Synopsis
X-box-binding protein 1 (XBP1) is a key modulator of unfolded protein response (UPR), which is involved in a wide range of pathological and physiological processes. The active/spliced form of XBP1 (XBP1s) messenger RNA is generated from unspliced form by IRE1 during UPR. However, post-translational modulation of XBP1s remains largely unknown. Here, we demonstrate that XBP1s is a target of acetylation and deacetylation mediated by p300 and SIRT1 respectively. p300 increases acetylation and protein stability of XBP1s, and enhances the transcriptional activity of XBP1s. SIRT1 deacetylates XBP1s and inhibits the transcriptional activity of XBP1s. Deficiency of SIRT1 enhances the XBP1s-mediated luciferase reporter activity in HEK293 cells and the upregulation of XBP1s target gene expression under ER stress in mouse embryonic fibroblasts (MEFs). Consistent with XBP1s favoring cell survival under ER stress, Sirt1−/− MEFs display a greater resistance to the ER stress-induced apoptotic cell death compared with Sirt1+/+ MEFs. Taken together, these results suggest that acetylation/deacetylation constitutes an important post-translational mechanism in controlling protein levels as well as transcriptional activity of XBP1s. This study provides a novel insight into molecular mechanisms by which SIRT1 regulates UPR signaling.
Keywords: acetylation, SIRT1, p300, unfolded protein response, X-box-binding protein 1, cell death
INTRODUCTION
X-box-binding protein-1 (XBP1), a basic-region leucine zipper (bZIP) transcription factor of the CREB/ATF protein family, is a key mediator of the unfolded protein response (UPR). When cells experience endoplasmic reticulum (ER) stress due to disturbance of ER homeostasis, UPR is activated to reestablish ER homeostasis or to induce apoptosis of overly-stressed cells [1, 2]. The UPR consists of three parallel signaling branches initiated by PKR-like endoplasmic reticulum kinase (PERK), insositol requiring enzyme-1 (IRE1), and activating transcription factor-6 (ATF6). IRE1, the most conserved proximal sensor of ER stress, executes unconventional splicing of XBP1 mRNA to generate the spliced form of XBP1 (XBP1s). The XBP1s mRNA encodes an active transcription factor that activates the expression of a wide variety of genes required for biogenesis of the protein secretory pathway, protein folding and secretion, as well as clearance of misfolded proteins from ER [3, 4]. Functionally, the IRE1/XBP1s signaling supports cell survival under ER stress [5].
XBP1 is implicated in a wide range of human physiological and pathological processes. The critical function of XBP1 was first shown by a gene knock-out mouse model in which Xbp1 deficient embryos died in uterus due to fatal anemia caused by liver hypoplasia and apoptosis [6]. XBP1s induces interleukin-6 production [7] and expands secretory apparatus in B-cell during their differentiation into plasma cells [8]. Moreover, XBP1s drives cancer pathogenesis, and it is required for survival of multiple myeloma (MM) cells [9]. XBP1s also plays an essential role in regulating adipogenesis and hepatic lipogenesis [10, 11]. Although the pathophysiological significance of XBP1s has been well-established, the molecular mechanisms in regulating XBP1s protein expression and its transcriptional activity remain to be elucidated.
The Silent Information Regulator 2 (SIR2) family is a group of histone deacetylases that are highly conserved among the species from prokaryotes to eukaryotes [12]. There are three distinct classes of histone deacetylases including the class I, class II, and the nicotinamide adenine dinucleotide (NAD+)-dependent class III histone deacetylase families [13]. SIRT1, the closest mammalian homologue to SIR2 in C. elegans, belongs to class III histone deacetylase, and is the best characterized SIR2 family member. It has been shown that SIRT1 also deacetylates non-histone proteins, including transcription factors, to modulate stress-responsive signaling pathways [14]. For instance SIRT1 deacetylates FOXO3, a member of the FOXO family of Forkhead transcription factors, that grant cells a greater resistance to oxidative stress [15]. In addition, SIRT1 represses p53 via deacetylating p53 to facilitate cellular recovery from genotoxic stress [16]. In C. elegans, SIR2 was shown to regulate UPR genes through an unidentified mechanism [17]. However, there is no evidence as yet to indicate whether SIRT1 is involved in regulating mammalian UPR, especially its proximal modulators such as XBP1s.
Acetylation/deacetylation of transcription factors is emerging as an important post-translational regulatory mechanism to modulate their protein expression and transcriptional activity [14]. Like XBP1s, ATF2 and ATF4 also belong to the CREB/ATF family [18, 19]. Both ATF2 and ATF4 have been shown to be acetylated by the histone acetyltransferase p300 [20, 21]. Sterol regulatory element-binding protein (SREBP), a transcription factor critical for lipid and sterol homeostasis in eukaryotes, has been shown to be a target of SIRT1-mediated deacetylation [22, 23]. Since XBP1s, ATF4 and SREBP1 are all ER stress-inducible molecules [24] and both XBP1s and SREBP1 share similar functions in regulating cellular lipid synthesis [25], we hypothesized that XBP1s is also a substrate for acetylase and deacetylase. In this study, we demonstrate that XBP1s can be acetylated and deacetylated mediated by p300 and SIRT1 respectively. These post-translational modifications regulate the protein level and transcriptional activity of XBP1s. Our study provides a novel mechanistic link between SIRT1 and the UPR signaling.
