Abstract
Killer immunoglobulin-like receptors (KIR) are expressed in a variegated, clonally restricted fashion on natural killer (NK) cells and are important determinants of NK cell function. Although silencing of individual KIR genes is strongly correlated with the presence of CpG dinucleotide methylation within the promoter, the mechanism responsible for silencing has not been identified. Our results show that antisense transcripts mediate KIR transcriptional silencing through a novel PIWI-like 28 base small RNA. Although PIWI RNA-mediated silencing of transposable elements within germ cells have been described, this is the first report that identifies a PIWI-like RNA in an immune somatic cell lineage and identifies a mechanism which may be broadly used in orchestrating immune development.
Introduction
Signaling through MHC class I-specific inhibitory receptors, such as KIR, are important not only to dampen activation signals, but also for the acquisition of NK cell function and the establishment of self-tolerance during development. Despite significant promoter homologies within the KIR locus, individual KIR genes are regulated independently and probabilistically (1–4). The question of how KIR transcriptional patterns are established in NK cells is largely unresolved. A thorough analysis of the complete 2 kb intergenic region upstream of the KIR3DL1 gene revealed that the promoter proximal to the translational start site has bidirectional activity, and a uni-directional distal promoter exists upstream of the conventional proximal promoter (5, 6). Polyadenylated antisense transcripts and full-length distal transcripts were previously cloned from several KIR genes, leading to the hypothesis that transcription across proximal KIR promoters may be involved in establishing epigenetic marks that influence gene expression (7). In support of this hypothesis, we show that a novel 28 base PIWI-like RNA processed from KIR3DL1 antisense transcripts is essential for transcriptional silencing.
Materials and Methods
NK cell and CD34+ HPC isolation
The use of all human tissue was approved by the Committee on the Use of Human Subjects in Research at the University of Minnesota, and informed consent was secured in accordance with the Declaration of Helsinki. CD56+ NK cells were column-isolated from peripheral blood using magnetic beads (Miltenyi Biotech). KIR3DL1 subsets were isolated by staining CD56+ NK cells with PE-labeled DX9 antibody (Bectin-Dickinson [BD]) and separating PE-positive and -negative populations using anti-PE magnetic beads (Miltenyi). CD34+ HPCs were isolated from umbilical cord blood by double-column positive selection using anti-CD34 microbeads (Miltenyi).
Quantitative RT-PCR for KIR transcripts and bisulfite DNA analysis
For the quantification of the KIR3DL1 antisense transcripts, cDNA synthesis was carried out at 55°C using a KIR3DL1/S1-specific RT primer. A Taqman primer and probe set was used for KIR3DL1 antisense transcript amplification. Primer probe sets for KIR3DL1 distal transcripts and KIR coding transcripts have been published previously (8, 9). Additional primers used in this study are shown in Supplementary Table I. Bisulfite sequencing analysis of the KIR3DL1 promoter was performed as previously described (2).
Periodate oxidation/β-elimination reaction
The small RNA fraction from 2×106 NK cells was isolated using the PureLink™ miRNA Isolation Kit (Invitrogen). For periodate oxidation/β-elimination, we followed published methods (10).
Retroviral and lentiviral vector designs
Lentiviral vectors were created by MluI/BamHI restriction digest and oligo annealing using pELNS with an H1 pol III promoter. Oligo sequences are shown in Supplementary Table I. All murine stem cell virus (MSCV)-enhanced green fluorescent protein (eGFP) vector constructs were created using the Gateway® cloning system (Invitrogen). Primer sequences are shown in Supplementary Table I.
CD34+ HPC viral transduction and in vitro NK cell differentiation
CD34+ cells isolated from umbilical cord blood were transduced with retrovirus or lentivirus and cultured for 21 days on the EL08-1D2 stromal line (11). The culture medium and cytokines for NK cell differentiation are published (8).
Flow cytometry and antibodies used
Cell sorting was performed on the FACSAria (BD Biosciences), and phenotypic analysis was performed on the FACSCalibur (BD) using CellQuest Pro Software (BD). The antibodies used in this study were APC-conjugated NCAM16.2 (CD56) and CD34, and PE-conjugated DX9 (anti-CD158e), EB6 (anti-CD158a/h) and GL183 (CD158b/j) (BD).
