SUMMARY
Cellular processes often depend on stable physical associations between proteins. Despite recent progress, knowledge of the composition of human protein complexes remains limited. To close this gap, we applied an integrative global proteomic profiling approach, based on chromatographic separation of cultured human cell extracts into more than one thousand biochemical fractions which were subsequently analyzed by quantitative tandem mass spectrometry, to systematically identify a network of 13,993 high-confidence physical interactions among 3,006 stably-associated soluble human proteins. Most of the 622 putative protein complexes we report are linked to core biological processes, and encompass both candidate disease genes and unnanotated proteins to inform on mechanism. Strikingly, whereas larger multi-protein assemblies tend to be more extensively annotated and evolutionarily conserved, human protein complexes with 5 or fewer subunits are far more likely to be functionally un-annotated or restricted to vertebrates, suggesting more recent functional innovations.
INTRODUCTION
Protein complexes are stable macromolecular assemblies that perform many of the diverse biochemical activities essential to cell homeostasis, growth and proliferation. Comprehensive characterization of the composition of multi-protein complexes in the sub-cellular compartments of model organisms like yeast, fly, worm and bacteria have provided critical mechanistic insights into the global modular organization of conserved biological systems (Hartwell et al., 1999), accelerated functional annotation of uncharacterized proteins via guilt-by-association (Hu et al., 2009; Oliver, 2000), and facilitated understanding of both evolutionarily conserved and disease-related pathways (Vidal et al., 2011). How the ~20,000 or so proteins encoded by the human genome are partitioned into heteromeric “protein machines” remains an important but elusive research question, however, as less than one fifth of all predicted human open reading frames are currently annotated as encoding subunits of protein complexes in public curation databases (Ruepp et al., 2010)
Loss of function mutations in genes encoding the subunits of protein complexes typically give rise to similar phenotypes, or, through genetic interaction, amplify the phenotypic effects of other alleles in functionally linked sets of genes. Identifying the membership of protein complexes, therefore, addresses a crucial layer in the hierarchical functional organization of biological systems that links the core biochemistry of a functioning cell to the general physiology of an organism and is fundamental to deciphering the relationship between genotype and phenotype. While bioinformatics analyses have been used to predict evolutionarily conserved human protein-protein interactions (PPIs) on a large-scale (Ramani et al., 2008; Rhodes et al., 2005), most of these associations remain to be verified experimentally.
Affinity purification of tagged exogenous proteins coupled with tandem mass spectrometry (AP/MS) is an effective method for isolating and characterizing the composition of stably-associated human proteins in experiments ranging from dozens to hundreds of different 'baits' (Behrends et al., 2010; Bouwmeester et al., 2004; Ewing et al., 2007; Hutchins et al., 2010; Jeronimo et al., 2007; Mak et al., 2010; Sardiu et al., 2008; Sowa et al., 2009). Likewise, immunoprecipitation can be used to systematically isolate endogenous human protein complexes from human cell lines (Malovannaya et al., 2011). Nevertheless, the limited availability of high-quality antibodies or sequence-verified cDNA clones suitable for targeted protein complex enrichment precludes scale-up required for the unbiased assessment of the molecular association networks underlying human cells. Hence, despite considerable successes in the comprehensive identification of protein complexes in model organisms (Butland et al., 2005; Gavin et al., 2006; Gavin et al., 2002; Guruharsha et al., 2011; Ho et al., 2002; Hu et al., 2009; Krogan et al., 2006; Kuhner et al., 2009), clone-based protein purification techniques remain challenging for proteome scale-studies of physical interaction networks in mammalian cells. Conversely, although traditionally used to isolate discrete complexes with specific assayable biochemical properties (e.g., enzymatic activity), classical biochemical fractionation procedures have been used to resolve biological mixtures as a means of ascertaining the collective composition of human protein complexes present in certain sub-cellular compartments (Ramani et al., 2008; Wessels et al., 2009).
Here, we have combined extensive, scaled-up biochemical fractionation with in-depth, quantitative mass spectrometric profiling and stringent computational filtering to resolve and identify endogenous, soluble, stably-associated human protein complexes present in cytoplasmic and nuclear extracts generated from cultured cells. While the resulting reconstructed high-quality physical interaction network shows strong overlap with existing curated and experimentally derived sets of annotated protein complexes, it contains many predicted subunits and previously unreported complexes with specific functional, evolutionary and disease-related biological attributes. To our knowledge, this resource represents the largest experimentally-derived catalog to date of human protein complexes from cell culture, measured using a single standardized assay, and a reliable first draft reference of the basic physical wiring diagram of a human cell.
RESULTS
High-throughput complex fractionation and detection by tandem mass spectrometry
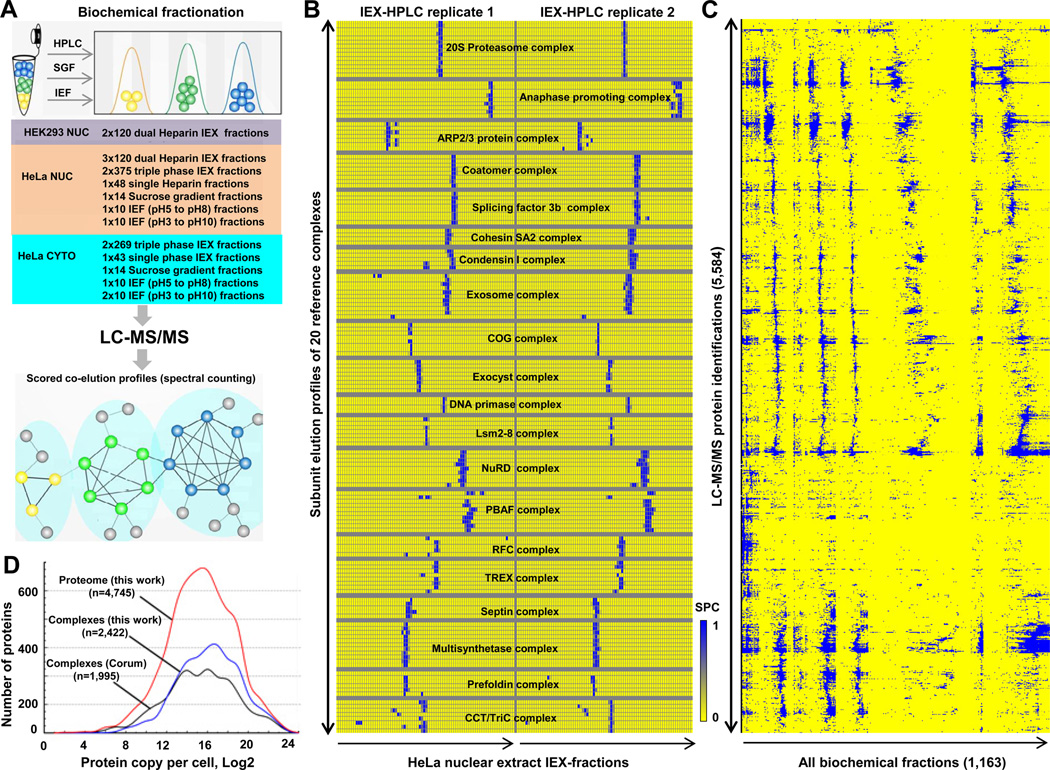
To isolate human protein complexes in a sensitive and unbiased manner, we subjected cytoplasmic and nuclear soluble protein extracts isolated from human HeLa S3 and HEK 293 cells, grown as suspension and adherent cultures respectively, to extensive, complementary biochemical fractionation procedures. These two widely-studied laboratory cell lines have been used as models of human cell biology for many decades (Graham et al., 1977; Masters, 2002), providing a rich biological context for interpreting the resulting proteomic data. Stably-interacting proteins that co-fractionated together were identified subsequently by nano-flow liquid-chromatography-tandem mass spectrometry (LC-MS/MS). We optimized our entire experimental pipeline, illustrated schematically in Figure 1A, using a multi-pronged strategy to minimize two major confounding issues: limited dynamic range (i.e., preferential detection of high abundance components) and ‘chance’ co-elution (i.e., co-fractionation of functionally-unrelated proteins).