EXPERIMENTAL
Reagents and antibodies
Dulbecco’s modified eagle medium (DMEM), fetal bovine serum (FBS), L-glutamine, MEM nonessential amino acid mix (NEAA), 200×pen/strep antibiotics, Lipofectamine reagent, Rhodamine Red™-X labeled anti-mouse antibody (R6393) and Alexa Fluor 488 labeled anti-rabbit antibody (A11034) were purchased from Invitrogen (San Diego, CA, USA). Trichostatin A was purchased from Cayman Chemical (Ann Arbor, MI, USA). EX-527 was purchased from Tocris Bioscience (Ellisville, MO, USA). Thapsigargin, tunicamycin, DTT, cycloheximide, protease inhibitor cocktail, trypan blue (T8154), anti-β-actin antibody (A5316), anti-Flag M2 antibody (F1804), anti-FLAG® M2 affinity gel (A2220) and FLAG peptide (F3290) were from Sigma-Aldrich (St. Louis, MO, USA). Anti-acetylated lysine antibody (#9441) and anti-Caspase-3 antibody (#9662) were from Cell Signaling Technology (Beverly, MA, USA). Anti-SIRT1 (sc-15404) and anti-XBP1 (sc-7160) were from Santa Cruz Technology (Santa Cruz, CA, USA). HRP-conjugated IgG secondary antibodies were from GE Healthcare Life Sciences (Little Chalfont, UK). Protein G agarose beads and polyvinylidene difluoride (PVDF) membranes were from Millipore (Bedford, MA, USA).
Cell culture
HEK293, HEK293T, and Cos-7 cells were grown in DMEM supplemented with 10% FBS and 1×pen/strep antibiotics. Sirt1+/+ and Sirt1−/− mouse embryonic fibroblasts (MEFs) [26] were kindly provided by Dr. Paul Robbins (University of Pittsburgh, Pittsburgh, PA, USA) and grown in DMEM supplemented with 10%FBS, 1×NEAA, 2 mM L-glutamine and 1×pen/strep antibiotics.
Plasmids and transfection
Expression construct for XBP1s was generated by inserting mouse XBP1s coding regions with or without a Flag tag into pQCXIP based vector (Clontech, Mountain View, CA). HA-p300 construct was kindly provided by Dr. Ming Hu (University of South Florida, Tampa, FL, USA). Flag-SIRT1 construct was obtained from AddGene (http://www.addgene.org) [15]. XBP1s-mediated transcriptional activity was measured using the 5×UPRE luciferase reporter (a kind gift from Randal J. Kaufman, University of Michigan, Ann Arbor, MI, USA). SIRT1 shRNA plasmid was constructed by inserting a double-stranded oligonucleotide containing 5′-GAAGTTGACCTCCTCATTGT-3′ [27] into the BglII/XhoI sites of pSUPER.neo+gfp vector (OligoEngine, Seattle, WA, USA). Transfection of cells was performed using Lipofectamine reagent.
Detection of acetylation of XBP1s and in vitro deacetylation assay
HEK293 cells overexpressing Flag-XBP1s were lysed in the lysis buffer (20 mM Tris HCl pH7.5, 150 mM NaCl, 1% Triton-X100,1 mM EGTA, 1mM EDTA, 50 mM NaF, 1 mM β-glycerophosphate, 2.5 mM sodium pyrophosphate, 1 mM orthovanadate, 1× protease inhibitor cocktail, 10 mM nicotinamide, 1mM TSA). Cell extracts were subjected to immunoprecipitation with anti-Flag M2 antibody. The immune complexes eluted from Protein G agarose beads were analyzed by Western blotting with an anti-acetylated lysine antibody. The stripped membranes were re-probed with the rabbit polyclonal antibody to XBP1.
For the in vitro deacetylation assay, acetylated XBP1s was purified form HEK293T cells transfected with Flag-XBP1s and p300. To maximize levels of acetylation, 10 mM nicotinamide was added to the medium for 5 hours before cell harvest. SIRT1 was purified from HEK293T cells transfected with Flag-SIRT1. After immunoprecipitation using anti-FLAG M2 affinity gel, epitope products were eluted using Flag peptide. The eluted acetylated XBP1s was then incubated in reaction buffer (25 mM Tris-HCl pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1mM MgCl2, 0.2 mM PMSF, 0.5 mM DTT) at 30°C for 1 hour in the presence or absence of SIRT1 and 5 mM NAD+. The acetylated and total amounts of XBP1s were then measured by Western blotting.
Luciferase assay
HEK293 cells were seeded into 24-well plates. After transfection and indicated treatment, cells were harvested using 1×passive lysis buffer (Promega). XBP1s transcriptional activity was measured by dual luciferase assay with normalization using a pRL-CMV Control Reporter Vector (Promega).
Western blotting and co-immunoprecipitation
Protein samples were obtained with the lysis buffer. Nuclear proteins were extracted using CelLytic™ NuCLEAR™ Extraction Kit (Sigma). Approximately 10 μg samples were loaded per lane in the SDS-PAGE gels. The separated proteins were transferred to PVDF membranes. The following primary antibodies were used: anti-XBP1 (1:1000), anti-Caspase 3 (1:1000), anti-SIRT1 (1:1000) and β-actin (1:20000). Signals were detected using HRP-conjugated secondary antibodies (1:2000) and ECL™ reagents (GE Healthcare Life Sciences). Band intensity measurements were done using the TotalLab100 software (Nonlinear Dynamics, Durham, NC). For the co-immunoprecipitation of XBP1s and SIRT1, we transfected Flag-XBP1s or an empty vector into HEK293 cells. Cell lysates were immunoprecipitated using anti-FLAG® M2 affinity gel and then analyzed for endogenous SIRT1 with an anti-SIRT1 antibody.
