Abstract
Insulators prevent promiscuous gene regulation by restricting the action of enhancers and silencers. Recent studies have revealed a number of similarities between insulators and promoters, including binding of specific transcription factors, chromatin-modification signatures and localization to specific subnuclear positions. We propose that enhancer-blockers and silencing barrier-insulators might have evolved as specialized derivatives of promoters and that the two types of element use related mechanisms to mediate their distinct functions. These insights can help to reconcile different models of insulator action.
Eukaryotic chromosomes are a mosaic of accessible euchromatic and inaccessible heterochromatic domains. Gene regulation occurs in the context of such domains and is mediated by elements such as enhancers and silencers. Insulators are DNA elements that insulate genes located in one chromatin domain from promiscuous regulation by enhancers or silencers in neighbouring domains (FIG. 1). Understanding insulator function is therefore necessary for a full understanding of gene regulation.
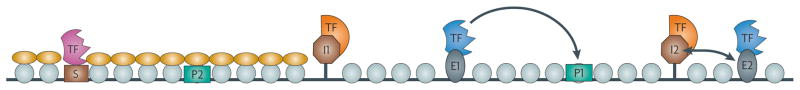
Figure 1. Chromatin domains and regulatory elements.
The relationships among silencers, enhancers, promoters and insulators are shown. Light blue circles represent nucleosomes and yellow ovals represent silencing proteins, such as heterochromatin protein 1 (HP1) and Sir3. Silencer elements (S) are sites of initiation of heterochromatin, which spreads and encompasses promoters (P2 in the diagram), silencing transcription. The I1 insulator functions to restrict the spread of heterochromatin. An enhancer (E1) that is present in an active chromatin domain flanked by insulators (I1 and I2) and that is bound by a transcription factor (TF) is able to communicate with a promoter (P1) in the same domain, whereas another enhancer (E2) is unable to communicate with promoter P1 because of an intervening insulator (I2).
Here, we explore the structural, functional and mechanistic relationships between insulators and another, better-understood class of gene regulatory element, promoters. Recent studies in systems as diverse as yeast, Drosophila melanogaster and vertebrate cells have shown many similarities between different types of insulators as well as between insulators and promoters. Here, we review these data and the new insights that they provide into the molecular steps involved in insulation and the role of compartmentalization in gene insulation. We propose that insulators may have evolved from a class of promoters that bind specific transcription factors with specific molecular properties, utilizing conserved mechanisms to mediate different forms of insulation.
Silencers, enhancers and insulators
Transcriptionally silenced heterochromatin initiates at either repetitive DNA or specific sequences called silencers to silence genes1,2. The silencing elements recruit either the RNAi machinery or specific transcription factors to initiate heterochromatin formation. Heterochromatin contains hypoacetylated histones in all species studied, and in many species it also contains histones and DNA methylated at specific residues. The initiating elements enlist enzymes that modify the chromatin to create binding sites for repressor proteins, leading to the recruitment of these proteins and the subsequent spread of heterochromatin over several tens to hundreds of kilobase pairs (FIG. 2). Heterochromatic loci cluster at the nuclear periphery, creating silencing foci rich in repressor proteins. Such clustering increases the stability of transcriptional silencing, most likely by a ‘pinball effect’ in which a repressor protein released from one DNA site is immediately bound by a neighbouring site.
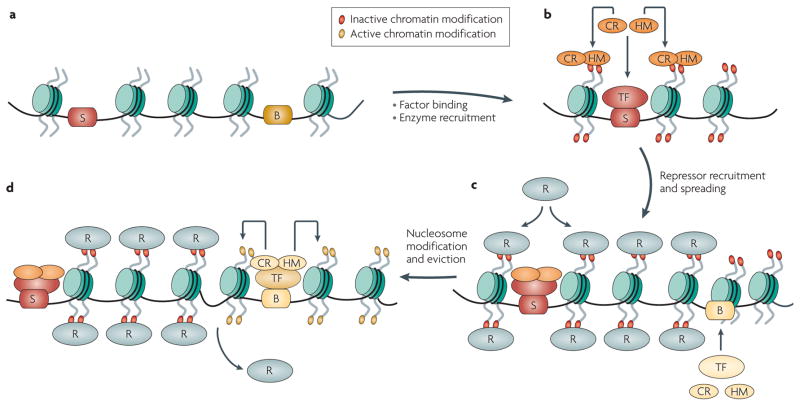
Figure 2. Establishment of silencing and barrier activity.
a,b|Silencing elements (S) recruit specific transcription factors (TF), which in turn recruit chromatin remodellers (CR) and histone-modifying enzymes (HM) that cooperate to modify chromatin and create binding sites in nucleosomes for repressor proteins. c,d|The binding and spreading of the repressor proteins (R) along the chromatin fibre results in the formation of a silenced heterochromatic domain. Barrier elements (B) bind a distinct set of transcription factors that recruit enzyme complexes, which modify nucleosomes with ‘active’ histone marks and evict nucleosomes. This creates a discontinuity in the chromatin fibre and thereby restricts the spread of repressor proteins.
