5.1 ± 0.2 | 1223 ± 63 | 128 ± 2 |
| WAT11 | + | 16.4 ± 0.4 | 15.9 ± 0.25 | 18.6 ± 0.6 | 11.6 ± 0.8 | 4258 ± 24 | 475 ± 3 |
| WAT11 | − | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
MCOD, ECOD, MROD, and EROD activities were determined fluorometrically using 16 pmol of CYP76B1 per 2-mL assay. Demethylation of CTUDM and IPUDM was tested using 14C-labeled substrates with approximately 3 pmol of CYP76B1 in the 200-μL assays. Conversion was estimated from the sum of the two polar metabolites detected after TLC analysis. Data are means ± sd of two to four determinations.
n.d., Not detected.
Characterization of the Phenylurea Metabolites
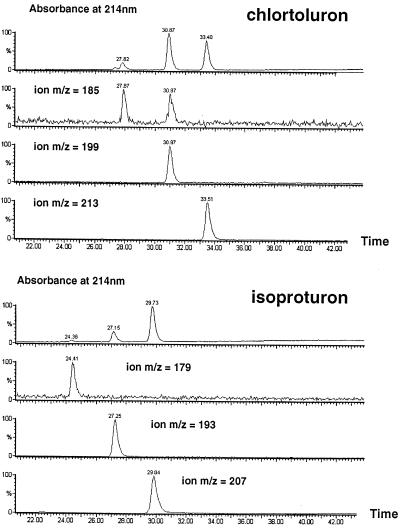
Microsomes from AP-treated Jerusalsem artichoke tuber, when incubated with NADPH and chlortoluron, produce principally mono- and di-N-dealkylated metabolites and minor amounts of the ring-methyl hydroxylated compound (Fonné, 1985). It was unclear whether these different metabolites were generated by a single or several P450 enzymes. The products of CYP76B1-dependent metabolism of chlortoluron and isoproturon were thus extensively characterized. Both TLC and HPLC analysis of the metabolites, including cochromatography with reference standards (not shown), suggested a sequential conversion of the herbicide to mono- and di-dealkylated derivatives. This was further confirmed by liquid chromatography-MS analysis (Fig. 3). For both chlortoluron and isoproturon, time-dependent and sequential formation of m/z −14 and −28 molecular ions was observed. To confirm the capacity of CYP76B1 to catalyze the double dealkylation of chlortoluron, the mono-demethylated derivative was purified by TLC from large-scale incubations of transformed yeast microsomes with NADPH and 14C-chlortoluron. In the absence of chlortoluron, its mono-N-demethylated derivative was actively dealkylated by CYP76B1.
Figure 3.
Products resulting from the CYP76B1-dependent metabolism of chlortoluron and isoproturon. Approximately 8 pmol of CYP76B1 in WAT11 yeast microsomes was incubated for 60 min at 30°C with 500 μm substrate, 500 μm NADPH, 1 mm Glc-6-P, and 0.5 unit of Glc-6-P dehydrogenase in 200 μL of 0.1 m sodium phosphate, pH 7.4. Twenty microliters of the acidified and centrifuged incubation medium was directly analyzed by liquid chromatography-MS using a gradient of 3% to 60% B (100% acetonitrile, 0.08% TFA) in A (water, 0.1% TFA) in 65 min. Absorbance was monitored at 214 nm.
From fluorescence emission spectra of the products of alkoxycoumarin and alkoxyresorufin metabolism with maxima at 460 (excitation at 380 nm) and 585 nm (excitation at 530 nm), respectively, it was assumed that all compounds were O-dealkylated to form umbelliferone or 7-hydroxyresorufin. These products were, however, not further characterized.
Catalytic Parameters of the CYP76B1-Dependent Reactions
Methoxy- and ethoxyresorufins were the best substrates of CYP76B1, with very high kcat/Km of 237 and 165 min−1 μm−1, respectively, in microsomes from the WAT11 yeast strain (Table II). The second-best substrate was chlortoluron, the first dealkylation proceeding significantly faster than the second. The first demethylation of isoproturon was slower than that of chlortoluron. 7-Ethoxycoumarin and 7-methoxycoumarin turned out to be the poorest substrates of CYP76B1. In terms of turnover number, phenyl-ureas were the fastest metabolized substrates of CYP76B1 (kcat of 803 and 147 min−1 for the first demethylation of chlortoluron and isoproturon, respectively), the second demethylation (kcat = 46 min−1 for monodemethyl-chlortoluron) being the rate-limiting step for total detoxification of the herbicides.