Results
Distinct intergenic transcriptional profiles in KIR3DL1− and KIR3DL1+ NK cells
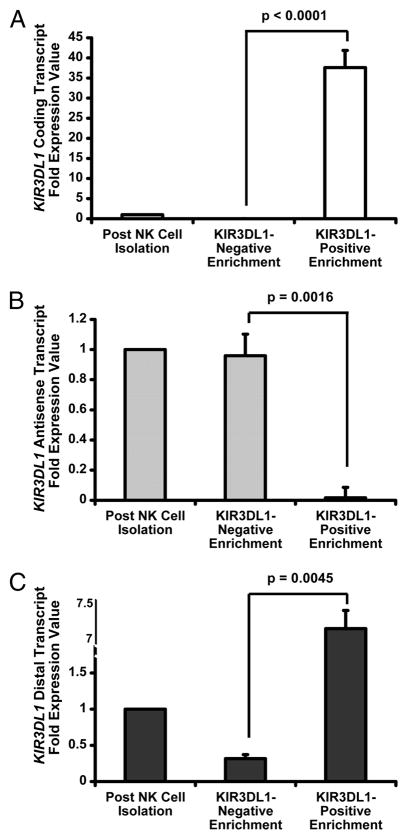
To determine how expression of KIR3DL1 gene transcripts (Fig. S1) relates to receptor expression, we isolated CD56+ NK cells from the peripheral blood of healthy donors and separated KIR3DL1− and KIR3DL1+ NK cell populations. As expected, the expression of KIR3DL1 coding transcripts strongly correlated with surface receptor expression (Fig. 1A). In contrast, KIR3DL1 antisense transcript expression was inversely correlated with surface receptor expression. Virtually all of the KIR3DL1 antisense transcripts detected in mature CD56+ NK cells were confined to the KIR3DL1− subset (Fig. 1B). KIR3DL1 distal transcripts were detected at high levels within the KIR3DL1+ NK cell population, which is consistent with a previous analysis of the distal promoter (8). Interestingly, low levels of KIR3DL1 distal transcript were present in the KIR3DL1− NK cell population (Fig. 1C). Therefore, the potential for creation of double-stranded RNA is restricted to cells lacking receptor expression.
Figure 1. KIR3DL1 Antisense Transcripts are Predominantly Expressed in NK Cells Lacking KIR3DL1 Surface Expression.
qRT-PCR measurement of relative KIR3DL1 (A) coding, (B) antisense, and (C) distal transcript levels in enriched KIR3DL1− and KIR3DL1+ NK cell populations normalized against transcript levels in total CD56+ NK cells. Data was pooled from 3 independent experiments, and error bars represent the standard error value between experiments. P values comparing transcript expression between isolated NK cell populations were derived using a Student’s t test.
Identification of double-stranded RNA across the KIR3DL1 promoter
We performed an S1 nuclease protection assay using RNA purified from CD56+ peripheral blood NK cells to determine whether KIR3DL1 distal and antisense transcripts form double-stranded RNA (dsRNA). Sequencing analysis of cloned products led to the identification of a 228 bp dsRNA that extends from position −18 to −306 bp relative to the KIR3DL1 mRNA transcriptional start site. The 5′ boundary of the cloned dsRNA region (−306) coincides precisely with the transcriptional start site of the KIR3DL1 distal transcript, consistent with a direct role of the distal promoter in the generation of dsRNA (Fig. S2).
A novel 28 base small RNA is processed from KIR antisense transcripts
To test whether small RNAs are processed within the dsRNA region, expression vectors producing sense and antisense KIR3DL1 transcripts spanning the promoter region were transfected into HEK293 cells to generate dsRNA. 24 hours post-transfection total RNA was isolated, and the small RNA fraction was enriched. A cDNA library was generated and screened with a probe containing the entire KIR3DL1 dsRNA region. A single 28 base antisense small RNA was identified in each of three independent small RNA libraries generated. The specific generation of this small RNA from dsRNA was confirmed in additional experiments in which KIR3DL1 sense- or antisense-expressing vectors were transfected separately into HEK293 cells, and the small RNA was not detected (Fig. S2). We also identified a 28 base RNA processed from the KIR2DL1 antisense transcript (Fig. S3). To confirm that the 28 base RNA is not generated as a cell line artifact, we harvested CD56+ NK cell small RNAs from multiple donors and were able to isolate the KIR3DL1 28 base RNA using a PCR-based cloning strategy.