Figure 1. Integrative co-fractionation strategy used to identify human soluble protein Complexes.
A- Cell extracts were extensively fractionated using different biochemical techniques (IEX, ion exchange chromatography; IEF, isoelectric focusing; SGF, sucrose density gradient centrifugation). Co-eluting proteins were identified by mass spectrometry and a co-elution network generated by calculating profile similarity (see Extended Experimental Procedures). B- Co-fractionation (IEX-HPLC) profiles of annotated subunits of 20 representative human protein complexes from HeLa nuclear extract. Shading indicates spectral counts recorded by LC-MS/MS. C- Hierarchical clustering of 5,584 proteins identified by LC-MS/MS. D- Protein abundance levels corresponding to components of our identified co-eluting proteins (red line), reconstructed complexes (blue) or annotated CORUM complexes (black) estimated from the reported HeLa proteome (Nagaraj et al., 2011). See also Figure S1 and Table S1.
To address the former concern, we performed extremely deep biochemical fractionations by employing multiple orthogonal separation techniques to better resolve distinct protein complexes. As a primary separation technique, we employed non-denaturing high-performance multibed ion-exchange chromatography (IEX-HPLC) using four different empirically optimized analytical column combinations (see Experimental Procedures) and shallow salt gradients unlikely to perturb non-ionic protein associations (Havugimana et al., 2007). In parallel, we applied complementary sucrose gradient centrifugation and isoelectric focusing technologies to capture salt-sensitive protein assemblies. In total, we collected 1,163 different fractions in a total of eight nuclear and five cytosolic extract fractionation experiments (see Table S1 for details), which were each subjected to label-free shotgun sequencing (duplicate LC-MS/MS analyses) using highly sensitive ion trap-based mass spectrometers (see Experimental Procedures).
We identified 5,584 distinct human proteins (Figure 1C; estimated theoretical false-discovery rate of 1% at both the protein and peptide level based on a statistical model (Kislinger et al., 2003); see Experimental Procedures for details). Despite the underrepresentation of membrane proteins in the starting cell extracts, this coverage encompasses about half of the experimentally-verified human proteome (Figure S1B) (Nagaraj et al., 2011). This included 989 proteins detected exclusively in nuclear fractions (of which 376 were annotated transcription or chromatin-related factors), and 1,006 with links to human disease (e.g. annotated in a public database like OMIM). Only 1,632 (29%) of the identified proteins had biochemical annotations as subunits of previously reported protein complexes (corresponding to 64% of all existing human protein entries) in the CORUM curation database (Figure S1C; (Ruepp et al., 2010)). Due to the extensive fractionation, we observed minimal bias in terms of protein abundance beyond that reported for previously annotated complexes or the experimentally-defined human proteome (Figure 1D).
Next, to minimize the possibility of chance co-elution, rather than simply identifying the proteins present in each fraction, we quantified variation in protein abundance based on the observed patterns of spectral counts recorded across all of the collected fractions to determine the extent to which pairs of proteins co-eluted. As shown in Figure 1B, these experimental profiles were highly reproducible (i.e., average Spearman rank correlation coefficients greater than 80% between replicate experiments; Figure S2), even using alternate methods of mass spectrometric quantification (i.e., extracted MS1 peak intensities were largely consistent with spectral counting; Figure S2 D). To objectively evaluate the biochemical data, we calculated a stringent summary statistic, termed the co-apex score, for each pair of proteins identified LC-MS/MS by determining the number of fractionation experiments in which the proteins showed maximum (modal) abundance in the same exact peak fraction.
To assess the effectiveness of our co-fractionation approach, we performed an initial validation by examining the co-elution profiles and co-apex scores obtained for a reference set of 20 well-known human protein complexes reported in CORUM. As illustrated by the representative HeLa nuclear extract IEX-HPLC profiles shown in Figure 1B, the subunits of these complexes typically co-eluted in the same biochemical fractions. Of the 155 components detected by mass spectrometry, most (85%; 499/585) of the detected subunit pairs of the reference complexes had high co-apex similarity scores (i.e., co-eluted together in at least two or more experiments), validating the overall efficacy of the fractionation procedures we used to isolate native protein complexes and the general correctness of the protein identification and quantification pipeline.
Reconstruction of a high-confidence co-complex interaction network
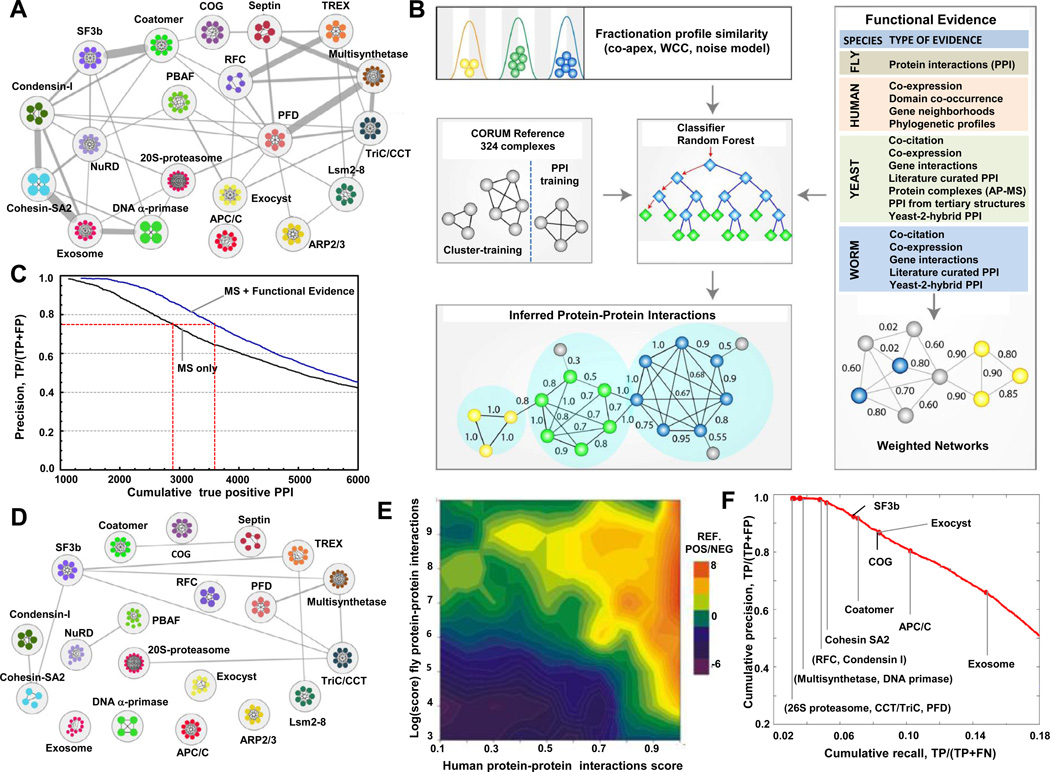
Despite the consistency in co-elution of annotated complex members, certain functionally distinct complexes occasionally exhibited overlapping chromatographic elution profiles (e.g. splicing factor 3b and Coatomer complexes; Figure 2A), presenting a potential source of spurious interactions. While this artifact was minimized to a certain degree by performing multiple independent fractionation experiments, we used an integrative computational approach to further improve deconvolution (Figure 2B). Since physically-interacting co-complexed proteins often perform related biological functions (Alberts, 1998) and are often evolutionarily co-conserved (Hartwell et al., 1999), we devised a machine learning procedure (Figure 2B; see Experimental Procedures for details) to score and select higher-confidence physical interactions based on both the experimentally measured co-elution profiles and the existence of additional supporting functional-association evidence inferred from correlated evolutionary rates (Tillier and Charlebois, 2009) and functional genomics datasets compiled for H. sapiens, S. cerevisiae, D. melanogaster and C. elegans (see Table S6 for details).