Table II.
Catalytic parameters of the reactions catalyzed by CYP76B1 in the presence of different P450 reductases
| Activity | Coexpressed Reductase | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μm | min−1 | min−1 μm−1 | ||
| MCOD | Yeast | 899 | 13 | 0.01 |
| ATR1 | 808 | 31 | 0.04 | |
| ECOD | Yeast | 74 | 10 | 0.13 |
| ATR1 | 60 | 30 | 0.5 | |
| MROD | Yeast | 0.23 | 15 | 64 |
| ATR1 | 0.17 | 41 | 237 | |
| EROD | Yeast | 0.49 | 6 | 12 |
| ATR1 | 0.17 | 28 | 165 | |
| IPUDM | Yeast | 361 | 40 | 0.11 |
| ATR1 | 289 | 147 | 0.51 | |
| CTUDM | Yeast | 202 | 376 | 1.8 |
| ATR1 | 179 | 803 | 4.5 | |
| Monodemethyl-CTUDM | ATR1 | 81 | 46 | 0.57 |
The CYP76B1 concentrations in the MCOD, ECOD, MROD and EROD fluorometric assays were 1.5 or 2.1 nm, depending on the coexpressed ATR1 or yeast reductases. In the CTUDM assays, the CYP76B1 concentrations were 1.7 nm (ATR1) and 8 nm (yeast), respectively. It was 12 nm for the determination of the catalytic parameters of monodemethyl-CTUDM, and 1.7 nm (ATR1) or 20 nm (yeast) in the IPUDM assays. Products were quantified by direct TLC analysis of 100 μL of the acidified incubation medium after 6 min of incubation at 30°C for the CTUDM and IPUDM assays, and after 9 min of incubation for the monodemethyl-CTUDM.
Influence of the P450 Reductases on CYP76B1 Activity
Preliminary results (Table I) indicated that the differences in catalytic activity were larger than the differences in P450 expression in the three yeast strains. Accordingly, the catalytic parameters of xenobiotic metabolism were determined with CYP76B1 expressed in both W(R) and WAT11 microsomes (Table II). The nature of the coexpressed reductase did not influence the Km of the reactions to any great extent, although Km measured in the presence of the Arabidopsis reductase ATR1 were consistently lower than those determined in presence of yeast reductase. However, large changes in kcat were observed depending on the origin of the coexpressed reductase. The maximum velocities of oxygenation of all six substrates were about 3 times higher in the presence of ATR1. Catalytic parameters were also measured in the presence of the other Arabidopsis reductase ATR2 with methoxyresorufin as the substrate. They were not significantly different (Km = 0.185 μm; kcat = 43 min−1) from those determined with microsomes from WAT11.
The observed variations in kcat could result from differences in reductase expression in the W(R) and WAT yeast strains. We thus compared the velocities of NADPH-dependent Cyt c reduction in microsomes from the three strains. The reductase activity in microsomes from 76B1W(R) (0.74 ± 0.057 μmol min−1 mg−1) was higher than in microsomes from transformed WAT11 and WAT21 (0.61 ± 0.023 and 0.60 ± 0.01 μmol min−1 mg−1, respectively). Differences in kcat observed in our experiments are thus likely to result from faster electron transfer between plant enzymes, or from a plant reductase-induced change in CYP76B1 conformation or stability.
Other Phenylurea Herbicides
Metabolism of both chlortoluron and isoproturon suggests that other herbicides of the class of phenylureas could be substrates as well. Therefore, we tested the binding of a set of related molecules in the active site of CYP76B1. The binding of substrates into the catalytic site of a P450 usually induces a shift of the absorption maximum, redox potential, and iron spin state (Raag and Poulos, 1989), resulting from the displacement of the water molecule that serves as the sixth ligand of the heme iron. Substrate binding is easily detected by difference spectroscopy, resulting in the formation of a so-called type I spectrum with a maximum at 390 nm and a trough at 420 nm (Jefcoate, 1978). The variation of the difference absorbance ΔA390–420 versus substrate concentration allows the calculation of an apparent dissociation constant (KS). A high ΔAmax induced by the ligand is usually an indication of correct ligand positioning for catalysis. Therefore, the high ΔAmax induced by the binding of the molecules in Table III suggests that a similar positioning of the dimethylurea function close to heme iron and favorable to catalysis is maintained for many phenyl-ureas. Affinity for the active site seems to be largely governed by the substituents on the phenyl ring, the presence of ring-deactivating halogens being more favorable than methyl or other potentially activating substituents. Replacement of one of the N-methyls by a hydroxymethyl group also seems to favor the binding of the molecules in the active site of CYP76B1.