Promoters are sites of accurate initiation of transcription mediated by transcription factors and various chromatin-remodelling and -modifying enzymes3. These proteins replace canonical histones with specific variants, evict nucleosomes and modify histones to enable transcription initiation. Enhancers positively regulate promoters in a distance- and orientation-independent manner4,5 and have binding sites for ubiquitous and tissue- or cell-specific transcription factors, which mediate activation by increasing the probability and/or rate of transcription initiation from a promoter by opening the chromatin domain and/or aiding in the recruitment and release of the transcription machinery. Enhancers are usually separated from their promoters by thousands of base pairs, with some even residing on separate chromosomes, but during gene activation these elements are in close three-dimensional (3D) proximity in the nucleus, clustering at RNA polymerase II (RNAPII) foci that are often referred to as transcription factories or active chromatin hubs6. Several models have been proposed to explain how an enhancer finds and communicates with its promoter4. Once an enhancer finds its promoter, the two elements interact directly through enhancer- and promoter-bound factors4, leading to gene activation (FIG. 3a). An enhancer only interacts with one promoter at a time, but the association is reversible7 and the strength of these interactions is likely to affect the stability and duration of this communication, and consequently transcription from the promoter. Furthermore, whereas enhancers can activate promoters over long distances, enhancers generally activate the nearest promoter preferentially.
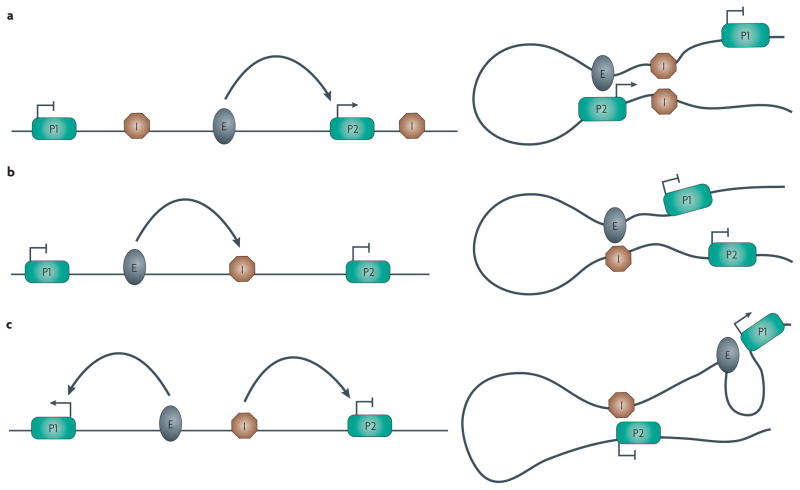
Figure 3. Interactions among enhancers, promoters and enhancer-blocking insulators.
Potential interactions between regulatory elements are shown. a|A pair of enhancer-blocking insulators (I) interact pairwise with each other, placing the enhancer (E) in a loop with a promoter (P2) to enable transcription activation, while isolating a second promoter (P1) in a separate loop. b|An enhancer-blocker functions by directly sequestering an enhancer, therefore disrupting its ability to interact with a promoter. c|The enhancer-blocker can also interact directly with a promoter, preventing it from interacting with the enhancer, which can freely interact with a second promoter.
The lack of promiscuity in long-range regulation by silencers and enhancers suggests that stringent mechanisms maintain the specificity of their effects. This specificity is likely to be mediated partly by complementary interactions between proteins at these elements and partly by the sequestration of sequences into silencing centres or transcription factories. However, important additional specificity is imposed by insulators, which restrict the activity of enhancers and silencers (FIGS 2,3). Several examples of chromosomal translocations that delete insulators and fuse domains and result in new patterns of gene expression have been identified, as have mutations in insulators that cause developmental defects8. Insulators that disrupt communication between the enhancer and its promoter when positioned between the two are called enhancer-blockers (FIG. 3b,c), and insulators that are located between a silencer and a promoter and protect the promoter from silencing are called barriers9,10 (FIG. 2). Although some native insulators possess one or the other activity, there are examples of insulators, such as the mouse short interspersed element (SINE), the chicken HS4 element and the D. melanogaster gypsy and SF1 insulators, that possess both11–14.