Figure 2. Denoising the biochemical co-elution network and generation of high-confidence physical interactions.
A- Biochemical co-fractionation network of 20 reference complexes with co-elution co-apex scores ≥2. Nodes represent protein subunits (colors reflect complex membership), while edges represent interactions (thickness proportional to the number of shared co-apexes). B- The biochemical data was combined with weighted functional association evidence using a random forest classifier and a training set of reference complexes (CORUM) to filter out spurious connections and infer a high-confidence interactome. The PPI and predicted clusters were evaluated with independent functional criteria to ensure high-quality. Arrows represent data flow, blue diamonds are attributes in the decision tree vector and green diamonds (leafs) are the final result (positive or negative). C- Cumulative precision-prediction rank curves for the LC-MS/MS data alone and after integration with genomic evidence. Incorporation of the functional evidence increased both precision (reduced false positives) and recall (more true positives). D- Network of 20 reference complexes after filtering with functional evidence. E- Overall correlation (Spearman r=0.40; n=11,675) of our scored human PPI with corresponding interaction scores reported for orthologous fly PPI from which validated, high confidence complexes were derived (Guruharsha et al., 2011). Heatmap shows prediction accuracy (log ratio of CORUM reference positives to negatives), with high-scoring pairs in both studies highly enriched for positives. F- Precision-recall curve showing performance reconstructing withheld reference CORUM complexes highlighted by red dots at the threshold at which half of the protein pairs per complex are recovered. See also Figure S5 and Table S2.
First, for each of the 13 fractionation experiments, we calculated correlation measures between all possible pairs of proteins to capture their tendency to co-elute. In addition to the co-apex summary statistic, to account for mass spectrometry sampling error, we devised a weighted cross-correlation function to account for slight variation in the protein profiles measured in each experiment. To account for low spectral values, we also generated a Poisson noise model before calculating Pearson correlation scores, deeming the co-elution profiles of protein pairs measured with low spectral counts as less predictive of genuine physical interactions (Figure S5). Only protein pairs with a correlation score of at least 0.5 by at least one of these measures in one or more experiments were considered for further analysis, reducing the total number of pairs from over 15 million initially to the roughly 800,000 pairs with reasonable biochemical evidence.
To improve the assignment of interaction probabilities, we also exploited the predictive power of correlated protein evolutionary rates (Tillier and Charlebois, 2009), mRNA co-expression, and domain co-occurrence, and, via orthology, of fly protein-protein interactions (based on binary yeast two-hybrid assay studies) and extensive physical and functional associations reported previously for yeast and worm (see Experimental Procedures) (Lee et al., 2011). The discriminatory power of the procedure was further improved by, penalizing those interactions which lacked independent supporting evidence – and which were thus more likely to correspond to cases of 'chance' co-elution – by integrating evidence from these functional association data (Figure 2B). A feature selection algorithm was used to select the most informative datasets (Table S2) in addition to the biochemical correlation scores, and the resulting features were used to estimate the probability of interaction to protein pairs using a cross-validated random forest classifier.
For training, we used the CORUM curated set of human protein complexes as our base reference, filtered for those complexes reported before based on biochemical methods. As many CORUM complexes are highly overlapping due to redundancy in existing annotations, we combined complexes sharing subunits (Simpson coefficient > 0.5 between complexes). We used half of the resulting 324 nonredundant reference complexes (Table S3) as the training set for co-complex probability prediction, defining gold standard positive interactions as pairs of proteins in the same complex and inferring gold standard negatives between proteins in different complexes. [The other half of the reference complexes was withheld for subsequent use as an independent training set for cluster optimization, as described below].
Although the biochemical data was a pre-requisite for scoring, the performance curves shown in Figure 2C indicate that the inclusion of the additional functional genomic information substantially increased recall at the same level of precision compared to classifiers based on the profiling data alone. Moreover, the integration of this additional supporting functional evidence removed the bulk of spurious, inter-complex interactions (Figure 2D). Another advantage of our bioinformatic pipeline is that the results of the feature selection algorithm (Table S2) can be explored to examine the impact of each dataset. For example, we find generally that sets of smaller biochemical fractionations using different separation techniques, while individually yielding a higher PPI false discovery rate, collectively provided more information on protein complex composition than deeper fractionations using a single separation method.
As an alternate measure of reliability, we compared our scored human protein interactions to a recently reported network of Drosophila co-complex protein interactions (Guruharsha et al., 2011), which had not been used for build the classifier. Strikingly, despite using vastly different experimental methods and scoring schemes, we observed a remarkably good overall correlation (Spearman r=0.40; n=11,675 orthologs mapped using Inparanoid). Even after removing interactions supported by alternate Drosophila data, high-scoring fly pairs matched high-scoring pairs in our analysis and were strongly enriched for reference positive cocomplex members (Figure 2E).
Finally, in order to remove any remaining false positive interactions, we further denoised our co-complex dataset by pruning loosely connected interactions using a computational diffusion procedure calibrated by protein co-localization semantic similarity scores (Pesquita et al., 2009; Yang et al., 2012) to enforce local network topologies more consistent with annotated complexes from the withheld portion of the reference Corum complexes (see Experimental Procedures). Benchmark precision and recall versus the holdout set of known reference complexes (Figure 2F) were significantly higher than those reported for a smaller, recently published set of affinity-purified human protein complexes (Hutchins et al., 2010) validating the reliability of our scoring procedure.
Applying a PPI score threshold of 0.75, which corresponds to an estimated false discovery rate of 21.5% (i.e. well below the roughly ~40% reported for AP/MS-based analyses of protein complexes in model organisms (Gavin et al., 2006; Krogan et al., 2006; Kuhner et al., 2009)), we thus derived a high-confidence set of 13,993 co-complex interactions among 3,006 unique human proteins (Table S2), most of which (8,691 PPI) have not been reported before (i.e., are not publicly annotated). It is worth reiterating that all of these physical interactions were directly supported by the experimental biochemical co-fractionation data; the addition of functional data and denoising served only to flag candidates lacking either functional support or topological support within the network (Table S2). The interaction probability scores may be underestimated, however, because the reference ‘gold standards’ used for learning are imperfect (Jansen and Gerstein, 2004).
Construction and validation of protein complexes from the probabilistic interaction network
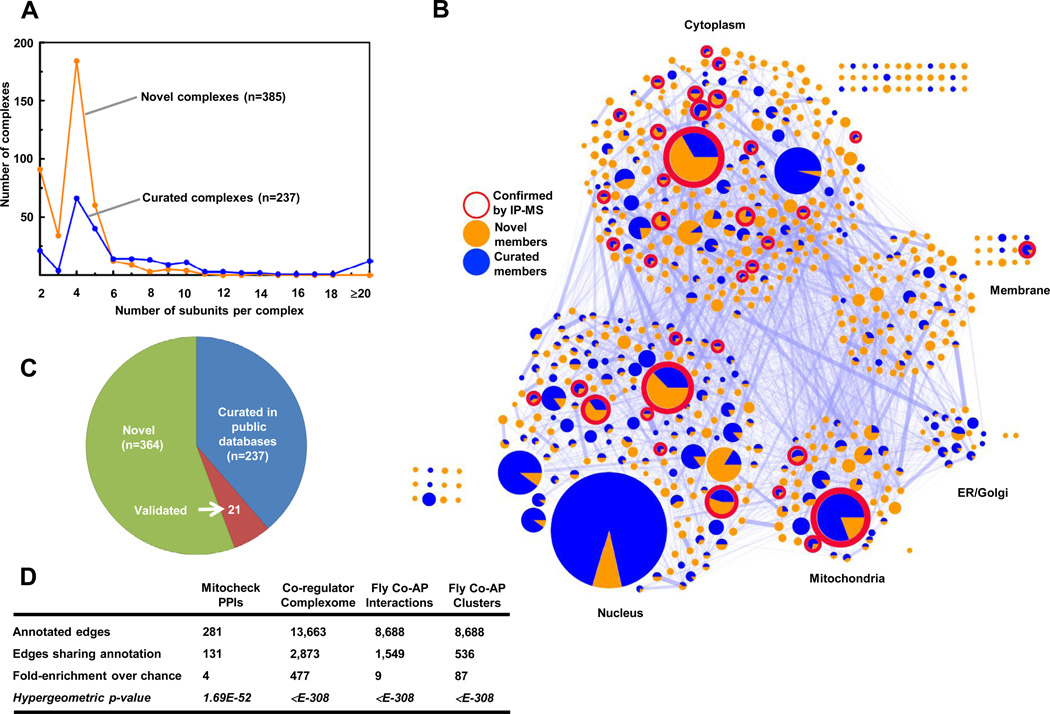
In order to define complex membership, we partitioned the high-confidence probabilistic physical interaction network using the cluster growth algorithm ClusterONE (Nepusz et al., 2012), which outperformed other clustering methods on the denoised PPI network (Table S5). In total, the clustering predicts 622 discrete putative complexes encompassing 2,634 distinct proteins (Table S3). Complex membership size distribution approximated an inverse power law with a median of 4 subunits (Figure S4A). The majority (62%; 385/622) of the complexes have not been annotated (i.e., only 237 are currently curated in a public database like CORUM; Figures 3A, C). Although the fraction of curated components varies, we also recapitulated 258 previously reported complexes (Figure 3C), including several well-known membrane-associated complexes, such as the Coat Protein I and II (COPI/II) vesicle transport complexes which shuttle cargo between the Golgi and endoplasmic reticulum. Strikingly, most (67%; 335) of the 500 smaller putative complexes with 5 or fewer components, including the bulk (74%; 83) of the 112 predicted heterodimers, have never been curated before (Figure 3C).