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 20 min and then permeabilized with 0.1% Triton X-100 for 5 min. Cells were washed and incubated in 10% normal goat serum solution for 20 min. Primary antibodies anti-XBP1 (1:200) and anti-Flag (1:500) were used to stain XBP1s and Flag-SIRT1, respectively, for 60 min. Rinsed cells were loaded with Alexa Fluor 488 labeled anti-rabbit antibody (1:200) and Rhodamine Red™-X labeled anti-mouse antibody (1:200) for 45 min. After the extensive PBS washing, samples were mounted and observed under an Olympus Fluoview 500 confocal microscope (Olympus America, Centerville, PA).
RNA extraction and real-time PCR
Total RNA was isolated using Trizol® reagent according to the manufacturer’s instructions. Reverse transcription was conducted using a reverse transcription system kit (Promega, Cat.A3500). The aliquot of the product cDNA was used for real-time PCR with iQ™ SYBR Green Supermix and iCycler iQ PCR Detection System (Bio-Rad Laboratories). 18S rRNA was applied as an internal control for data analysis. The nucleotide sequences of primers used for PCR are as follows: 18S rRNA 5′-cgcttccttacctggttgat-3′ and 5′-gagcgaccaaaggaaccata-3′; Gadd34 5′-gagattcctctaaaagctcgg-3′ and 5′-cagggacctcgacggcagc-3′; Grp78/BiP 5′-catggttctcactaaaatgaaagg-3′ and 5′-gctggtacagtaacaactg-3′; Ero1α 5′-tcagtggaccaagcatgatga-3′ and 5′-tccacatactcagcatcggg-3′; Sec61a 5′-ctatttccagggcttccgagt-3′ and 5′-aggtgttgtactggcctcggt-3′; Edem1 5′-aagtctcaggagctcagagtcattaa-3′ and 5′-cgatctggcgcatgtagatg-3′; spliced Xbp1 5′-gagtccgcagcaggtg-3′ and 5′-gtgtcagagtccatggga-3′; total Xbp1 5′-aagaacacgcttgggaatgg-3′ and 5′-actccccttggcctccac-3′.
Assay for cell viability
Cells were treated with tunicamycin for 24 h and were stained with trypan blue. The number of live and dead cells was counted using a hemacytometer under the microscope.
Statistical analysis
All the experiments were repeated at least 2 times. Data are presented as means S.E.M. Statistical analyses between groups were done with Student’s t test using Graphpad (http://www.graphpad.com).
RESULTS
XBP1s is a target of acetylation
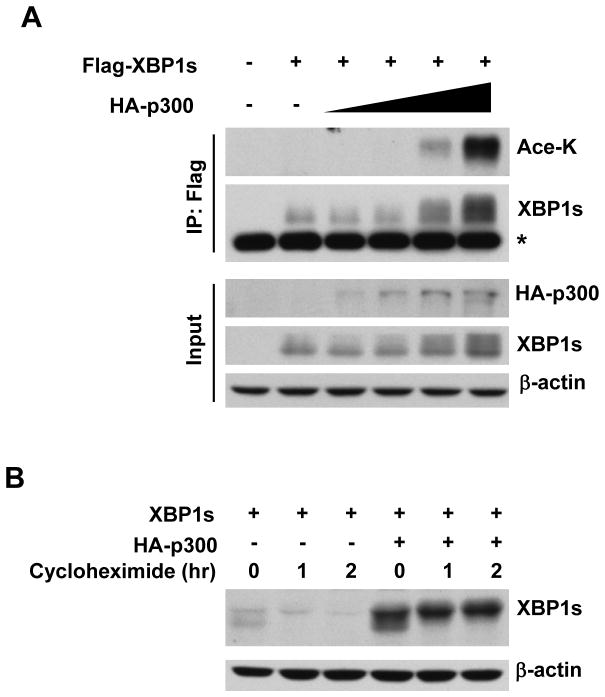
To determine if XBP1s is a target for p300-mediated acetylation, we transfected Flag-XBP1s with increasing amounts of p300 into HEK293 cells. It was found that overexpression of p300 increased the acetylation of XBP1s and promoted XBP1s protein level in a dose-dependent manner (Fig. 1A). We then determined whether p300 regulates XBP1s protein levels via modulating its protein stability. With cycloheximide blocking new protein synthesis, we observed a quick degradation of XBP1s in the absence of p300, while co-transfection with p300 significantly raised the basal protein levels of XBP1s and strongly stabilized XBP1s protein (Fig. 1B). These results demonstrate that XBP1s is subjected to p300-mediated acetylation.
Figure 1. XBP1s is a target of acetylation.
(A) HEK293 cells were transfected with Flag-XBP1s and increasing amount of p300. Cell lysates were analyzed by immunoprecipitation and Western blotting. Ace-K, acetylated lysine. *, non-specific band. (B) Equal amount of XBP1s was cotransfected with p300 or a control vector into HEK293 cells. Cells were harvested for lysates after cycloheximide (100 μg/ml) treatment for indicated time period. Cell lysates were analyzed by Western blotting.