Barrier-insulators are related to promoters
Barrier-insulators were originally identified based on the presence of DNase I hypersensitive sites located at transitions between open and condensed chromatin domains9. Reporter-based assays helped to identify the factors that associate with these elements and the general properties of barriers, but until recently the mechanism by which they block the spread of heterochromatin was unclear.
Barriers and promoters affect local chromatin structure
In Saccharomyces cerevisiae, the silenced HMR domain is restricted from spreading by an insulator element. Molecular dissection of this element led to the demonstration that its insulating activity is due to a tRNA gene promoter that functions as a barrier9. This promoter blocks the spread of hypoacetylated, silenced heterochromatin that is bound by Sir proteins. Other RNAPIII promoters have since been shown to function as barriers in mice and Schizosaccharomyces pombe, in which they restrict the spread of hypoacetylated, histone 3 lysine 9 trimethylated (H3K9me3), Swi6-bound heterochromatin, suggesting a conserved function for RNAPIII promoters as barrier elements14–16. Insulation at these barriers is mediated by the RNAPIII transcription factors TFIIIC and TFIIIB but is independent of transcription15,17,18. This ability to restrict the spread of heterochromatin is not limited to RNAPIII promoters in yeast; strong RNAPII promoters can also insulate genes in S. cerevisiae and S. pombe, again without requiring transcription9.
Whereas certain gene promoters function as barriers in yeast, barriers are distinct elements in higher eukaryotes but share molecular features with promoters. The β-globin locus in chicken cells is adjacent to a heterochromatin domain and is insulated from silencing by the HS4 insulator, which binds the factors CCCTC-binding factor (CTCF), upstream stimulatory factor 1 (USF1) and vascular endothelial zinc finger 1 (VEZF1) (FIG. 4a). These proteins coordinate distinct activities, which collectively result in robust barrier function. Although CTCF alone is not capable of protecting transgenes from silencing19, it blocks enhancers and helps to recruit the locus to specific compartments in the nucleus, and these processes are thought to aid in insulation20. USF1 recruits specific chromatin-modifying enzymes that acetylate and methylate histones, which aids barrier activity21,22, whereas VEZF1 restricts the spread of DNA methylation and generates a structure at the HS4 insulator that has the characteristics of a CpG island promoter23.
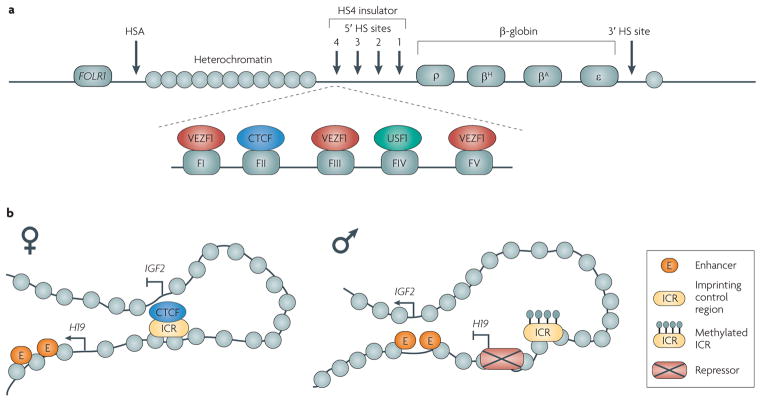
Figure 4. Vertebrate loci at which insulator function is well-studied.
a|The chicken HS4 insulator. A 16-kb heterochromatin domain is located between the folate receptor 1 (FOLR1) gene and the β-globin locus. The HS4 insulator separates the heterochromatin from the active β-globin gene domain. The insulator contains binding sites (FI to FV) for the proteins vascular endothelial zinc finger 1 (VEZF1), CCCTC-binding factor (CTCF) and upstream stimulatory factor 1 (USF1). CTCF is necessary for enhancer-blocking activity and tethering the element to the nucleolus, whereas VEZF1 and USF1 are required for barrier activity. HSA and 3′ HS are DNase I hypersensitive sites with enhancer-blocking activity. b|Enhancer-blocking activity at the imprinted insulin-like growth factor 2 (IGF2)–H19 locus. On the maternal allele, CTCF binds the imprinting control region (ICR) and interacts with the IGF2 promoter, blocking activation by the upstream enhancers, which are then able to interact with the H19 gene. On the paternal allele, the CTCF-binding sites in the ICR are methylated and CTCF is unable to bind the ICR. The enhancers can then activate the IGF2 gene while repressors bind H19. Part a is modified, with permission, from REF. 19 © (2002) National Academy of Sciences USA.