Figure 3. Global validations of the map of high confidence human protein complexes.
A- Complex size distribution of the 622 inferred complexes. B- Network of predicted human protein complexes proportioned according to subunit number and displaying existing curations, validation status by AP/MS (Malovannaya et al., 2011), and PPI connectivity (proportioned edge width). C- Proportions of annotated complexes in public repositories (CORUM, PINdb, REACTOME, HPRD) or independently experimentally-verified. D- Enrichment analysis showing overlap with large-scale APMS datasets generated for human (Hutchins et al., 2010; Malovannaya et al., 2011) and (via orthology) fly (Guruharsha et al., 2011). See also Table S3.
Both independent experimental validation based on more traditional immunoprecipitation or co-affinity purification methods and orthology mapping support at least 21 of these putative complexes (i.e., not in any reference database) (Table S3; see Supplemental Information for details). For example, Guruharsha et al. recently reported 299 co-complex interactions based on pull-down experiments of 43 affinity-tagged human proteins present in 41 of our complexes, of which 143 interactions map precisely to our predicted complexes, representing a 47.8% validation rate (which may be an underestimate as Guruharsha et al. do not report human interactions that fall outside the fly interologs examined in their study). Likewise, the results of Malovannaya et al. , who used large-scale immunoprecipitation to isolate native human protein complexes, show excellent agreement to 123 of our complexes (i.e., Benjamini-corrected hypergeometric p ≤ 0.05), including 42 (34%) of our complexes that are not curated in CORUM (Figure 3B and Table S3). Figure 3D summarizes the highly significant overlap of our inferred complexes with these fully independent datasets, with enrichments ranging from 4- to 477-fold over chance, thus broadly and systematically validating our network of derived human protein complexes.
By design, insoluble membrane-associated (hydrophobic) protein complexes were largely missed in this study, while the proteins assigned to complexes had a higher average transcript abundance (Figure S2A–B). Moreover, in an effort to control the false positive rate, our conservative clustering algorithm, ClusterONE, underweighted small clusters of size 2 or 3 for lack of sufficient association evidence, likely contributing to the prominence of complexes with 4 subunits in Figure 3A. But we did not observe any significant bias toward negative (pI ≤ 7) or positive (pI ≥ 7) charge as compared to complexes curated in CORUM (Figure S4B).
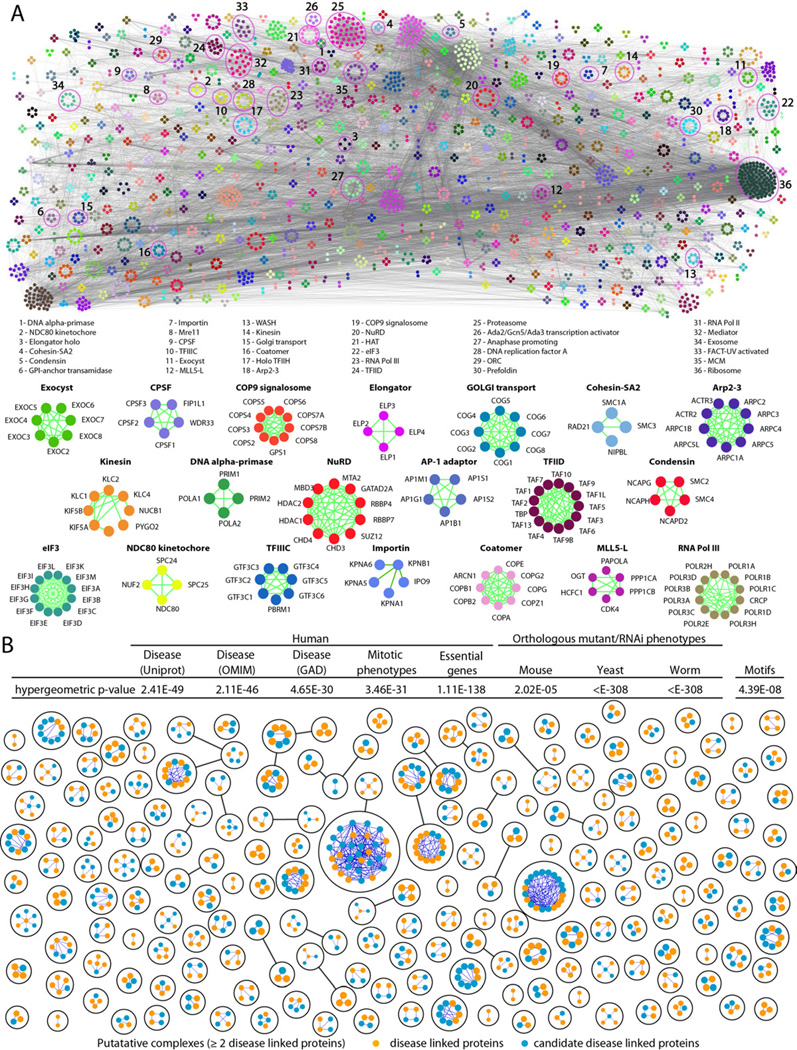
Figure 4 shows the broad functional diversity of the predicted complexes (a navigable map is available online for close visualization of individual clusters and their supporting cocomplex interactions). Consistent with biological expectation (Hartwell et al., 1999; Lage et al., 2007; Oliver, 2000; Vidal et al., 2011), the subunits of the complexes were significantly enriched for related biological functions, transcriptional regulatory motifs, and pathological processes (Figure 4B, inset table). Compared to the entire set of identified proteins, the clustered proteins also showed enrichment for post-translation modifications linked to cellular regulation, like acetylation (Benjamini-corrected p ≤ 10−41) and phosphorylation (p ≤ 10−5). Many of the complexes are linked to core cellular processes, such as mRNA splicing (p ≤ 10−15) or transcription (p ≤ 10−5), that either are essential in human (p ≤ 10−138) or which have RNAi-induced phenotype in cell culture (e.g. cell division arrest, p ≤ 10−31) or are associated, via orthology, with similar mouse, yeast or worm mutant phenotypes (Figure 4B, inset table; see Table S4 for details).
Figure 4. Global map of high confidence human protein complexes.