Table III.
Binding parameters of the spectra induced upon fixation of phenylureas in the active site of CYP76B1
The CYP76B1 concentration was 60 nm in the assays. Difference spectra were recorded after addition of increasing concentrations of ligands to the sample cuvette. An equal volume of the herbicide solvent DMSO was added to the reference. Ks and ΔAmax at saturating ligand concentrations were calculated from the double-reciprocal plots of ΔA(390–420nm) versus substrate concentrations.
DISCUSSION
Oxidative N-demethylation of phenylureas, described by Frear and coworkers as early as 1969, together with kaurene hydroxylase, were the two first P450-dependent reactions characterized in higher plants. This pioneer work supported the idea that several phenylurea derivatives might be substrates of the same P450 enzyme, and that this P450 might be able to carry out the double dealkylation of the molecules. The involvement of P450 in the Ndemethylation of chlortoluron was first demonstrated using microsomes from Jerusalem artichoke tuber treated with PB (Fonné, 1985). Subsequent reports indicated that at least one other P450, found mostly in cereals, metabolized chlortoluron via hydroxylation on its ring-methyl (Ryan et al., 1981; Fonné-Pfister and Kreuz, 1990; Mougin et al., 1990). Both double N-dealkylation and ring-methyl hydroxylation of the herbicide lead to inactive compounds (Ryan et al., 1981; Ryan and Owen, 1982). Plant tolerance to phenylurea and selectivity of these herbicides thus rely on the presence and relative expression of at least two P450 oxygenases.
We show here that CYP76B1 catalyzes the mono- and di-N-demethylation of both chlortoluron and isoproturon. Other phenylureas bind to its active site with high affinity. Their positioning close to the heme iron is favorable to catalysis. Such binding parameters, together with preliminary analysis of metabolites (not shown), indicate that CYP76B1 dealkylates most phenylureas, including the methoxylated forms such as monolinuron or chlorbromuron. The Ks of CYP76B1 for diuron (20 μm) is very similar to the Km previously reported for diuron demethylation (15 μm) in cotton seedling hypocotyl microsomes (Frear et al., 1969). This suggests that CYP76B1 may be the Jerusalem artichoke ortholog of the phenylurea dealkylase originally characterized by Frear et al. (1969) in cotton.
A few other P450s have previously been reported to metabolize chlortoluron. Recombinant human CYP1A1 (Shiota et al., 1994) and CYP3A4 (Mehmood et al., 1995) have low regiospecificity and catalyze both N-demethylation and ring-methyl hydroxylation of the herbicide. Although turnover numbers were not reported, available data indicate moderate efficiency. Two yeast-expressed plant enzymes, CYP73A1 (Pierrel et al., 1994) and CYP81B1 (Cabello-Hurtado et al., 1998a), were also reported to catalyze ring-methyl hydroxylation of chlortoluron. In both cases, however, the reaction was extremely slow. This contrasts with the very fast N-demethylation observed with CYP76B1. Turnover rates found for the first demethylation of chlortoluron and isoproturon (803 and 147 min−1) compare favorably to the turnover measured with physiological substrates. Even the second demethylation, which is the slow step for complete detoxification of chlortoluron, proceeds with a relatively high kcat of 46 min−1. CYP76B1 is thus expected to be a major phenylurea metabolizing P450 in higher plants, and probably plays a significant role in the detoxification and selectivity of this broad class of PSII inhibitors. Phenyl-ureas have been widely used for selective weed control in cereal crops and are considered serious environmental contaminants. It may thus provide a strategy for bioremediation of contaminated sites.