Although HS4-bound CTCF does not have barrier activity, other CTCF- bound insulators can function as barriers, protecting transgenes from silencing in a CTCF-dependent manner24,25. Furthermore, CTCF flanks some silenced H3K27me3 (REF. 26) and lamin-associated domains27, which suggests that this protein may aid in barrier activity in an insulator-specific manner.
The D. melanogaster Suppressor of hairy wing (SU(HW)) protein, the primary function of which seems to be enhancer blocking, also has the ability to act as a barrier11, and several studies in yeast, D. melanogaster and human cells have shown that artificial recruitment of specific transcription activators, such as Reb1 and Gal4, to DNA elements is sufficient to block the spread of heterochromatin9. Insulation at these sites is a consequence of direct competition between the spreading heterochromatin and transcription-factor binding coupled with chromatin remodelling at the barrier17.
Nuclear organization and barrier function
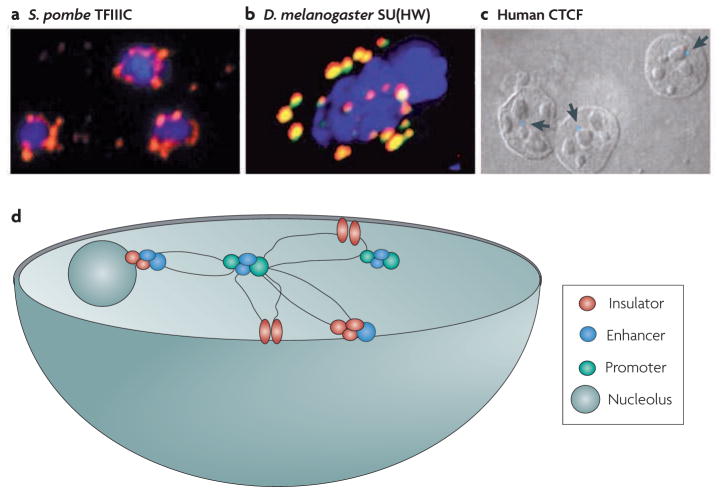
The organization of the chromatin fibre in the nucleus is also thought to have a role in barrier function. For example, immunofluorescence studies have shown that TFIIIC-mediated barriers in S. pombe coalesce at specific foci15 (FIG. 5a). The HS4 barrier is similarly tethered at specific foci by CTCF and its interacting proteins20 (FIG. 5c). In mammalian cells, cohesins, which are involved in higher-order folding of chromatin, are recruited to specific sites by CTCF28, and although it is not known whether this is required for barrier function, a role for cohesins in barrier function has been documented at TFIIIC barriers in yeast29,30. These findings suggest that barriers may function in part by sequestering loci to specific compartments in the nucleus. Although the significance of this subnuclear localization is not clear, it might promote insulation by placing the insulator in a microenvironment that is rich in specific factors. Whereas silencers and active genes are often clustered in the nucleus6,31, their subnuclear localization is mutually exclusive. As some promoters function as barriers, transcription factories and barriers might be expected to be coincident and should also be excluded from silencing foci. Additional fluorescence and in situ hybridization studies, coupled with circularized chromosome conformation capture (4C) and carbon-copy chromosome conformation capture (5C) analyses32, should help to resolve these questions.
Figure 5. Insulators and three-dimensional organization in the nucleus.
a|Clustering of transcription factor IIIC (TFIIIC) in Schizosaccharomyces pombe. b|Clustering of Suppressor of hairy wing (SU(HW)) in Drosophila melanogaster. c|The interaction of CCCTC-binding factor (CTCF) insulator sites with the nucleolus in human cells. The arrows point to the HS4 insulator interacting with the nucleolus, as visualized by fluorescence in situ hybridization (FISH). d|Model of interactions among insulators, enhancers, promoters and subnuclear structures in the nucleus. Insulators interact with each other and with other regulatory elements to partition the chromosome into structural and functional domains. Image a is reproduced, with permission, from REF. 15 © (2006) Elsevier. Image b is reproduced, with permission, from REF. 53 © (2000) Elsevier. Image c is reproduced, with permission, from REF. 20 © (2004) Elsevier. Image d is adapted, with permission, from REF. 37 © (2001) Annual Reviews.