A- Schematic of the global network of inferred human soluble protein complexes (colored by membership), with representative examples and supporting PPI highlighted. B- Putative complexes with 2 or more components with human disorder associations annotated in UniProt (The UniProt Consortium, 2011), Online Inheritance of Man (OMIM)(Hamosh et al., 2005) or the Genetic Association Database (GAD)(Becker et al., 2004). Inset table shows highly significant interaction overlap (i.e., shared annotated edges) with phenotypic datasets that reveals protein subunits of the same predicted human complex tend to exhibit similar disease and genetic associations in human populations (see Extended Experimental Procedures), RNAi phenotypes in cell culture (Neumann et al., 2010), mutational and RNAi phenotypes in other species (via orthology), and shared transcriptional regulatory motifs (Xie et al., 2005). See also Figure S4C, and Table S4.
Clinical and biological implications of the reconstructed human protein complexes
Consistent with this strong tendency for proteins in the same complex to be affiliated with similar mutational and RNAi phenotypes, subunits of the predicted human protein complexes were much more likely than chance (p ≤ 10−46) to have links to a documented clinical pathology (Figure 4B, see Table S4 for details), with disease-associated proteins distributed broadly amongst the complexes (Figure 4B and Figure S4C). Closer examination of the interaction sub-networks comprising known human disease genes with genes that currently lack annotation or which have not previously been associated with any human disorders (Figure 4B) highlights the utility of the map.
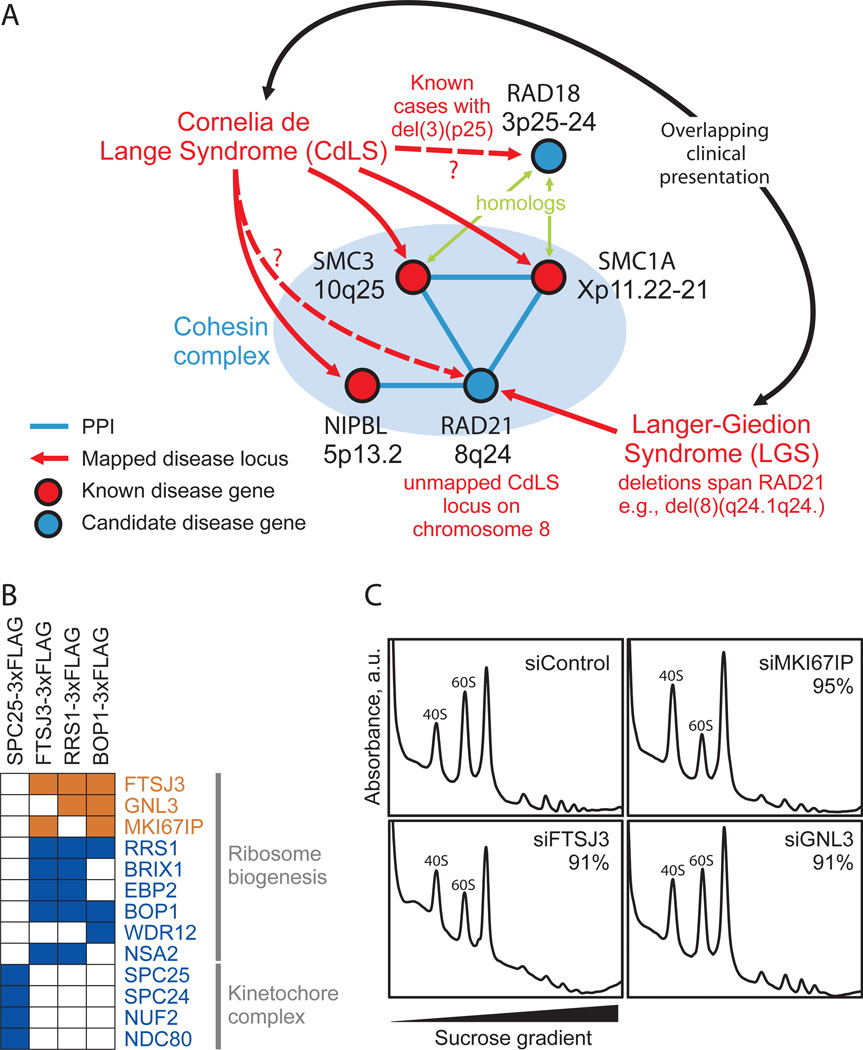
One such example is shown in Figure 5A, illustrating the case of the human developmental disorder Cornelia de Lange syndrome (CdLS). Mutations in three subunits of the cohesin complex (SMC1A, SMC3, NIPBL) have been linked to CdLS (Pie et al., 2010), implicating an additional component (RAD21) as a candidate CdLS locus, and consistent with at least one unmapped CdLS locus residing on chromosome 8 (DeScipio et al., 2005). The link to RAD21 provides a likely explanation for the occasional overlap of Langer-Giedion Syndrome (LGS) clinical presentation with CdLS, as all LGS patients are at least partially defective for RAD21 [see e.g. (McBrien et al., 2008; Wuyts et al., 2002)]. Similarly, RAD18, a homolog of SMC3 and SMC1A, may play a role in CdLS, consistent with unmapped CdLS deletions within chromosome 3p25 (DeScipio et al., 2005). Reports coinciding with the preparation of this manuscript confirm that RAD21 mutations do indeed lead to a CdLS-like syndrome (Deardorff et al., 2012), supporting the use of the complex map to prioritize promising candidate genes for human diseases.
Figure 5. Membership in complexes predicts protein function and disease associations.
A- Three of four proteins mapped to the cohesin complex account for roughly half of cases of the human congenital disorder Cornelia de Lange syndrome (Pie et al., 2010), implicating the fourth component, RAD21, as a candidate disease gene. This association may explain similarities in clinical presentation between CdLS and Langer-Giedion syndrome, as the latter patients routinely harbor RAD21 deletions, e.g. (McBrien et al., 2008; Wuyts et al., 2002). B- Confirmation of ribosome biogenesis candidate (orange) associations with annotated components (blue) by AP/MS analysis of tagged proteins (top). Colored squares indicate validation (see Extended Experimental Procedures). C- Polysome profiling after siRNA targeting in tissue culture supports functional roles in ribosome biogenesis for three candidate proteins. Knockdown of MKI67IP, FTSJ3, and to a lesser extent GNL3, results in 60S ribosomal subunit biogenesis defects manifested by a reduced ratio of free 60S to 40S ribosomal subunits during gradient sedimentation as compared to control. Percentages indicate siRNA knockdown efficiency as measured by qRT-PCR.
Similarly, participation in the same complex suggests shared functions; the map can thus be used to predict new biochemical functions for proteins and other types of functions. We experimentally validated one such case for a ribosome-associated sub-complex containing BOP1, RRS1, GNL3, EBP2, FTSJ3, and MK1671P, first confirming the interactions by affinity tagging/purification and mass spectrometry (Figure 5B). BOP1, EBP2, and the yeast ortholog of RRS1 are known to participate in maturation of the large 60S ribosomal subunit, suggesting the other factors likewise engage in ribosome assembly, consistent with the nucleolar localizations of GNL3, FTSJ3, and MKI67IP. Supporting a role in ribosome biogenesis, short interfering RNA knockdowns of FTSJ3, MKI67IP and, to a lesser extent, GNL3 perturbed 60S formation in cell culture, decreasing the ratio of free 60S to 40S subunits (Figure 5C). Taken together, these data support roles in ribosome biogenesis for these proteins, and confirm the utility of the map for identifying biological functions.