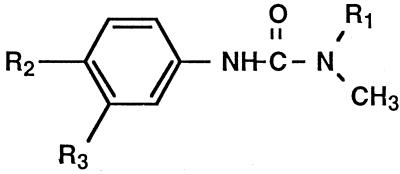
The physiological function of CYP76B1 remains an intriguing question. Phenylurea demethylase activity was found in many plants, both dicots and monocots (Frear et al., 1969; Ryan et al., 1981; Fonné, 1985; Fonné-Pfister and Kreuz, 1990; Mougin et al., 1990; Frear et al., 1991). In cotton, activity was detected in leaves and roots and was highest in etiolated hypocotyls (Frear et al., 1969). Several P450 families may contribute to phenylurea metabolism in different organs or different plant species. It is also possible that several 76 orthologs, isoforms, or variants participate to a different extent in the metabolism of phenylureas. The first two CYP76 sequences were isolated from eggplant (Toguri et al., 1993) without any assignment of a possible function. A recent search of the sequence data banks reveals the presence of 4 CYP76 genes clustered on the chromosome II of Arabidopsis. The CYP76 family thus appears widespread and diversified in higher plants. All of the identified substrates of CYP76B1 (Fig. 4) are planar aromatic molecules larger than simple phenylpropanoids. The most closely related P450 enzymes are CYP75 and CYP93, which are known or implied to metabolize flavonoids or isoflavonoids (Holton et al., 1993; Schopfer and Ebel, 1998). Molecules with a flavonoid-like structure are thus good potential substrates of CYP76. Only naringenin has been tested so far and was not metabolized by CYP76B1.
Figure 4.
Reactions catalyzed by CYP76B1. Resorufin is 7-hydroxyresorufin.
Our aim was to isolate plant P450s with high xenobiotic-metabolizing capacities. We expected such P450s to be strongly inducible by exogenous chemicals. In the case of CYP76B1, this assumption proved to be correct. If the physiological substrate turns out to be a flavonoid, strong induction by foreign chemicals could be a response to oxidative stress related to the antioxidant properties of such molecules. Alternatively, PB, AP, or Mn2+ could be mimicking endogenous signals that specifically trigger events of plant development or defense mechanisms. In this case, selective induction of CYP76B1 will be more difficult to explain. However, it is possible that enzymes corresponding to downstream portions of the secondary metabolism are less restricted with respect to the selectivity of their active site, and submitted to more specific regulation than P450s with more pleiotropic functions in the upper parts of the pathways.
ACKNOWLEDGMENTS
We would like to thank Drs. D. Pompon and P. Urban for providing the W(R), WAT11, and WAT21 yeast strains and the pYeDP60 expression vector. We would also like to thank M.F. Castaldi for her technical assistance and R. Kahn for the linguistic improvement of the manuscript.
Abbreviations:
- AP
aminopyrine
- B(a)P
benzo(a)pyrene
- CTUDM
chlortoluron demethylase
- ECOD
7-ethoxycoumarin O-deethylase
- EROD
7-ethoxyresorufin O-deethylase
- Flav
flavone
- IPUDM
isoproturon demethylase
- MCOD
7-methoxycoumarin O-deethylase
- MROD
7-methoxyresorufin O-deethylase
- NA
naphthalic anhydride
- P450
Cyt P450
- PB
phenobarbital
Footnotes
This work was supported in part by the Convention Groupement de Recherches et d'Etudes sur les Génomes/Institut National de la Recherche Agronomique “Complémentation de la levure par des gènes de plantes.” Y.B. was supported by the Ministère de la Recherche et de l'Enseignement Supérieur, S.N. by a fellowship in toxicology from the European Science Foundation, and F.C.H. by a postdoctoral grant from the Spanish Ministerio de Agricultura, Pesca y Alimentacíon.
The accession number for the CYP76B1 sequence described in this article is Y0992.
LITERATURE CITED
- Barrett M. Metabolism of herbicides by cytochrome P450 in corn. Drug Metab Drug Interact. 1995;12:299–315. doi: 10.1515/dmdi.1995.12.3-4.299. [DOI] [PubMed] [Google Scholar]
- Batard Y, LeRet M, Schalk M, Zimmerlin A, Durst F, Werck-Reichhart D. Plant J. 1998;14:111–120. doi: 10.1046/j.1365-313x.1998.00099.x. [DOI] [PubMed] [Google Scholar]