The KIR3DL1 28 base small RNA contains a protective group at its 3′ terminus
The only known species of small RNAs in the 25–30 nucleotide range are piRNAs, which are implicated in the germline silencing of transposons (12). Mammalian piRNAs are 2′-0-methylated at their 3′ terminal ribose, while siRNAs and microRNAs have terminal hydroxyl groups at both the 2′ and 3′ positions (13). To determine whether the KIR3DL1 28 base small RNA has a 3′ terminal modification, a periodate oxidation/β-elimination reaction was performed on total RNA from CD56+ peripheral blood NK cells mixed with synthetic control RNA lacking any 3′-terminal modifications. Only RNAs containing both 2′ and 3′ hydroxyl groups react with NaIO4, and β-elimination shortens NaIO4-reacted RNA by one nucleotide, leaving a 3′-monophosphate terminus. Both the KIR3DL1 28 base RNA and control RNA were cloned out of the total reacted RNA and sequenced. As expected, the majority (8/10) of control RNA sequences were shortened by one nucleotide. In contrast, all 10 sequences for the KIR3DL1 28 base RNA were full-length (Table I). Therefore, the KIR3DL1 28 base RNA contains a protective group at either the 2′ or 3′ ribose position of its 3′ terminus that renders it resistant to periodate oxidation/β-elimination.
Table 1.
The KIR3DL1 28 bp RNA is Protected at the 3′ Terminus from Periodate Oxidation/β-Elimination*
| RNA sequence without periodate oxidation/β-elimination (5′-3′) | RNA sequence with periodate oxidation/β-elimination (5′-3′) | # of clones | |
|---|---|---|---|
| Spiked Control RNA (no 3′-terminal-O-methylation) | UGACGAAUGCACGUAAUGCAGUGUAU | UGACGAAUGCACGUAAUGCAGUGUA UGACGAAUGCACGUAAUGCAGUGUAU |
8/10 2/10 |
| KIR3DL1 28 base RNA | ACAUGGCUUCCUGGAAAUUGCUCUCACU | ACAUGGCUUCCUGGAAAUUGCUCUCACU | 10/10 |
Control RNA lacking a 3′-terminal 2′-O-methylation group was mixed with total RNA from peripheral blood CD56+ NK cells. Periodate oxidation and β-elimination reactions were carried out on pooled RNA. The treated control RNA and KIR3DL1 28 base RNAs were then converted to cDNA, cloned, and sequenced.
KIR3DL1 antisense transcripts inhibit KIR expression, and the 28 base PIWI-like RNA sequence is necessary and sufficient for function
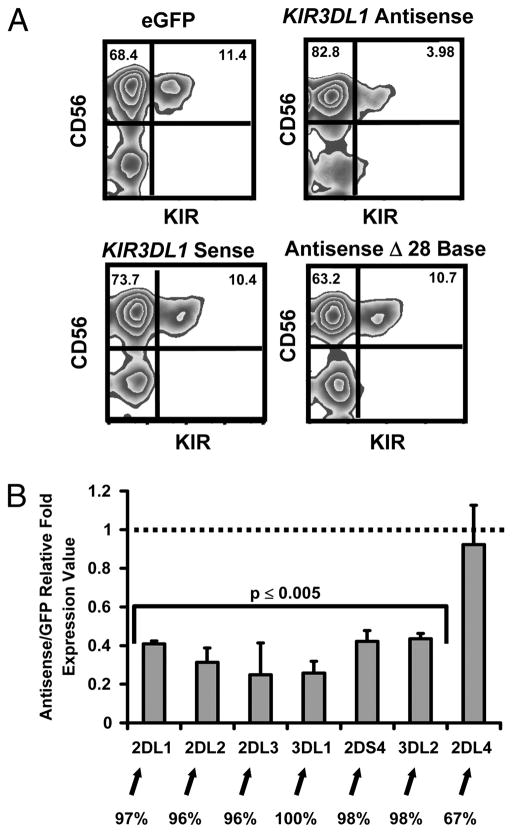
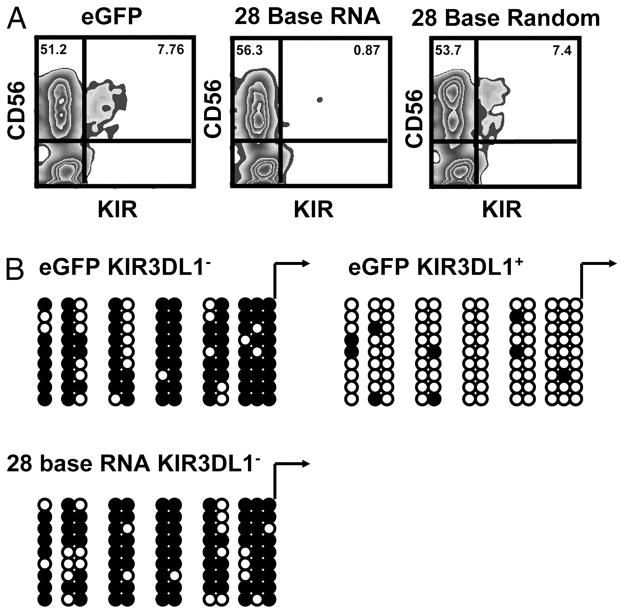
We transduced primary CD34+ hematopoietic progenitor cells with a retroviral vector expressing the full-length KIR3DL1 antisense transcript and differentiated these cells into mature CD56+ NK cells. Over-expression of KIR3DL1 antisense transcripts led to an approximately 70% reduction in KIR expression. This effect was dependent upon transcript orientation, as over-expression of sense transcripts homologous to the KIR3DL1 promoter did not lead to any reduction in KIR expression. Importantly, over-expression of KIR3DL1 antisense transcripts with the 28 base RNA sequence removed did not result in any reduction in KIR expression, implying that the processed KIR3DL1 28 base PIWI-like RNA is necessary for silencing (Fig. 2A). We also transduced HPCs with a lentiviral vector expressing the 28 base PIWI-like RNA. Compared with eGFP and scramble controls, over-expression of the 28 base PIWI-like RNA led to an approximately 90% reduction in KIR expression (Fig. 3A).