Conservation of human protein complexes
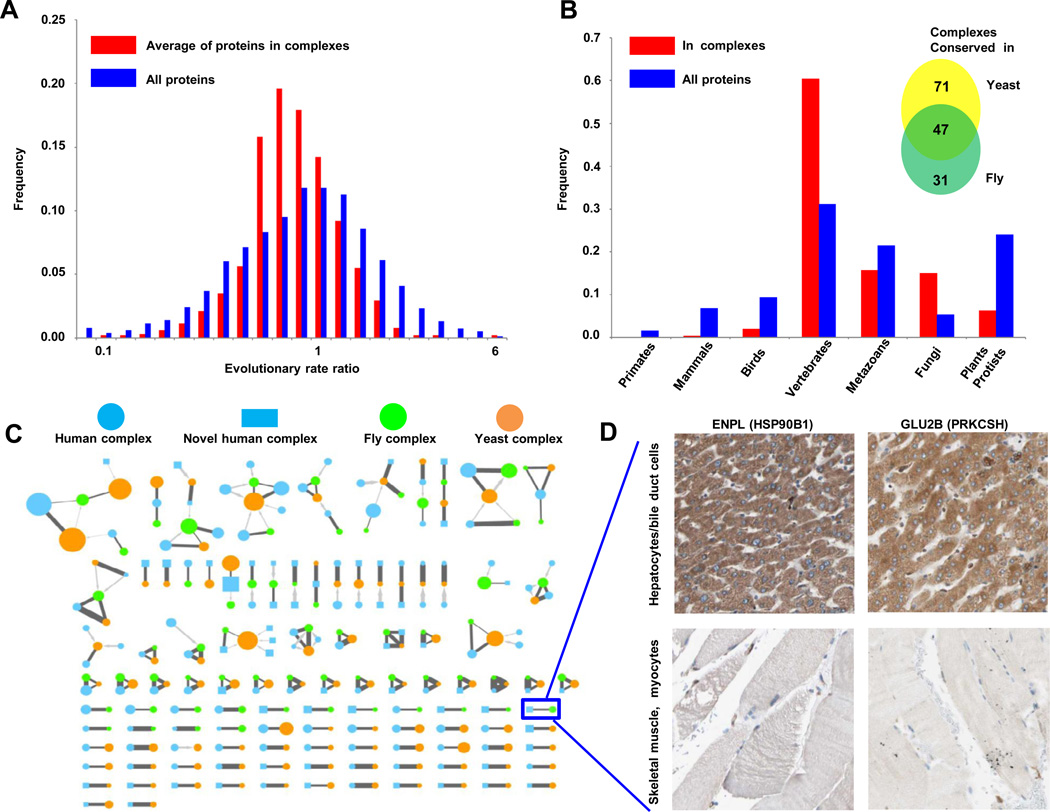
Estimates based on sequence similarity across orthologs indicate that the components of the complexes we detect are generally more ancient and have higher conservation on average than most human proteins (Figure 6A; see Table S3 for details). Using orthology relationships derived from well established sources and calculating evolutionary rates and ages for all human proteins as a base distribution for gauging the emergence of complexes (see Extended Experimental Procedures), many complexes appear to be quite ancient and slowly evolving (Figure 6B). Strikingly, however, most (60%; 376/622) human complexes likely arose with vertebrates i.e., orthologs not present in invertebrates or fungi (Table S3). Hence, our analyses suggest a major shift/expansion in the ancestral protein interaction network coincident with the emergence of vertebrates.
Figure 6. Evolutionary conservation of protein complexes.
A- Components of predicted human complexes evolved more slowly, calculated as the average of evolutionary rate ratios, compared to the entire set of expressed proteins (see Extended Experimental Procedures). B- Pronounced spike in number of complexes originated with the emergence of vertebrates. X-axis shows increasingly inclusive orthologous groups in the phylogeny of eukaryotes. C- Human complexes conserved in fly (Guruharsha et al., 2011), and yeast (Babu et al., 2012)(see Table S3 and Extended Experimental Procedures). Nodes represent complexes (human, blue; fly, green; yeast, orange), with size proportional to subunit number. Reciprocal best matches shown as dark grey edges, non-reciprocal as lighter grey directed edges, with edge thickness proportional to Sorensen-Dice overlap of complex members. Human complexes absent from public databases (putative complexes) are drawn as rectangles, the remaining as circles. D- Similar tissue-specific expression patterns support a functional association between interacting proteins ENPL and GLU2B, whose orthologs were reported to interact in fly (Guruharsha et al., 2011). Panels show representative antibody staining in normal tissue biopsies collected and reported by the Human Protein Atlas (Uhlen et al., 2010)(www.proteinatlas.org). See also Figure S3 and Table S3.
Given the availability of experimentally-derived networks of fly and yeast protein complexes, we could directly examine the evolutionary conservation of protein complexes across animals by comparing our network of human complexes with the extensive maps of 556 fly protein complexes recently reported for D. melanogaster (Guruharsha et al., 2011) and 720 yeast protein complexes documented for S. cerevisiae (Babu et al., 2012). Roughly one quarter (24%; 149/622) of the predicted human protein complexes showed statistically significant overlaps with complexes reported for these models (inset, Figure 6B; see Table S3 for details), with half of the subunits having clear orthologs (Figure 6C); the remaining components presumably represent genuine differences or incomplete orthology annotations.
The functional significance of un-annotated ancestral human complexes supported by conservation in yeast or fly (Table S3 and Figure 6) warrants further investigations. At least one such complex, a multi-subunit tRNA-splicing ligase (Popow et al., 2011), was characterized recently. The interaction between DDX1 and C14orf166 was detected at high confidence both in our dataset (probability score 0.899) and in the Guruharsha et al. fly co-complex data, while the other respective associated complex subunits likewise show significant overlap (Benjamini-corrected P-value 1.1×10−7). Additional examples of complex conservation are similarly supported by independent experimental evidence, e.g. such as the matching tissue specificities of the putatively interacting proteins endoplasmin and glucosidase 2β (Figure 6D), which form an uncharacterized complex conserved in both the fly and human maps.
Functional enrichment analysis of ancient complexes in comparison to vertebrate-specific ones also reveals intriguing biological trends. For example, we expected ancient, core cellular functions to be depleted among vertebrate-specific complexes. Consistent with this expectation, we find proteins associated with the ribosome (p ≤ 10−67, 113 proteins) and RNA polymerase II (p ≤ 10−27, 45 proteins) to be highly enriched only among conserved complexes. However, we also observe several notable variations from this hypothesis. For example, compared to the genomic background, mitochondrial proteins are more highly enriched among proteins assigned to vertebrate complexes than among those assigned to conserved complexes: 159 vertebrate proteins have a mitochondrial GO BP annotation (p ≤ 10−31), vs. only 81 proteins assigned to conserved complexes (p ≤10−5). Similarly, proteins annotated as being part of the splicing apparatus are enriched in both conserved (p ≤ 10−33 ; 63 proteins) and vertebrate complexes (p ≤ 10−11, 43 proteins), which is consistent with an ancient function gaining additional complexity in vertebrates (e.g. increased alternative splicing). Our study therefore offers a unique perspective into the functional conservation and diversification of protein complexes across animals.
Protein abundance, ubiquity, and complex subunit stoichiometries
Consistent with the documented origins of the HeLa and HEK293 cells analyzed in this study, the complexes we identified were significantly enriched for epithelial markers (p ≤ 10−183; UniProt tissue annotations). Explicit comparison of results across the two cell lines used in this study provided little evidence for tissue-specific or cell-type specific complexes (see Supplementary Information). Most proteins were detected in both cell line fractionations, consistent with the similar protein and mRNA expression patterns observed in these cell lines (Figure S1), while the few proteins detected uniquely in one cell line or the other did not preferentially assort into tissue-specific complexes (Figure S2). The vast majority of complex components are universally expressed in 11 cancer cell lines (Geiger et al., 2012) (Figure S3A) and show high and largely invariant expression in an mRNAseq study of 16 normal human tissues (EBI accession no. E-MTAB-513) (Figure S3B). Indeed, complex subunits are considered near ubiquitous (p ≤ 10−11; PIR tissue specificity annotations), and are expressed in the top quartiles of 1,045 of 7,067 neoplastic and normal tissue CGAP EST libraries (1% FDR), including normal kidney (p ≤ 10−39), muscle (p ≤ 10−20), liver (p ≤ 10−12), brain (p ≤ 10−20), vascular (p ≤ 10−30), bone (p ≤ 10−15), and embryonic tissue (p ≤ 10−31). Consistent with this, genes encoding complex subunits also tend to share common upstream transcriptional regulatory motifs (p ≤ 10−8) (Figure 4B, inset table). Proteins mapped to complexes showed no major bias in abundance over the complete set of human proteins identified by mass spectrometry (Figure 1D).