The molecular details from yeast and chickens allow us to propose a core mechanism for barrier function, around which elaborations have been added in metazoans. Barriers recruit specific transcription factors, such as USF1 and TFIIIC, to mediate nucleosome eviction or rapid turnover of histones coupled with histone modifications in their immediate vicinity21,33–35. This leads to a local environment that is unfavourable to the propagation of heterochromatin9. Barriers also harness the ability of specific factors, such as CTCF, to tether loci to nuclear substructures to aid in insulation, and in vertebrates additional factors, such as VEZF1, help to restrict the spread of DNA methylation. This model is similar to processes observed at promoters during gene activation and suggests that common mechanisms might underlie activation and barrier insulation. Given the conserved mechanism in yeast and chicken cells, similar mechanisms might also function in other species and at other barrier elements, although this generalization awaits confirmation.
Enhancer-blockers and promoters
Reporter-based assays have identified many enhancer-blockers in D. melanogaster and vertebrate cells, which has led to two main models of insulator function (FIG. 3). One model is based on the observation that enhancers interact with promoters (FIG. 3a) and suggests that the enhancer-blocker acts as a decoy by directly interacting with the enhancer (FIG. 3b), thereby precluding fruitful interactions between the enhancer and the promoter. A second model is based on the observation that enhancer-blocking insulators interact with one another to form chromatin loops (FIGS 3a,5d). This model posits that insulators interact with each other and with structures in the nucleus to partition the chromatin fibre into loops, such that enhancers in one loop do not or cannot activate promoters in a different loop36,37.
The recent identification of promoters with enhancer-blocking ability coupled with the genome-wide mapping of enhancer-blocking proteins and histone modifications suggest that the two models described above could be unified. This leads us to suggest that enhancer-blockers might use mechanisms similar to those used by barriers to mediate insulation.
Promoters as enhancer-blockers
Although many barrier-insulators have been mapped to proximal promoters, until recently it was unclear whether the same was also true for enhancer-blockers. Promoters with paused RNA polymerases flank the D. melanogaster Bithorax gene cluster, and these promoters were recently shown to possess enhancer-blocking activity, which is dependent on the transcription factor Negative elongation factor (NELF)38. In addition, a SU(HW)-binding site located between the yellow and achaetescute genes was also shown recently to both function as an enhancer-blocker and map close to the proximal promoter of a non-coding RNA39. Finally, the D. melanogaster enhancer-blockers special chromatin structure (scs) and scs′ have been known to map at or near promoters9, although it is unclear whether transcription is necessary for enhancer blocking or whether other mechanisms, such as localized chromatin remodelling or tethering of the loci to specific nuclear compartments, are crucial for insulation.
Mapping of insulator-binding proteins
Enhancer-blockers in D. melanogaster use many proteins for insulation, six of which were recently mapped across the genome: dCTCF (a distant sequence homologue of mammalian CTCF), SU(HW), Boundary element-associated factor (BEAF), GAGA factor (GAF), Modifier of MDG4 (MOD(MDG4)) and CP190, which does not bind DNA but is required for enhancer blocking by some of the other factors40–44. SU(HW) binds primarily at intergenic regions and often colocalizes with MOD(MDG4). Although dCTCF, BEAF and CP190 are also present at some intergenic sites, a large fraction of these proteins map at promoters or near transcription start sites of highly transcribed genes, and they often colocalize together. Some BEAF sites also colocalize with the transcription factor NELF, which is present at promoters with paused RNA polymerases40, although the mechanistic interplay among these proteins at potential insulator sites has yet to be determined. Further analyses indicate that BEAF, dCTCF, CP190 and SU(HW) are enriched at sites that flank enhancers and their target promoters or between enhancers and their non-target promoters; therefore these proteins separate genes with differing expression profiles44, which is consistent with the idea that these sites are enhancer-blocking insulators. These findings indicate that enhancer-blocking insulators use specific transcription factors for insulation, which is analogous to the situation at barrier-insulators.
Although promoters and promoter-bound factors can block enhancers in D. melanogaster, it is currently unknown whether mammalian promoters can or do function as enhancer-blocking insulators. Most vertebrate enhancer-blockers identified to date are standalone insulators that require CTCF and its interacting proteins for enhancer blocking (both at their native loci and in reporter-based systems), although a recent report showed that TFIIIC-bound SINE elements can also function as enhancer-blockers14.