- Batard Y, Schalk M, Pierrel MA, Zimmerlin A, Durst F, Werck-Reichhart D. Regulation of the cinnamate 4-hydroxylase (CYP73A1) in Jerusalem artichoke tubers in response to wounding and chemical treatments. Plant Physiol. 1997;113:951–959. doi: 10.1104/pp.113.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batard Y, Zimmerlin A, Le Ret M, Durst F, Werck-Reichhart D. Multiple xenobiotic-inducible P450s are involved in alkoxycoumarins and alkoxyresorufins metabolism in higher plants. Plant Cell Environ. 1995;18:523–533. [Google Scholar]
- Benveniste I, Gabriac B, Durst F. Purification and characterization of the NADPH-cytochrome P450 (cytochrome c) reductase from higher-plant microsomal fraction. Biochem J. 1986;235:365–373. doi: 10.1042/bj2350365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Bozac K, Zimmerlin A. Plant cytochrome P450. Phytochemistry. 1994;37:1491–1506. doi: 10.1016/s0031-9422(00)89567-9. [DOI] [PubMed] [Google Scholar]
- Batard FY, Salaün J, Cabello-Hurtado P, Durst F, Pinot F, Werck-Reichhart D. Cloning, expression in yeast and functional characterization of CYP81B1, a plant P450 which catalyzes in-chain hydroxylation of fatty acids. J Biol Chem. 1998a;273:7260–7267. doi: 10.1074/jbc.273.13.7260. [DOI] [PubMed] [Google Scholar]
- Cabello-Hurtado F, Durst F, Jorrin JV, Werck-Reichhart D (1998b) Coumarins in Helianthus tuberosus: characterization, induced accumulation and biosynthesis. Phytochemistry (in press)
- Duggleby RG. Regression analysis of nonlinear Arrhenius plots: an empirical model and a computer program. Comput Biol Med. 1984;14:447–455. doi: 10.1016/0010-4825(84)90045-3. [DOI] [PubMed] [Google Scholar]
- Fonné R (1985) Intervention du cytochrome P450 des végétaux supérieurs dans l'oxydation de composés exogènes: l'aminopyrine et le chlortoluron. PhD thesis. Université Louis Pasteur, Strasbourg, France
- Fonné-Pfister R, Kreuz K. Ring-methyl hydroxylation of chlortoluron by an inducible cytochrome P450-dependent enzyme from maize. Phytochemistry. 1990;9:2793–2796. [Google Scholar]
- Fonné-Pfister R, Simon A, Salaün JP, Durst F. Xenobiotic metabolism in higher plants: involvement of microsomal cytochrome P450 in aminopyrine N-demethylation. Plant Sci. 1988;55:9–20. [Google Scholar]
- Frear DS. Wheat microsomal cytochrome P450 monooxygenases: characterization and importance in the metabolic detoxification and selectivity of wheat herbicides. Drug Metab Drug Interact. 1995;12:329–357. doi: 10.1515/dmdi.1995.12.3-4.329. [DOI] [PubMed] [Google Scholar]
- Frear DS, Swanson HR, Tanaka FS. N-demethylation of substituted 3-(phenyl)-1-methylureas: isolation and characterization of a microsomal mixed function oxidase from cotton. Phytochemistry. 1969;8:2157–2169. [Google Scholar]
- Frear DS, Swanson HR, Thalacker FW. Induced microsomal oxidation of diclofop, triasulfuron, chlorsulfuron and linuron in wheat. Pestic Biochem Physiol. 1991;41:274–287. [Google Scholar]
- Gabriac B, Werck-Reichhart D, Teutsch GH, Durst F. Purification and immunocharacterization of a plant cytochrome P450: the cinnamic acid 4-hydroxylase. Arch Biochem Biophys. 1991;288:302–309. doi: 10.1016/0003-9861(91)90199-s. [DOI] [PubMed] [Google Scholar]
- Guengerich FP (1995) Human cytochromes P450 enzymes. In PR Ortiz de Montellano, ed, Cytochrome P450: Structure, Mechanism, and Biochemistry, 2nd Ed. Plenum Press, New York, pp 367–390
- Higashi K, Nakashima K, Karasaki Y, Fukunaga M, Mizuguchi Y. Activation of benzo(a)pyrene by microsomes of higher plant tissues and their mutagenesis. Biochem Internat. 1981;2:373–380. [Google Scholar]
- Holton T, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu CY, Farcy E, Srevenson TW, Cornish EC. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature. 1993;366:276–279. doi: 10.1038/366276a0. [DOI] [PubMed] [Google Scholar]
- Jefcoate CR. Measurement of substrate and inhibitor binding to microsomal cytochrome P450 by optical-difference spectroscopy. Methods Enzymol. 1978;52:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- Mehmoud Z, Kelly DE, Kelly SL (1995) Metabolism of the herbicide chlortoluron by human cytochrome P4503A4. Chemosphere 11/12: 4515–4529 [DOI] [PubMed]
- Moreland DE, Corbin FT, Fleischmann TJ, McFarland JE. Partial characterization of microsomes isolated from mung bean cotyledons. Pest Biochem Physiol. 1995;52:98–108. [Google Scholar]