SIRT1 deacetylates XBP1s
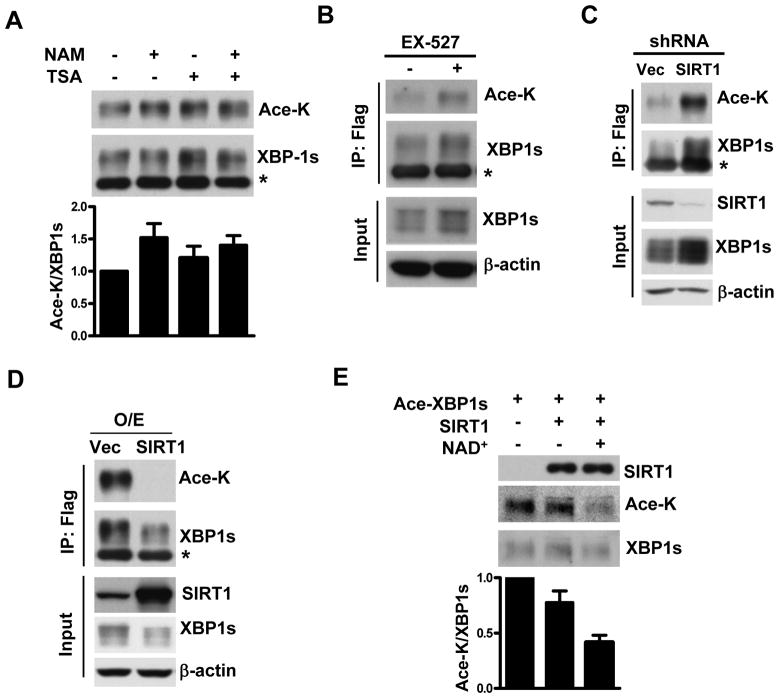
Since acetylation and deacetylation are coupled biological processes, we determined if XBP1s is also subjected to deacetylation. Histone deacetylases consist of the class I, class II, and the nicotinamide adenine dinucleotide (NAD+) dependent class III histone deacetylase family [13]. In order to determine which class of histone deacetylases could be involved in regulating XBP1s, we examined the effects on XBP1s acetylation of trichostatin A (TSA) and nicotinamide (NAM), two inhibitors for histone deacetylases class I/II and class III, respectively. We found that NAM enhanced XBP1s acetylation, whereas TSA alone did not affect XBP1s acetylation much. We calculated the ratio of acetylated vs. total XBP1s to indicate the extent of acetylation (Fig. 2A). This result suggests that it is the class III histone deacetylase that plays a dominant role in regulating XBP1s deacetylation. Since SIRT1 is the closest mammalian homologue of class III histone deacetylase Sir2 in C. elegans, we tested whether SIRT1 regulates XBP1s deacetylation via three strategies as follows:
Figure 2. XBP1s is deacetylated by SIRT1.
(A and B) HEK293 cells were transfected with Flag-XBP1s and p300. Cells were incubated for 4 hours in the absence or presence of TSA (5μM), NAM (10 mM), or EX-527 (1 μM). Cell lysates were analyzed by immunoprecipitation and Western blotting. *, non-specific band. The extent of acetylation was defined by the ratio of acetylated vs. total XBP1s from duplicate experiments (Fig.2A, lower panel). (C) HEK293 cells were transfected with Flag-XBP1s and p300 together with a pSuper-shRNA vector targeting SIRT1 or its empty vector. Cell lysates were analyzed by immunoprecipitation and Western blotting. *, non-specific band. (D) HEK293 cells were transfected with Flag-XBP1s and p300 together with or without SIRT1 expressing construct. O/E, overexpression. Cell lysates were analyzed by immunoprecipitation and Western blotting. *, non-specific band. (E) The acetylated Flag-XBP1s was purified from 293T cells transfected with Flag-XBP1s and p300. The Flag-SIRT1 was purified from 293T cells separately transfected with Flag-SIRT1. Purified acetylated XBP1s was incubated in the presence or absence of SIRT1 and/or NAD+. The acetylated and total amounts of XBP1s were determined by Western blotting and the extent of acetylation was defined by the ratio of acetylated vs. total XBP1s from duplicate experiments.
First, we studied the effect of SIRT1specific inhibitor EX-527 on XBP1s acetylation status. It was found that EX-527 increased the acetylation of XBP1s and caused accumulation of XBP1s protein (Fig. 2B). In addition, we evaluated the effects of SIRT1 knock-down on XBP1s acetylation. It was noticed that co-transfection with a shRNA expression vector targeting SIRT1appreciably increased XBP1s acetylation in HEK293 cells (Fig. 2C).
Second, we determined whether overexpression of SIRT1 could lead to deacetylation of XBP1s. XBP1s and p300 were co-transfected either with or without SIRT1 into HEK293 cells. It was shown that overexpression of SIRT1 completely diminished the p300-mediated acetylation of XBP1s (Fig. 2D).
Third, we performed an in vitro deacetylation assay, using purified acetylated XBP1s and SIRT1, to determine if XBP1s is a direct target for SIRT1. As expected, the purified SIRT1 deacetylated XBP1s protein in an NAD+-dependent manner (Fig. 2E). These results demonstrate that SIRT1 is a bona fide NAD+-dependent deacetylase for XBP1s and can directly deacetylate XBP1s.