CTCF is a multifunctional protein involved in gene regulation. To gain a better understanding of its function, its distribution was mapped in mammalian cells26,28,45. Consistent with a role in enhancer blocking, the majority of sites map to intergenic regions but, surprisingly, a substantial proportion (~20%) also map to the proximal promoters of genes46, a pattern similar to the D. melanogaster enhancer-blocking proteins. Although the mere presence of a binding site does not imply functional significance and experimental validation of these putative insulators is currently lacking, the observations that the intergenic sites colocalize with cohesins and flank clusters of co-regulated genes or active chromatin domains have led to the suggestion that most of these sites are likely to be enhancer-blockers47. By contrast, the role of promoter-bound CTCF remains nebulous; in the context of insulator function it is unclear whether the promoter-bound CTCF sites function independently as enhancer-blockers or in cooperation with intergenic sites. Given the vast number of interacting partners of CTCF48, the potential for differential regulation of the various CTCF sites through its interacting partners remains a distinct possibility.
The chromatin structure flanking CTCF-binding sites is very similar whether these sites are in intergenic regions or at proximal promoters. Most mammalian CTCF-bound sites are, like promoters, nucleosome-free, regardless of their position43,49. The histone-modification signatures flanking the binding sites are also similar to those observed at promoters that have mammalian CTCF sites, in that they are enriched for H2A.Z and specifically methylated histones but lack p300. This pattern is also found at proximal promoters26,45,50,51. It should be noted that some of the modifications flanking CTCF sites are enhancer-specific, and it is entirely possible that the 3D clustering of insulators with promoters or enhancers may contribute to the modification patterns observed at these sites.
The similarities between promoters and insulators suggest that the transcription factors bound to these intergenic putative enhancer-blocking insulators may use molecular mechanisms that are similar to those that occur during transcriptional activation at promoters. At promoters, nucleosome eviction and histone modifications enable stable binding of transcription factors while simultaneously disrupting the packaging of the chromatin fibre. This increases chromatin accessibility and flexibility, thereby allowing enhancer–promoter interactions. Given the similarities in the molecular signatures at enhancer-blockers and promoters, localized disruption at the insulator may also be important for enhancer blocking. Chromodomain-helicase-DNA-binding protein 8 (CHD8), a protein with homology to chromatin remodellers, interacts with and is required for CTCF-dependent enhancer blocking34, although it is unknown whether chromatin remodelling mediated by CHD8 is required for insulation. Similarly, the D. melanogaster SF1 insulator is directly or indirectly affected in chromatin remodeller mutants52. These data are consistent with a scenario in which enhancer-blockers use various chromatin enzymes in insulation, although further studies are necessary before firm models can be built.
Enhancer-blockers and chromatin loops
As mentioned above, many active promoters coalesce in the nucleus at transcription foci6. Similarly, many enhancer-blockers also interact with each other or cluster in the nucleus at ‘insulator bodies’ (FIG. 5). The scs and scs′ insulators interact directly with each other through protein–protein interactions to organize chromatin loops. One of the best characterized D. melanogaster insulators is the gypsy insulator, which binds SU(HW) and other proteins. These proteins cluster in the nucleus to form insulator bodies and organize the chromatin into loops (FIG. 5b). The formation of these loops correlates with insulation53,54, although the presence of gypsy DNA sequences in these bodies has recently been questioned55.
Besides these studies, a vast body of functional analyses in D. melanogaster53,56,57 has revealed that like enhancer–promoter interactions, insulator–insulator interactions occur in pairwise combinations and are specific. For example, either one or three SU(HW)-containing insulators located between an enhancer and a promoter blocked the enhancer, but a pair of insulators were ‘neutralized’ and could not block the enhancer. These data are consistent with the suggestion that insulators interact with one another in organizing chromatin loops, although these interactions have not yet been shown to be necessary for enhancer blocking58.
Insulators do not only interact with each other. DamID experiments revealed that the D. melanogaster Fab7 insulator interacts with the Abdominal B (AbdB) promoter57, and a recent chromosome conformation capture (3C) study has shown that a synthetic insulator of the interferon-β1 (IFNB1) gene interacts with an enhancer59 (FIG. 3). These data suggest that enhancer-blockers interact with promoters or enhancers, possibly sequestering these elements into non-productive interactions, consistent with the decoy model. One of the best-characterized native enhancer-blockers in mammalian cells is the imprinting control region (ICR) that is located between the tandem insulin-like growth factor 2 (IGF2) and H19 imprinted genes, which are regulated by an enhancer that lies downstream of H19 (FIG. 4b). On the maternal allele, the enhancer activates the H19 gene while CTCF binds the ICR insulator and blocks the long-range communications between the enhancer and the IGF2 promoter60. Mutational analyses have shown that CTCF binding to the ICR is necessary for insulation. 3C and 4C analysis further demonstrate that the ICR-bound CTCF mediates numerous long-range interactions with the IGF2 promoter and other regulatory elements on the same and different chromosomes, revealing a role for CTCF in forming chromatin foci60. A similar role for CTCF has been elucidated at the apolipoprotein gene cluster, where a CTCF-bound insulator segregates the C3 enhancer from the apolipoprotein A1 (APOA1) gene and enables the enhancer to activate the APOA4 and C3 genes, which are present in the same loop61.