Figure 2. KIR Antisense Transcripts Negatively Regulate KIR Gene Expression.
KIR3DL1 antisense and sense transcripts were separately cloned into the MSCV-eGFP vector. An additional KIR3DL1 antisense MSCV-eGFP construct lacking the 28 base segment was also generated. Retroviral supernatants were used to transduce CD34+ cells, which were developed into NK cells. (A) FACS plots representative of 4 experiments showing the surface KIR expression for cells transduced with each construct. (B) qRT-PCR comparison of KIR gene expression in cells transduced with either the KIR3DL1 Antisense vector or the control eGFP vector. Values are presented as a fold ratio with 1 equal to KIR expression observed with eGFP-only vector. Percent homology with the KIR3DL1 promoter is indicated below the graph. Data was pooled from 4 independent experiments, and error bars represent the standard error values between experiments. P values comparing KIR transcript expression between isolated NK cell populations were derived using a Student’s t test.
Figure 3. Expression of the 28 Base RNA correlates with methylation of the KIR3DL1 Proximal Promoter.
Lentiviral vectors designed to over-express eGFP alone or, the 28 base PIWI-like RNA or a random 28 base RNA were transduced into CD34+ cells and developed into NK cells. (A) FACS plots representative of 3 experiments showing the surface KIR expression for cells transduced with each construct. (B) Bisulfite sequencing analysis of the DNA methylation status of CpG dinucleotides within the KIR3DL1 proximal promoter from KIR3DL1− and KIR3DL1+ cells from eGFP control cultures (upper panel) and KIR3DL1− cells from 28 base RNA over-expressing cultures (lower panel). Blackened circles represent non-converted, methylated cytosines. Open circles represent unmethylated cytosines.
The mRNA expression levels for each gene were reduced 4–5 fold in cells over-expressing KIR3DL1 antisense transcripts. Interestingly, expression of the KIR2DL4 gene, which shares only 67% sequence identity with the KIR3DL1 promoter and is constitutively expressed by mature CD56+ NK cells (1), was not affected by over-expression of the KIR3DL1 antisense transcript (Fig. 2B). The silencing effect of the KIR3DL1 antisense on other KIR genes with significant promoter homologies is likely due to the presence of high levels of antisense transcript produced by the constitutive MSCV promoter throughout development.