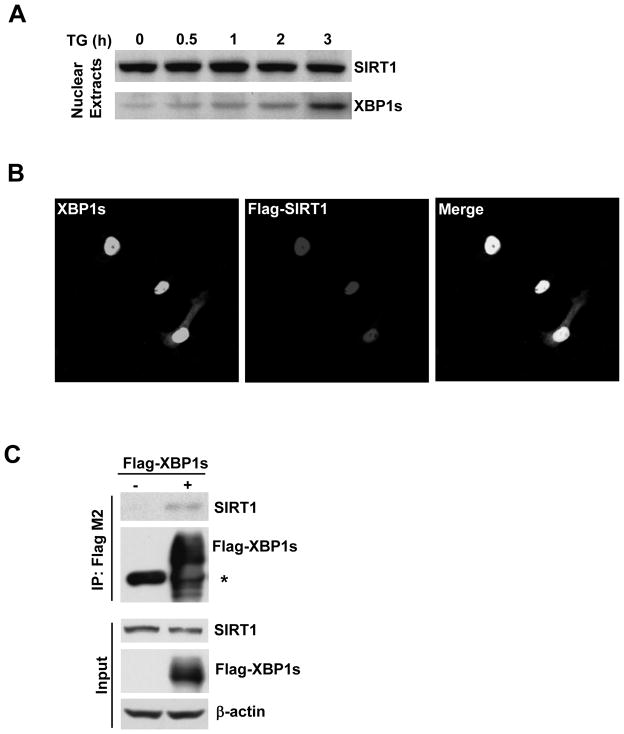
These findings promoted us to determine if XBP1s and SIRT1 co-localizes with, and furthermore physically binds to each other. As determined by Western blotting, endogenous XBP1s protein induced by ER stress activator thapsigargin accumulated within the nucleus of HEK293 cells. The endogenous SIRT1 was also detected in the nuclear compartment (Fig 3A). Immunofluorescence staining demonstrated that the overexpressed XBP1s and SIRT1 were co-localized within the nucleus of all the transfected Cos-7 cells (Fig. 3B). In addition, we found that there was a physical interaction between exogenous XBP1s and endogenous SIRT1 (Fig. 3C).
Figure 3. Physical interaction between XBP1s and SIRT1.
(A) Western blotting showing the presence of endogenous SIRT1 and XBP1s in the nuclear extracts HEK293 cells treated with thapsigargin (TG). (B) Cos7 cells were transfected with untagged XBP1s and Flag-SIRT1. Immunofluorescent staining for XBP1s and Flag-SIRT1 was performed using the anti-XBP1 and anti-Flag antibodies respectively. Confocal microscopy images at 60× magnification were presented to indicate the co-localization of XBP1s and SIRT1. (C) HEK293 cells were transfected with Flag-XBP1s or an empty vector. After immunoprecipitation using an anti-FLAG® M2 affinity gel, Western blotting was performed to detect endogenous SIRT1. *, non-specific band.
SIRT1 represses transcriptional activity of XBP1s
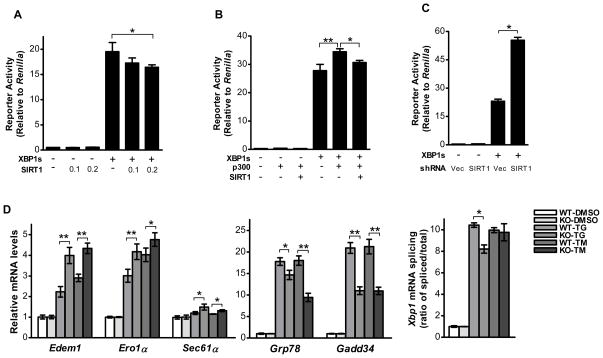
We next sought to assess whether SIRT1 regulates the transcriptional activity of XBP1s. We used a 5× UPRE luciferase reporter to determine the transcriptional activity of XBP1s. It was found that SIRT1 co-transfection significantly attenuated the XBP1s-mediated 5× UPRE luciferase reporter activity (Fig. 4A). SIRT1 also significantly compromised the stimulatory effect of p300 on XBP1s-mediated luciferase reporter activity (Fig. 4B). To further test the role of endogenous SIRT1, we used shRNA strategy to knock-down the endogenous SIRT1 in HEK293 cells. Co-transfection of the shRNA targeting SIRT1 significantly increased the transcriptional activity of XBP1s by 2.4 folds, compared with co-transfection of the control vector, while SIRT1 shRNA did not affect basal level 5×UPRE luciferase reporter activity (Fig. 4C). This data demonstrated that endogenous SIRT1 specifically regulates transcriptional activity of overexpressed XBP1s rather than exerting non-specific effects on the 5×UPRE luciferase reporter activity.
Figure 4. SIRT1 negatively regulates the transcriptional activity of XBP1s.
(A, B, and C) A 5×UPRE luciferase reporter was used to evaluate the transcriptional activity of XBP1s in HEK293 cells. To normalize the level of the experimental reporter activity, firefly luciferase value was divided by Renilla luciferase value from the same sample. (A) Cells were transfected with XBP1s or an empty vector together with SIRT1 at indicated doses. Asterisk indicates statistical significance determined by Student’s t test (*p<0.01). (B) Cells were transfected with XBP1s and SIRT1 either in the presence or the absence of p300. Asterisks indicate statistical significance determined by Student’s t test (*p < 0.05; **p < 0.01). (C) Cells were transfected with XBP1s and an SIRT1 shRNA expression plasmid. Asterisk indicates statistical significance determined by Student’s t test (*p < 0.01). (D) Both Sirt1 WT and KO MEFs were treated with 300 nM thapsigargin, or 2.5 μg/mL tunicamycin, or their corresponding vehicle for 5 hours. The mRNA levels of UPR target genes were evaluated by quantitative PCR. UPR-mediated Xbp1 mRNA splicing in Sirt1 WT and KO MEFs treated with ER stress agents was calculated based on the quantitative PCR results. Asterisks indicate statistical significance determined by Student’s t test (*p < 0.05, **p < 0.01).