These observations collectively indicate that the mechanism by which enhancer-blockers function is by modulating long-range interactions in the nucleus. Proteins, such as cohesins, that are recruited to enhancer-blocking insulators could help in this process, and the observations that CTCF recruits cohesins to chromosomes in mammalian cells28 and that mutations in cohesin subunits affect enhancer–promoter and insulator communication in D. melanogaster62 are consistent with this model.
Differentiating between models of enhancer blocking
One crucial difference between the loop model and the decoy model for enhancer-blocking function is that in the former, enhancer-blockers only function when located between the enhancer and the promoter, whereas in the latter model insulators should function whether they are located upstream of the enhancer or placed between the enhancer and the promoter. The observation that the effectiveness of enhancer blocking is increased when a second promoter is placed upstream of the enhancer58 suggests that the upstream promoter interacts with and sequesters the enhancer, and seems to increase the strength of the enhancer-blocking insulator. This is consistent with the flip-flop model of enhancer function7 and the decoy model of insulation. Elements that block enhancer function when located upstream of the enhancer are traditionally regarded as distinct from insulators and are referred to as silencers of enhancers, based primarily on their position with respect to the enhancer (these silencers should not be confused with silencers that generate heterochromatin). However, this distinction may not be as clear-cut as previously thought, as bona fide insulators are capable of this behaviour too: a recent screen in humans for enhancer-blockers and enhancer-silencers identified many DNA sequences that are able to act as both63. Furthermore, the well-studied HS4 insulator can partially inactivate enhancers from both positions, which suggests some overlap in the mechanisms of enhancer silencing and blocking.
Collectively, the data suggest that enhancer-blockers interact with each other or promoters and enhancers, and the consequences of these interactions are either gene activation or insulation. Insulator–insulator interactions form loops that would simultaneously neutralize the insulators and allow the enhancer to interact with and activate the promoter. Conversely, insulator–promoter or insulator–enhancer interactions would sequester these elements in non-productive interactions, therefore precluding gene activation and resulting in enhancer blocking (FIG. 3). To determine whether this scenario occurs, the locations of enhancers and promoters with respect to insulators during insulation will need to be determined. That is, do insulators cluster with the enhancer or promoter during insulation? Or are the enhancer and promoter in separate chromatin loops from each other rather than in proximity to the insulator during insulation?
Conclusions and perspectives
We have outlined common themes from an overview of the recent literature on insulators in different organisms. Most if not all insulators are nucleosome-free, flanked by specifically modified histones and maintained in this state by chromatin modifiers and remodellers. Most insulators use factors that have a propensity to mediate long-range interactions and cluster, and some insulators, whether barriers or enhancer-blockers, are promoters of genes.
We propose a model for insulation in which factors such as USF1 are important for their ability to recruit modifying and remodelling enzymes to evict nucleosomes and increase the accessibility and flexibility of the chromatin fibre. By comparison, proteins, such as CTCF and SU(HW), that are involved in long-range interactions (FIG. 5b,c) allow insulators to interact with each other and with other regulatory elements. In the context of barrier function, the removal of nucleosomes and the modification of flanking nucleosomes enable transcription factors to bind stably and simultaneously reduce the binding of heterochromatic proteins, therefore disrupting the spread of heterochromatin. Clustering of insulators would aid in this process by sequestering the insulator to a nuclear compartment rich in remodellers and modifiers. In the context of enhancer blocking, nucleosome eviction and specific chromatin modification would aid in the stable binding of factors and increase the flexibility and accessibility of the chromatin. Factors such as CTCF could orchestrate insulator–insulator, insulator–promoter or enhancer–promoter interactions through different interacting partners. Although this property might not be crucial for barrier function, it may be essential for enhancer blocking, which may explain why most vertebrate enhancer-blockers require CTCF for insulation.
Although promoters can satisfy models for insulator function, it is clear that not all promoters function as insulators. What features do some promoters have that allow them to insulate? During evolution, insulators may have been selected for specific properties, such as the ability to bind proteins that are capable of efficiently remodelling chromatin and creating nucleosome-free regions and/or the ability to mediate long-range interactions or form foci in the nucleus. If insulators evolved from promoters, then promoters that bind these factors may have been selected to become insulators precisely for these abilities. In the context of yeast tRNA insulators, the observation that pseudogenes that have maintained their ability to bind transcription factors and chromatin remodellers can function as insulators, even in the absence of RNAPIII, highlights a pathway by which insulators may have diverged from promoters to become standalone insulators17,18.