Antisense transcript expression correlates with CpG methylation within the KIR3DL1 promoter
Since the correlation between DNA methylation of KIR promoters and transcriptional silencing is well established (2, 3, 4), we analyzed the methylation status of the endogenous KIR3DL1 promoter region in cells that were transduced with either the eGFP control construct or the 28 base PIWI-like RNA and then differentiated along the NK lineage. CD56+ KIR3DL1+ and KIR3DL1− cells were sorted from eGFP cultures, and CD56+ KIR3DL1-negative cells were sorted from 28 base PIWI-like RNA cultures. Genomic DNA was extracted, and bisulfite sequencing was performed using primers specific for the KIR3DL1 promoter region (2). Consistent with previous studies, we observed very little methylation of the KIR3DL1 promoter in KIR3DL1+ cells and dense methylation in the KIR3DL1− cells from the eGFP control culture. There were too few KIR3DL1+ cells from 28 base PIWI-like RNA cultures for analysis. However, when we analyzed the KIR3DL1 promoter in sorted KIR3DL1− cells from 28 base PIWI-like RNA over-expressing cultures we observed dense methylation (Fig. 3B.). These results show that the 28 base PIWI-like RNA participates in KIR promoter methylation during human NK cell development.
Discussion
Three facts support the conclusion that the KIR small RNAs described herein belong to the piRNA family: (i) the 28 base size of the RNA, (ii) the presence of a protective group at the 3′ terminus of the RNA and (iii) the finding that the KIR small RNAs are processed exclusively from antisense transcripts. Although we have not yet identified the protein(s) that interact with the 28 base PIWI-like RNA, we have confirmed the expression of PIWIL4 in human NK cells (Fig. S4.).
During human NK cell development, CpG sites within KIR promoters are demethylated by a mechanism that remains to be elucidated. We propose that at a late stage in human NK cell development, transcription is initiated in a probabilistic fashion from the bi-directional promoters of clonally restricted KIR genes. If the promoter initiates in the forward direction, coding transcripts are generated, and the open epigenetic state of the promoter becomes “locked-in”. If the promoter initiates in the reverse direction, antisense transcripts are generated and quickly processed into the 28 base PIWIL-like RNA, which participates in cis in the maintenance or establishment of DNA methylation and stable, clonal silencing of the promoter (Fig. S5).
It may appear contradictory that distal transcript expression is higher in KIR+ cells (Fig. 1C), yet it is required for formation of dsRNA and processing of the 28 base PIWI-like RNA. This paradox may be explained by the observation that distal transcripts appear to have duel functions. If antisense transcripts from the bi-directional proximal promoter are present, distal transcripts are involved in formation of dsRNA for subsequent processing of the 28 base PIWI-like RNA. However, if antisense transcripts are not present, transcription from the distal promoter appears to promote transcription of KIR genes in response to IL-15 stimulation (8).
While there is significant homology in the 28 base PIWI-like RNA sequence amongst the clonally restricted KIR genes, we envision the shutdown of single genes to occur though the local action of chromatin-modifying enzymes that interact with the 28 base PIWI-like RNA at the promoter. We suggest that antisense transcripts are rapidly and efficiently processed into the 28 base PIWI-like RNA, which mediates epigenetic silencing without diffusion or transport away from the promoter.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant P01-CA-111412 (JSM, SKA) and R01-HL-55417 (JSM). This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E (SKA). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (SKA).
Footnotes
The authors have no financial conflicts of interest.
References
- 1.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in the killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 2.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, Carrington M, Trowsdale J, Lutz CT. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 4.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial Role of DNA Methylation in Determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 5.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, Anderson SK. Identification of bidirectional promoters in human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 6.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 7.Pascal V, Stulberg MJ, Anderson SK. Regulation of class I major histocompatibility complex receptor expression in natural killer cells: one promoter is not enough! Immunol Rev. 2006;214:9–21. doi: 10.1111/j.1600-065X.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- 8.Cichocki F, Hanson RJ, Lenvik T, Pitt M, McCullar V, Li H, Anderson SK, Miller JS. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann TL, McQueen KL, Guethlein LA, Parham P, Miller JS. A subpopulation of human peripheral blood natural killer cells that lacks inhibitory receptors for self-MHC is developmentally immature. Blood. 2007;110:578–586. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akbergenov R, Si-Ammour A, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins F, Jr, Hohn T, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oostendorp RA, Harvey KN, Kusadasi N, de Bruijn MF, Saris C, Ploemacher RE, Medvinsky AL, Dzierzak EA. Stromal lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood. 2002;99:1183–1189. doi: 10.1182/blood.v99.4.1183. [DOI] [PubMed] [Google Scholar]
- 12.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 13.Kirino Y, Mourelatos Z. Mouse PIWI-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.