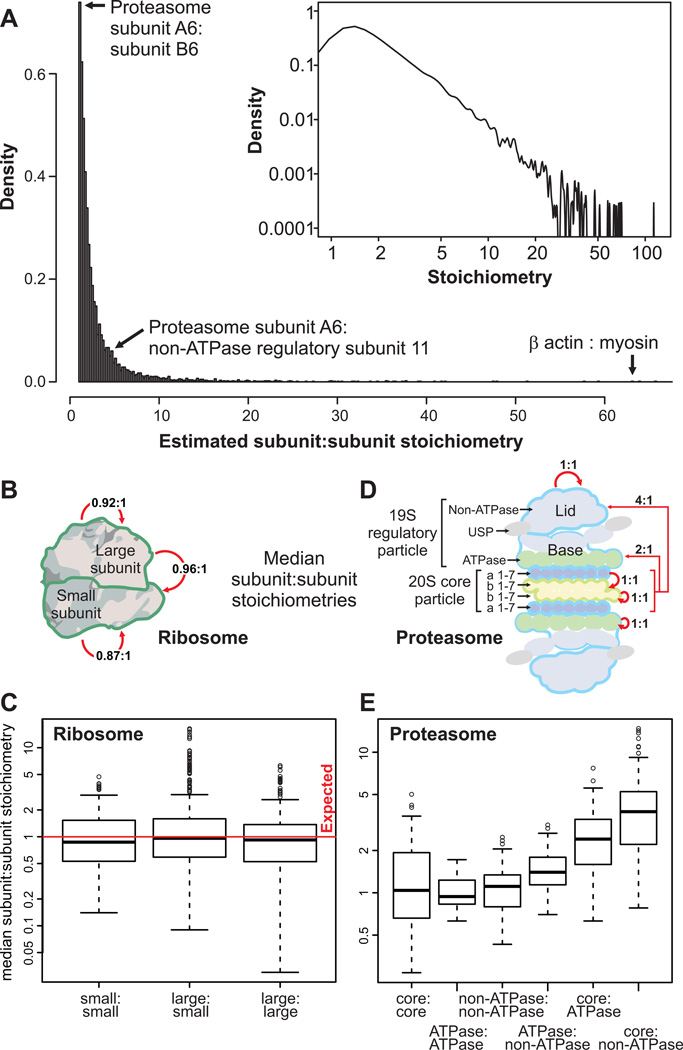
The pervasiveness of ubiquitously expressed protein complexes argues strongly for broad relevance to basic human cell biology. Although often co-expressed, the subunit stoichiometries of human protein complexes in vivo are largely unknown, and have never been systematically measured globally. Since all reconstructed complexes are supported by the same set of extensive experimental mass spectrometry data, we could estimate subunit stoichiometries based on the ratios of recorded spectral counts after correcting appropriately for protein size and composition (see Extended Experimental Procedures). While only approximate ratios were inferred and peaked around ~1:1 (Figure 7A), such as between known ribosomal subunits (Figures 7B, C), the results highlight intriguing deviations in subunit abundance (Table S2). An example drawn from the proteasome is illustrative: whereas the median stoichiometry of core alpha and beta enzymatic subunits is close to the expected 1:1 ratio, the median of stoichiometries of core to non-ATPase regulatory subunits deviated significantly at ~4:1 (Mann-Whitney p ≤ 10−16; Figures 7D, E). Hence, these data suggest a rich source of information about the physical organization of human proteins.
Figure 7. Protein complex stoichiometries.
A- Overall distribution of derived intra-complex component stoichiometries B, C- Estimated subunit stoichiometries within and between proteins of the large and small ribosome subunits agree on average with the expected 1:1 ratio. Boxes summarize first quartile, median and third quartiles, whiskers represent +/− 1.5 IQR and circles outliers. D, E- Estimated protein subunit stoichiometries within and between proteasomal proteins. Intra-subunit stoichiometries within the core, ATPase, or nonATPase regulatory subunits agree well with the expected 1:1 ratio, but stoichiometries observed between these complexes deviate significantly from 1:1 (ATPase:non-ATPase, Mann-Whitney p ≤ 10−3; core:ATPase, p ≤ 10−12; core:non-ATPase, p ≤ 10ȡ16). See also Table S2.
DISCUSSION
The biochemically-based interaction data obtained in this integrative proteomic study have enabled the identification of both 364 previously unannotated protein complexes (i.e., predicted complexes with no statistically significant match to complexes in public databases) encompassing 1,278 human proteins, many of which are linked to human disease, as well as unexpected components and interactions for well-studied, widely-conserved nuclear and cytoplasmic protein machineries, such as ribosome biogenesis, with clear biological implications. Most of the high-confidence protein interactions provided in this resource have not been previously reported in public interaction databases and hence motivate mechanistic investigations of specific biological systems.
Prior to this work, experimental knowledge regarding soluble protein complex membership in human cells has generally been ad hoc or focused on specific sub-cellular systems. Our relatively unbiased integrative approach, wherein biochemical evidence (cofractionation) of soluble native macromolecules was combined with genomic inferences (imputed functional associations) provides an inclusive snapshot of human protein complexes present under a standardized cellular context, thus serving as a reference against which future process- or cell-type specific or dynamic interaction datasets can be compared.
Information gleaned from orthology proved to be an important resource in separating true pairwise interactions from putative false positives, and in turn could reasonably be expected to bias our results toward conserved complexes. In fact, although we do find conserved complexes as expected, we also find a majority that are not conserved (in fly and yeast) and which seemingly have arisen with vertebrates (i.e., Figure 6B). The slower rate of evolution of the subunits we report for our protein complexes is also a feature of other human PPI networks, such as in CORUM, and thus our predictions of broad complex conservation, albeit incomplete, are not just artifacts of our methodology.
The fact we detected little evidence of tissue specificity for most of the derived human protein complexes, and few cell-type-specific components, likely reflects under-sampling by our mass spectrometry procedures, a common limitation of LC-MS/MS. At the level of predicted PPI (which are derived from multiple biochemical fractions), differences in the proteomic profiles generated for the two cell lines lie within the variance observed between biological replicates of the same cell line (Figure S1 and S2). Yet it is clear that differential interactomes and the contextual re-wiring of PPI networks are major determinants of cell behavior and phenotypes. The complexes we report undoubtedly undergo differential re-wiring in response to environmental, physiological, developmental or disease states. With further refinements to our experimental procedures, our interaction mapping strategy has the potential to interrogate changes in interaction space in a systematic manner in the future.
To enable exploitation of these data by the scientific community, we have generated a dedicated web database of human protein complexes (http://human.med.utoronto.ca) that contains all the data generated in this study in an easily navigated format. These include all of the supporting information for each of the pairwise protein interactions obtained through integration of our co-fractionation data with public genomic evidence, a list of the 5,584 proteins detected in each of the 1,163 biochemical fractions collected, and the subunit composition of the 622 putative protein complexes obtained through clustering of our generated high-confidence interaction network. This 'first pass' draft of the soluble, stably-associated human protein 'complexome' provides a glimpse into the global physical molecular organization of human cells, which is likely to be perturbed in pathological states.
EXPERIMENTAL PROCEDURES
Cell Culture and Extract Preparation
HeLa S3 (ATCC cat# CCL-2.2) and HEK293 (ATCC cat# CRL-1573) soluble nuclear and cytoplasmic protein extracts were prepared by conventional methods (see Extended Experimental Procedures). Prior to fractionation, lysates were treated with 100 U/mL Benzonase (Novagen Inc.) to remove nucleic acids and clarified by centrifugation to remove debris.