Next, we determined whether SIRT1 affects XBP1 target-gene transcription. For this purpose, we employed wild-type (WT, Sirt1+/+) and knock-out (KO, Sirt1−/−) mouse embryonic fibroblasts (MEFs) for Sirt1 and a pharmacological ER stress model. The ER stressor, either thapsigargin (TG) or tunicamycin (TM), induced mRNA expression of the UPR target genes, including Edem1, Ero1L, Sec61α, Bip and Gadd34 (Fig. 4D). Among them, Edem1, Ero1α and Sec61α are regarded as transcription targets of XBP1s [10, 28, 29], while Bip and Gadd34 are regarded as targets of ATF6 and PERK signaling respectively [30]. Intriguingly, ER stress triggered greater induction of XBP1s-dependent UPR target genes Edem1, Ero1α and Sec61α in Sirt1−/− MEFs than that in Sirt1+/+ MEFs. In contrast, the induction of ATF6 target gene Bip and PERK pathway target gene Gadd34 was less in Sirt1−/− MEFs than in Sirt1+/+ MEFs (Fig. 4D). Further, we noticed that Sirt1 −/− MEFs did not display higher Xbp1 mRNA splicing, as determined by the ratio of spliced vs. total Xbp1 mRNA (Fig. 4D), excluding the possibility that the stronger induction of XBP1s-target genes was caused by a compensatory over-activation of the IRE1/XBP1 pathway due to reduced ATF6 and/or PERK signaling. Taken together, these results demonstrated that SIRT1 deficiency specifically augments XBP1s-mediated gene expression rather than universally affecting gene expression of all three signaling branches of UPR in the same manner. In other words, these data supports that SIRT1 exerts a repressive regulation specific for XBP1s signaling branch of UPR.
SIRT1 sensitizes cells to ER stress-induced cell death
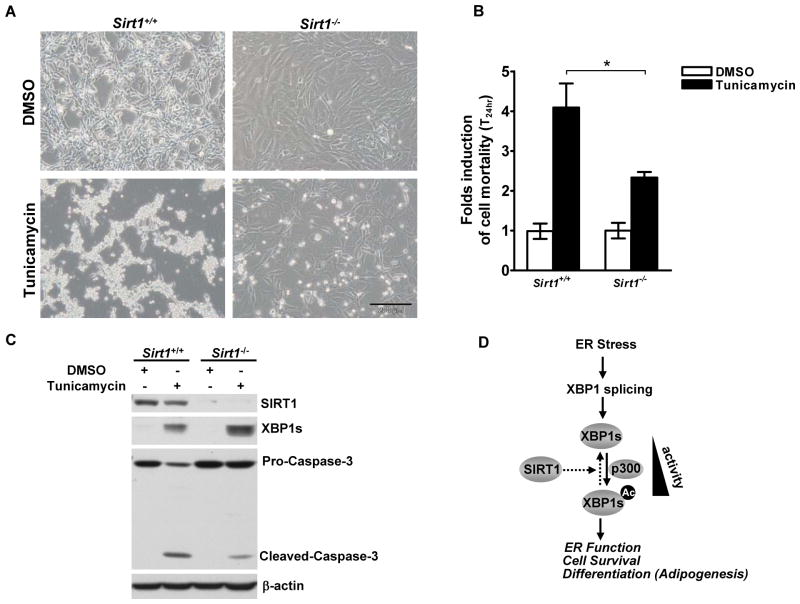
The IRE1/XBP1s signaling prevents activation of cell death pathways and determines the cell fate during sustained ER stress [5, 31, 32]. To evaluate the physiological significance of the enhanced XBP1s signaling in Sirt1 deficient cells, we measured cell viability in Sirt1+/+ and Sirt1−/− MEFs under ER stress induced by TM for 24 hours. As expected, ER stress induced cell death in both cell types (Fig.5A). Importantly, Sirt1−/− MEFs displayed approximately 50% less induction of cell death, compared with the WT control cells, as shown by a quantitative cell viability assay (Fig. 5B). Decreased cell apoptosis in Sirt1−/− cells can also be evidenced by decreased processing of pro-Caspase-3 to active-Caspase-3 in these cells compared with the WT cells (Fig. 5C). In agreement with greater resistance to ER stress-induced cell apoptosis, the Sirt1−/− MEFs displayed higher levels of XBP1s protein (Fig. 5C). These results suggest that SIRT1 sensitizes cells to ER stress-induced cell apoptosis possibly via repressing XBP1s signaling.
Figure 5. SIRT1 sensitizes ER stress-induced cell death.
(A, B, and C) Sirt1+/+ and Sirt1−/− MEFs were treated with 2.5 μg/mL tunicamycin or its vehicle (DMSO) for 24 hours. (A) Cell culture images were taken under a phase contrast microscope. Scale Bar, 200 μm. (B) The relative fold of cell mortality was assessed after trypan blue staining. Asterisk indicates statistical significance determined by Student’s t test (*p < 0.01). (C) Parallel cell lysates were analyzed for expression levels of SIRT1, XBP1s, Caspase-3 and β-actin by Western blotting. (D) Working model illustrating the role of SIRT1 in regulating XBP1s during UPR.
DISCUSSION
The UPR modulator XBP1 plays important roles in a wide range of human physiological and pathological processes [1, 2]. Although XBP1 transcription and mRNA splicing have been extensively studied [3], post-translational modifications of XBP1s remain unclear, except for a recent study revealing that XBP1s is subjected to SUMO modification [33]. We demonstrated, for the first time, that XBP1s is a target of acetylation and deacetylation mediated by p300 and SIRT1 respectively. During preparation of the manuscript, p300 was shown to physically interact with XBP1s but not with the unspliced form of XBP1 (XBP1u) [34]. Our results coupled with this finding suggest a role of p300 specific for XBP1s. We observed that overexpression of p300 increased acetylation of XBP1s and elevated its protein levels (Fig. 1), whereas SIRT1 deacetylated XBP1s and decreased its protein levels (Fig. 2D and 2E). Moreover, the p300-induced XBP1s protein levels resulted from the stabilization of XBP1s (Fig.1B), echoing the previous reports that acetylation stabilizes transcription factors such as SREBP1a [21, 23]. We further demonstrated that acetylation of XBP1s increases XBP1s-mediated 5×UPRE luciferase reporter activity (Fig. 4B). Since we have observed that increasing the protein levels of XBP1s by proteasome inhibitor, MG132 failed to stimulate XBP1s-mediated 5XUPRE luciferase reporter activity (data not shown). It is likely that acetylation regulates XBP1s transcriptional activity independent of its regulation of XBP1s protein levels. Another side of the same token, overexpression of SIRT1 repressed XBP1s-mediated luciferase reporter activity (Fig. 4A and 4B). Our study suggested that like SUMOylation, acetylation and/or deacetylation also constitute an important regulatory mechanism controlling XBP1s’ protein levels and its transcription function. However, further studies are necessary to identify the acetylated lysine(s) in XBP1s. Is it unique to XBP1s as compared to XBP1u? Does acetylation compete with SUMOylation for the same lysine(s) in XBP1s? How does the modification of the lysine(s) regulate the multiple functions of XBP1s? The answers to these questions are critical for elucidating the post-translational regulation of the UPR transcription factor XBP1s.