Additional studies on barriers in other species would be useful to prove the generality of the mechanisms of barrier function. Additional analyses of enhancer blocking, particularly the chromatin structure changes during insulation and the identification of cofactors necessary for enhancer blocking, would increase our understanding of how these elements function. Finally, greater effort needs to be devoted towards determining which of the many enhancer-blockers identified in mammalian cells also have barrier activity.
Acknowledgments
We would like to thank B. Cairns, O. Rando, M. Bulger, D. Clark, G. Hartzog, M. Oki, K. Noma, S. Henikoff, P. Geyer and G. Felsenfeld for comments on an early draft of this Review. We would also like to thank members of the Ro laboratory for comments and criticisms. We apologize for the selective citations, which are a consequence of space limitations. This work was supported by grants from the US National Institutes of Health to R.T.K. (GM078068) and J.R.R (T32-GM008646), and by grant 2008-16 from the GREAT Training Program of the University of California System-wide Biotechnology Research and Education Program to J.R.R.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/gene
FOLR1|IGF2|H19
FlyBase: http://flybase.org
achaete-scute|scs|scs′|yellow
UniProtKB: http://www.uniprot.org
BEAF|CP190|CTCF|GAF|MOD(MDG4)|SU(HW)|USF1|VEZF1
FURTHER INFORMATION
Rohinton T. Kamakaka’s homepage: http://research.pbsci.ucsc.edu/mcdb/chromatin
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 2.Pirrotta V, Gross DS. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol Cell. 2005;18:395–398. doi: 10.1016/j.molcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 5.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 7.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 9.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 10.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nature Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 11.Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumder P, et al. Diverse transcription influences can be insulated by the Drosophila SF1 chromatin boundary. Nucleic Acids Res. 2009;37:4227–4233. doi: 10.1093/nar/gkp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 14.Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 15.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 16.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 17.Valenzuela L, Dhillon N, Kamakaka RT. Transcription independent insulation at TFIIIC-dependent insulators. Genetics. 2009;183:131–148. doi: 10.1534/genetics.109.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simms TA, et al. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recillas-Targa F, et al. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc Natl Acad Sci USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 21.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol. 2007;27:7991–8002. doi: 10.1128/MCB.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson J, et al. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010;6:e1000804. doi: 10.1371/journal.pgen.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filippova GN, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Ottaviani A, et al. The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet. 2009;5:e1000394. doi: 10.1371/journal.pgen.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 28.Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- 29.Dubey RN, Gartenberg MR. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 2007;21:2150–2160. doi: 10.1101/gad.1583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nature Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 32.Vassetzky Y, et al. Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol Biol. 2009;567:171–188. doi: 10.1007/978-1-60327-414-2_12. [DOI] [PubMed] [Google Scholar]
- 33.Oki M, Kamakaka RT. Barrier function at HMR. Mol Cell. 2005;19:707–716. doi: 10.1016/j.molcel.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Dhillon N, et al. DNA polymerase ε, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J. 2009;28:2583–2600. doi: 10.1038/emboj.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 37.Gerasimova TI, Corces VG. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu Rev Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- 38.Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–1509. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soshnev AA, Li X, Wehling MD, Geyer PK. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 2008;4:e1000159. doi: 10.1371/journal.pgen.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol Cell Biol. 2009;29:3556–3568. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith ST, et al. Genome wide ChIP–chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol. 2009;328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartkuhn M, et al. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negre N, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 46.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie X, et al. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci USA. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 49.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin C, et al. H3.3/H2A.Z. double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nature Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Belozerov VE, Cai HN. Modulation of chromatin boundary activities by nucleosome-remodeling activities in Drosophila melanogaster. Mol Cell Biol. 2009;30:1067–1076. doi: 10.1128/MCB.00183-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 54.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 55.Golovnin A, et al. ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 2008;9:440–445. doi: 10.1038/embor.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 58.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 59.Nolis IK, et al. Transcription factors mediate long-range enhancer–promoter interactions. Proc Natl Acad Sci USA. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishiro T, et al. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorsett D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrykowska HM, Vockley CM, Elnitski L. Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Res. 2008;18:1238–1246. doi: 10.1101/gr.073817.107. [DOI] [PMC free article] [PubMed] [Google Scholar]