Biochemical Fractionation and Proteomic Analysis
We performed weak anion-exchange and mixed-bed ion exchange, both with and without a heparin pre-column to enrich for nucleic-acid binding proteins. In total 1,095 chromatography fractions were collected (see Extended Experimental Procedures). Isoelectric focusing was carried out on a MicroRotofor Liquid-Phase IEF cell (Bio-Rad) according to the manufacturer’s protocol, with 40 fractions collected across a pH range. Sucrose density gradient centrifugation was performed as previously described (Ramani et al., 2008), with 28 fractions collected.
Proteins were acid precipitated and trypsin digested, and the peptide mixtures fractionated and sequenced using nanoflow liquid chromatography–electrospray–tandem mass spectrometry. Spectra were collected on a LTQ linear ion trap (ThermoFisher Scientific )(majority) or LTQ Orbitrap Velos hybrid mass spectrometer and searched against a UniProt human target-decoy sequence database using SEQUEST (Eng et al., 2008)(see Extended Experimental Procedures). The LC-MS/MS identifications were filtered to a 1.0% protein and peptide theoretical FDR.
Bioinformatics Analyses
Protein co-fractionation networks were scored by correlation analysis (Pearson correlation, weighted cross-correlation, co-apex) based on the protein spectral counts recorded across each set of fractions (see Extended Experimental Procedures). Weighted networks were likewise constructed based on functional evidence reported in HumanNet (Lee et al., 2011) omitting human protein interaction data to minimize circularity that might bias our association predictions. A co-evolution network (Tillier and Charlebois, 2009) based on correlated evolutionary rates was built to account for additional associations not covered in HumanNet.
For the machine-learning classifier, we used the Fast Random Forest implementation in Weka (see Extended Experimental Procedures) to integrate all generated networks. Cross-validated decision trees were learned and benchmarked using independent training and test sets of CORUM reference complexes (Ruepp et al., 2010). We de-noised the network by using a diffusion procedure to delete interactions lacking network topology support, and by calibrating the diffused interaction scores with Gene Ontology (Cellular Component) normalized semantic similarity scores (see Extended Experimental Procedures).
Clusters were defined using the ClusterONE algorithm with parameter settings chosen to yield the highest Maximum Matching Ratio (Nepusz et al., 2012) between the predicted complexes and set of cluster-training complexes (see Extended Experimental Procedures). Stoichiometries calculation is shown in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Proteomic profiling identifies 13,993 physical interactions among 3,006 proteins
The interactions lead to a map of 622 complexes, many of which are highly conserved
The map reveals many unexpected biological and disease associations
ACKNOWLEDGEMENTS
We thank R. Isserlin, Z. Ni, H. Guo, D. Merico and A. Alpert for technical assistance, and J. Parkinson, G. Bader, A. Wilde and J. Greenblatt for critical suggestions. PCH was a recipient of a University of Toronto Open Fellowship, TN was supported by the Newton International Fellowship Scheme of the Royal Society, AE is an Ontario Research Chair, and SJW is a Canada Research Chair Tier 1. This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BB/F00964X/1; BB/K004131/1) and the Royal Society (NF080750) to AP, from the Canada Institutes of Health Research (MOP#82940) and the SickKids Foundation to SJW, from the National Institutes of Health, National Science Foundation, Cancer Prevention Research Institute of Texas, and Welch (F1515) and Packard Foundations to EMM, and from the Ontario Ministry of Research and Innovation to AE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, 5 figures, 6 tables, Supplemental References, and can be found with this article online at:
REFERENCES
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BDM, Vizeacoumar FJ, Burston HE, Snider J, Phanse S, et al. Interaction Landscape of Membrane Protein Complexes in Saccharomyces cerevisiae. Nature. 2012 doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Monnich M, Yan Y, Xu W, Gil-Rodriguez MC, et al. RAD21 Mutations Cause a Human Cohesinopathy. Am J Hum Genet. 2012;90:1014–1027. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeScipio C, Kaur M, Yaeger D, Innis JW, Spinner NB, Jackson LG, Krantz ID. Chromosome rearrangements in cornelia de Lange syndrome (CdLS): report of a deR3)t(3;12)(p25.3;p13.3) in two half sibs with features of CdLS and review of reported CdLS cases with chromosome rearrangements. Am J Med Genet A. 2005;137A:276–282. doi: 10.1002/ajmg.a.30857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Fischer B, Grossmann J, Maccoss MJ. A fast SEQUEST cross correlation algorithm. J Proteome Res. 2008;7:4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human proteinprotein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. A Protein Complex Network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Havugimana PC, Wong P, Emili A. Improved proteomic discovery by sample pre-fractionation using dual-column ion-exchange high performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847:54–61. doi: 10.1016/j.jchromb.2006.10.075. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hu P, Janga SC, Babu M, Diaz-Mejia JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Gerstein M. Analyzing protein function on a genomic scale: the importance of gold-standard positives and negatives for network prediction. Curr Opin Microbiol. 2004;7:535–545. doi: 10.1016/j.mib.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T, Rahman K, Radulovic D, Cox B, Rossant J, Emili A. PRISM, a generic large scale proteomic investigation strategy for mammals. Mol Cell Proteomics. 2003;2:96–106. doi: 10.1074/mcp.M200074-MCP200. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak AB, Ni Z, Hewel JA, Chen GI, Zhong G, Karamboulas K, Blakely K, Smiley S, Marcon E, Roudeva D, et al. A lentiviral functional proteomics approach identifies chromatin remodeling complexes important for the induction of pluripotency. Mol Cell Proteomics. 2010;9:811–823. doi: 10.1074/mcp.M000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- McBrien J, Crolla JA, Huang S, Kelleher J, Gleeson J, Lynch SA. Further case of microdeletion of 8q24 with phenotype overlapping Langer-Giedion without TRPS1 deletion. Am J Med Genet A. 2008;146A:1587–1592. doi: 10.1002/ajmg.a.32347. [DOI] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. Guilt-by-association goes global. Nature. 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- Pesquita C, Faria D, Falcao AO, Lord P, Couto FM. Semantic similarity in biomedical ontologies. PLoS Comput Biol. 2009;5:e1000443. doi: 10.1371/journal.pcbi.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pie J, Gil-Rodriguez MC, Ciero M, Lopez-Vinas E, Ribate MP, Arnedo M, Deardorff MA, Puisac B, Legarreta J, de Karam JC, et al. Mutations and variants in the cohesion factor genes NIPBL, SMC1A, and SMC3 in a cohort of 30 unrelated patients with Cornelia de Lange syndrome. Am J Med Genet A. 2010;152A:924–929. doi: 10.1002/ajmg.a.33348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, Martinez J. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- Ramani AK, Li Z, Hart GT, Carlson MW, Boutz DR, Marcotte EM. A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol Syst Biol. 2008;4:180. doi: 10.1038/msb.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM. Probabilistic model of the human protein-protein interaction network. Nat Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes-2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier ER, Charlebois RL. The human protein coevolution network. Genome Res. 2009;19:1861–1871. doi: 10.1101/gr.092452.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels HJ, Vogel RO, van den Heuvel L, Smeitink JA, Rodenburg RJ, Nijtmans LG, Farhoud MH. LC-MS/MS as an alternative for SDS-PAGE in blue native analysis of protein complexes. Proteomics. 2009;9:4221–4228. doi: 10.1002/pmic.200900157. [DOI] [PubMed] [Google Scholar]
- Wuyts W, Roland D, Ludecke HJ, Wauters J, Foulon M, Van Hul W, Van Maldergem L. Multiple exostoses, mental retardation, hypertrichosis, and brain abnormalities in a boy with a de novo 8q24 submicroscopic interstitial deletion. Am J Med Genet. 2002;113:326–332. doi: 10.1002/ajmg.10845. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Nepusz T, Paccanaro A. Improving GO semantic similarity measures by exploring the ontology beneath the terms and modelling uncertainty. Bioinformatics. 2012;28:1383–1389. doi: 10.1093/bioinformatics/bts129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.