Emerging evidence has implied that there exists intricate relationship between ER stress response and lifespan or aging [35, 36]. Previously, SIR2, a homologue of mammalian SIRT1 in C. elegans, has been shown to regulate the lifespan of C. elegans, and this effect has been associated with its capacity to regulate ER stress response genes [17]. SIRT1 has also been implicated in promoting longevity in mammals [14]. However, it remains unclear if SIRT1 regulates mammalian UPR signaling. Our study demonstrated that SIRT1 is a deacetylase for XBP1s. Overexpression of SIRT1 inhibited XBP1s’ activity (Fig. 4A, 4B). Conversely, knock-down of SIRT1 increased XBP1s’ activity (Fig. 4C). Furthermore, Sirt1 deficiency led to enhanced upregulation of XBP1 target gene expression and less apoptotic cell death in MEFs upon ER stress challenge. These results are innovative and provide a solid biochemical and functional connection between SIRT1 and the most conserved UPR signaling branch-IRE1/XBP1 signaling.
XBP1s has been shown to play a crucial role in pathogenesis of MM, a neoplastic disease of B cell origin. High level of XBP1s expression has been linked to rapid tumor expansion and a poor prognosis of MM disease [37, 38]. Inhibition of XBP1 splicing, by inhibiting IRE1α activity, increased apoptosis in MM cells in response to ER stress [39]. Intriguingly, several lines of works including our own data (unpublished data) showed that SIRT1 activator resveratrol also induced apoptotic cell death in MM cells [40, 41]. Although the detailed molecular mechanism underlying the therapeutic effects of resveratrol remains unclear, our current study demonstrating that SIRT1 represses XBP1s and sensitizes cells to ER stress-induced apoptosis suggests that resveratrol might act through activating SIRT1 and consequently inhibiting XBP1s signaling, an absolute requirement for MM tumor growth, to achieve its killing effects. It would be interesting to determine if SIRT1/XBP1s signaling pathway mediates the therapeutic effect of resveratrol on MM cells and other cancers that are responsive to resveratrol, such as breast cancer and colon cancer, which were also associated with an upregulation of XBP1 [42–44].
Furthermore, it was recently shown that XBP1s plays a critical role for adipogenesis via directing gene expression of C/EBPα, an important transcription factor required for adipogenesis [11]. Either overexpression of SIRT1 or XBP1s knock-down attenuates adipogenesis. In contrast, RNA interference of Sirt1 enhances adipogenesis [11, 45]. However, it remains unknown if there exists a functional connection between SIRT1 and XBP1s in regulating adipogenesis. Our results indicated that SIRT1 regulates XBP1s post-translationally and represses its transcriptional activity, suggesting that XBP1s might serve as an important downstream effector through which SIRT1 exerts its anti-adipogenic effects. Elucidation of the mechanism by which SIRT1 coordinates with XBP1s in regulation adipogenesis might provide a framework for the development of promising therapeutic strategies that target SIRT1/XBP1s signaling axis to treat human diseases with deregulated fat metabolism and/or UPR signaling, such as obesity and type II diabetes.
In summary, our study demonstrated that mammalian XBP1s is subjected to acetylation and deacetylation mediated by p300 and SIRT1 respectively. The acetylation/deacetylation constitutes an important post-translational mechanism controlling the protein stability and activity of XBP1s (Fig. 5D). The biochemical and functional linkage between SIRT1 and XBP1s would shed novel insights into the molecular mechanisms by which SIRT1 regulates UPR signaling and suggests SIRT1 as a potential therapeutic target for treating diseases where deregulation of UPR/XBP1s signaling is implicated.
Acknowledgments
We are grateful to Drs. Randy Kaufman, Paul Robbins, and Ming Hu for generously providing plasmids and cells. We thank Dr. Yi-Jiun Chen for constructing the XBP1s plasmids and other co-workers of Dr. Ouyang’s laboratory for technical supports and stimulating discussion. We also thank Mimi Li and Donna Gaspich for assistance in the preparation of the manuscript.
FUNDING
This work is supported by grants from the NIH/NIDCR (DE017439 to HJ.O.), American Association of Endodontists (HJ.O.), the start-up fund provided by the Department of Medicine, School of Medicine at University of Pittsburgh (HJ.O.) and National Natural Science Foundation of P.R. China (No. 30901677 to FM.W.).