- Mougin C, Cabanne F, Canivenc MC, Scalla R. Hydroxylation and N-demethylation of chlortoluron by wheat microsomal enzymes. Plant Sci. 1990;66:195–203. [Google Scholar]
- Persan MW, Schuler MA. Differential induction of cytochrome P450-mediated triasulfuron metabolism by naphthalic anhydride and triasulfuron. Plant Physiol. 1995;109:1483–1490. doi: 10.1104/pp.109.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel MA, Batard Y, Kazmaier M, Mignotte-Vieux C, Durst F, Werck-Reichhart D. Catalytic properties of the plant cytochrome P450 CYP73 expressed in yeast: substrate specificity of a cinnamate hydroxylase. Eur J Biochem. 1994;224:835–844. doi: 10.1111/j.1432-1033.1994.00835.x. [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environment. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Potter S, Moreland DE, Kreuz K, Ward E. Induction of cytochrome P450 genes by ethanol in maize. Drug Metab Drug Interact. 1995;12:317–327. doi: 10.1515/dmdi.1995.12.3-4.317. [DOI] [PubMed] [Google Scholar]
- Raag R, Poulos TL. The structural basis for substrate-induced changes in redox potential and spin equilibrium in cytochrome P450CAM. Biochemistry. 1989;28:917–922. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- Ryan PJ, Gross D, Owen WJ, Laanio TL. The metabolism of chlortoluron, diuron, and CGA 43057 in tolerant and susceptible plants. Pestic Biochem Physiol. 1981;16:213–221. [Google Scholar]
- Ryan PJ, Owen WJ (1982) The mechanism of selectivity of chlortoluron between cereals and grassweeds. Proc Br Crop Prot Conf-Weeds 317–323
- Sandermann H., Jr Higher plant metabolism of xenobiotics: the “green liver” concept. Pharmacogenetics. 1994;4:225–241. doi: 10.1097/00008571-199410000-00001. [DOI] [PubMed] [Google Scholar]
- Schopfer CR, Ebel J. Isolation of elicitor-inducible cytochrome P450s of soybean (Glycine max L.) using differential display of mRNA. Mol Gen Genet. 1998;258:315–322. doi: 10.1007/s004380050736. [DOI] [PubMed] [Google Scholar]
- Schuler MA. Rev Plant Sci. 1996;15:235–284. [Google Scholar]
- Shiota N, Nagasawa A, Sakaki T, Yabusaki Y, Ohkawa H. Herbicide-resistant plants expressing the fused enzyme between rat cytochrome P4501A1 (CYP1A1) and yeast NADPH-cytochrome P450 reductase. Plant Physiol. 1994;106:17–23. doi: 10.1104/pp.106.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutsch GH, Hasenfratz MP, Lesot A, Stoltz C, Garnier JM, Jeltsch JM, Durst F, Werck-Reichhart D. Isolation and sequence of a cDNA encoding the Jerusalem artichoke cinnamate hydroxylase, a major plant cytochrome P450 involved in the general phenylpropanoid pathway. Proc Natl Acad Sci USA. 1993;90:4102–4106. doi: 10.1073/pnas.90.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toguri T, Kobayashi O, Umemoto N. The cloning of eggplant seedling cDNA encoding proteins from a novel cytochrome P450 family (CYP76) Biochim Biophys Acta. 1993;1216:165–169. doi: 10.1016/0167-4781(93)90058-l. [DOI] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Chang TKH (1995) Hormonal regulation of liver cytochrome P450 enzymes. In PR Ortiz de Montellano, ed, Cytochrome P450: Structure, Mechanism, and Biochemistry, 2nd Ed. Plenum Press, New York, pp 391–417
- Werck-Reichhart D. Herbicide metabolism and selectivity: role of cytochrome P450. Proc Br Crop Prot Conf-Weeds. 1995;3:813–822. [Google Scholar]
- Werck-Reichhart D, Batard Y, Kochs G, Lesot A, Durst F. Monospecific polyclonal antibodies directed against purified cinnamate 4-hydroxylase from Helianthus tuberosus: immunopurification, immunoquantitation, interspecies cross-reactivity. Plant Physiol. 1993;102:1291–1298. doi: 10.1104/pp.102.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Gabriac B, Teutsch H, Durst F. Two cytochrome P450 isoforms catalyzing O-dealkylation of ethoxycoumarin and ethoxyresorufin in higher plants. Biochem J. 1990;270:729–735. doi: 10.1042/bj2700729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JP, Denison MS (1995) Induction of cytochrome P450 enzymes that metabolize xenobiotics. In PR Ortiz de Montellano, ed, Cytochrome P450: Structure, Mechanism, and Biochemistry, 2nd Ed. Plenum Press, New York, pp 367–390