Abbreviations
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- XBP1
X-box-binding protein 1
- XBP1s
spliced form of XBP1
- XBP1u
unspliced form of XBP1
- bZIP
basic-region leucine zipper
- CREB
cAMP responsive element binding protein
- ATF
activating transcription factor
- SIR2
Silent Information Regulator 2
- Ace-K
acetylated lysine
- PERK
PKR-like endoplasmic reticulum kinase
- IRE1
insositol requiring enzyme-1
- ATF6
activating transcription factor-6
- C/EBP
CCAAT/enhancer-binding protein
- DMEM
Dulbecco’s Modified Eagle Medium
- FBS
fetal bovine serum
- NEAA
nonessential amino acid
- IP
immunoprecipitation
- PCR
polymerase chain reaction
- TG
thapsigargin
- TM
tunicamycin
- MEF
mouse embryonic fibroblasts
- TSA
trichostatin A
- NAM
nicotinamide
- NAD
nicotinamide adenine dinucleotide
- PVDF
polyvinylidene difluoride
- WT
wild-type
- KO
knock-out
- MM
multiple myeloma
References
- 1.Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida H. Unconventional splicing of XBP-1 mRNA in the unfolded protein response. Antioxid Redox Signal. 2007;9:2323–2333. doi: 10.1089/ars.2007.1800. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 5.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 7.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 8.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, Munshi N, Horner J, Ivanova EV, Protopopov A, Anderson KC, Tonon G, DePinho RA. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon HS, Ott M. The ups and downs of SIRT1. Trends Biochem Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 16.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 17.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Clauss IM, Chu M, Zhao JL, Glimcher LH. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 1996;24:1855–1864. doi: 10.1093/nar/24.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry. 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lassot I, Estrabaud E, Emiliani S, Benkirane M, Benarous R, Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J Biol Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 22.Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li X, Dyson NJ, Hart AC, Naar AM. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giandomenico V, Simonsson M, Gronroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol Cell Biol. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci USA. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton JD. Physiology. Unfolding lipid metabolism. Science. 2008;320:1433–1434. doi: 10.1126/science.1159651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, deCabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 28.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 30.Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 31.Iwakoshi NN, Pypaert M, Glimcher LH. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J Exp Med. 2007;204:2267–2275. doi: 10.1084/jem.20070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A. HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction. PLoS Biol. 2010;8:e1000410. doi: 10.1371/journal.pbio.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Qi L. SUMO modification regulates the transcriptional activity of XBP1. Biochem J. 2010;429:95–102. doi: 10.1042/BJ20100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng L, Liu YP, Sha H, Chen H, Qi L, Smith JA. XBP-1 couples endoplasmic reticulum stress to augmented IFN-beta induction via a cis-acting enhancer in macrophages. J Immunol. 2010;185:2324–2330. doi: 10.4049/jimmunol.0903052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naidoo N. The endoplasmic reticulum stress response and aging. Rev Neurosci. 2009;20:23–37. doi: 10.1515/revneuro.2009.20.1.23. [DOI] [PubMed] [Google Scholar]
- 36.Tatar M. SIR2 calls upon the ER. Cell Metab. 2005;2:281–282. doi: 10.1016/j.cmet.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura M, Gotoh T, Okuno Y, Tatetsu H, Sonoki T, Uneda S, Mori M, Mitsuya H, Hata H. Activation of the endoplasmic reticulum stress pathway is associated with survival of myeloma cells. Leuk Lymphoma. 2006;47:531–539. doi: 10.1080/10428190500312196. [DOI] [PubMed] [Google Scholar]
- 38.Bagratuni T, Wu P, Gonzalez de Castro D, Davenport EL, Dickens NJ, Walker BA, Boyd K, Johnson DC, Gregory W, Morgan GJ, Davies FE. XBP1s levels are implicated in the biology and outcome of myeloma mediating different clinical outcomes to thalidomide-based treatments. Blood. 2010;116:250–253. doi: 10.1182/blood-2010-01-263236. [DOI] [PubMed] [Google Scholar]
- 39.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhardwaj A, Sethi G, Vadhan-Raj S, Bueso-Ramos C, Takada Y, Gaur U, Nair AS, Shishodia S, Aggarwal BB. Resveratrol inhibits proliferation, induces apoptosis, and overcomes chemoresistance through down-regulation of STAT3 and nuclear factor-kappaB-regulated antiapoptotic and cell survival gene products in human multiple myeloma cells. Blood. 2007;109:2293–2302. doi: 10.1182/blood-2006-02-003988. [DOI] [PubMed] [Google Scholar]
- 41.Boissy P, Andersen TL, Abdallah BM, Kassem M, Plesner T, Delaisse JM. Resveratrol inhibits myeloma cell growth, prevents osteoclast formation, and promotes osteoblast differentiation. Cancer Res. 2005;65:9943–9952. doi: 10.1158/0008-5472.CAN-05-0651. [DOI] [PubMed] [Google Scholar]
- 42.Shajahan AN, Riggins RB, Clarke R. The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 2009;22:241–246. doi: 10.1358/dnp.2009.22.5.1378631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujimoto T, Yoshimatsu K, Watanabe K, Yokomizo H, Otani T, Matsumoto A, Osawa G, Onda M, Ogawa K. Overexpression of human X-box binding protein 1 (XBP-1) in colorectal adenomas and adenocarcinomas. Anticancer Res. 2007;27:127–131. [PubMed] [Google Scholar]
- 44.Delmas D, Rebe C, Lacour S, Filomenko R, Athias A, Gambert P, Cherkaoui-Malki M, Jannin B, Dubrez-Daloz L, Latruffe N, Solary E. Resveratrol-induced apoptosis is associated with Fas redistribution in the rafts and the formation of a death-inducing signaling complex in colon cancer cells. J Biol Chem. 2003;278:41482–41490. doi: 10.1074/jbc.M304896200. [DOI] [PubMed] [Google Scholar]
- 45.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]