Abstract
Corticotropin releasing hormone is a neurotransmitter in the inferior olive complex of marsupials and mammals. The ontogeny of corticotropin releasing hormone gene expression in the rat inferior olive has not been described. Using in-situ hybridization histochemistry in 25 animals, we established the developmental profile of the peptide’s messenger ribonucleic acid in the postnatal rat. CRH-messenger RNA was first detectable in two of four animals on the fifth postnatal day. Subsequently, gene expression increased linearly with age: by day 14, CRH was expressed in all olivary complex nuclei, and the distribution and relative abundance on day 18 were comparable to those in the adult. The developmental profile of CRH-mRNA in the rat inferior olive differs from those in the mouse and opossum, and from the pattern in the rat hypothalamus, suggesting species- and site-specificity of the peptide’s function.
Keywords: corticotropin releasing hormone, development, messenger RNA, inferior olive, rat, in-situ hybridization
The inferior olive is the source of climbing fiber afferents to the cerebellar Purkinje cells.11,20 The development of the olivo-cerebellar pathway has been delineated using both anatomical and immunocytochemical techniques.1,8,23,26 A number of neuropeptides are expressed in the developing inferior olive neurons, either transiently or to adulthood.18,21,25,26 Corticotropin releasing hormone and its messenger RNA (mRNA) have been demonstrated in the inferior olive of adult opossum,10 rat,22,28 rabbit,11 cat19 and primates.12,28 The developmental profile of CRH-mRNA has been established in the mouse17 and opossum.9 In this study, we examined the developmental profile of CRH-mRNA in the inferior olive during the first three postnatal weeks in the rat.
EXPERIMENTAL PROCEDURES
Animals and study design
Time-pregnant Sprague–Dawley rats were obtained from Zivic-Miller (Zelienople, PA), kept on a 12 hr light/dark cycle and given access to unlimited lab chow and water.4,30 Time of birth of pups was determined every 12 hr, and day of birth was considered day 0. Litters were culled to 12 pups on the first postnatal day.
Pups (n=3 per age group) were sacrificed by rapid decapitation on postnatal days 1, 2, 3, 5 (n=4), 7, 10, 14 and 18. Several litters were used, so that the minimal number of remaining pups per litter was eight. Rats were sacrificed within 45 sec of disturbance,29 between 8 and 9:30 a.m. to avoid potential diurnal variability in CRH-mRNA abundance.27 Brains were removed onto dry ice and stored at −80°C. After removal of olfactory bulbs, brains were placed on a chuck with medulla up and sectioned coronally.24 Sections (20 µm) were mounted on gelatin-coated slides and stored at −80°C.4,5,29,30
Preparation of CRH oligonucleotide
A 60-nucleotide synthetic probe corresponding to the codons for the 20 COOH-amino acids of CRH16 was generated using an Applied Biosystems DNA Synthesizer.29,30 The probe was labeled on the 3′ end with S35-dATP (New England Nuclear) using terminal deoxynucleotidyl transferase deoxynucleotidyl transferase (Bethesda Research Lab.), as previously described.4,5,30 The control probe consisted of a similar length sense-strand, similarly generated and labeled. Specific activity of probes was 5–8×108 cpm/µg.
In-situ hybridization (ISH) and quantitation procedures
ISH has been described in detail.4,5,29,30 Briefly, slides were brought to room temperature, air-dried and fixed for 20 min in 4%-buffered paraformaldehyde. Following a graded ethanol treatment, sections were exposed to acetic anhydride-triethanolamine, then dehydrated through 100% ethanol. Sections were subjected to prehybridization for 1 hr at 40°C, in hybridization buffer lacking dextran sulfate and probe. Hybridization was for 20 hr at 40°C in a humidity chamber. Hybridization buffer consisted of: 50% formamide, 4× SSC (1× SSC is 0.15 M NaCl in 0.015 M sodium citrate, pH=7), 0.5 g/ml sheared, single-stranded salmon sperm DNA, 25 µg/ml yeast tRNA, 100 mM DTT, 5× Denhardt’s solution and 10% dextran sulfate. The reaction mixture contained probe (5–8×105 cpm) in a volume of 0.03 ml per section. Serial washes (2× SSC for 15 min × 4 at 40°C, 1× and 0.4× SSC for 30 min each at room temperature) were followed by dehydration and apposition to film (Hyperfilm B-Max, Amersham, U.K.).
Sections at the level of anterior, middle and caudal inferior olive24 were used for semi-quantitative image analysis. Anatomic landmarks were verified by cresyl violet staining, and each point was derived from four to eight sections from three individual rats. Quantitation and statistical analyses have been described.4,30 Briefly, optical density (OD) was determined over specific olivary nuclei and over dorsolateral medulla, as background, using the MCID software image analysis system (Imaging Research, Ontario, Canada). Ratios of olive to medulla OD were determined as a measure of the abundance of CRH-mRNA, thus eliminating background variability with age.
RESULTS
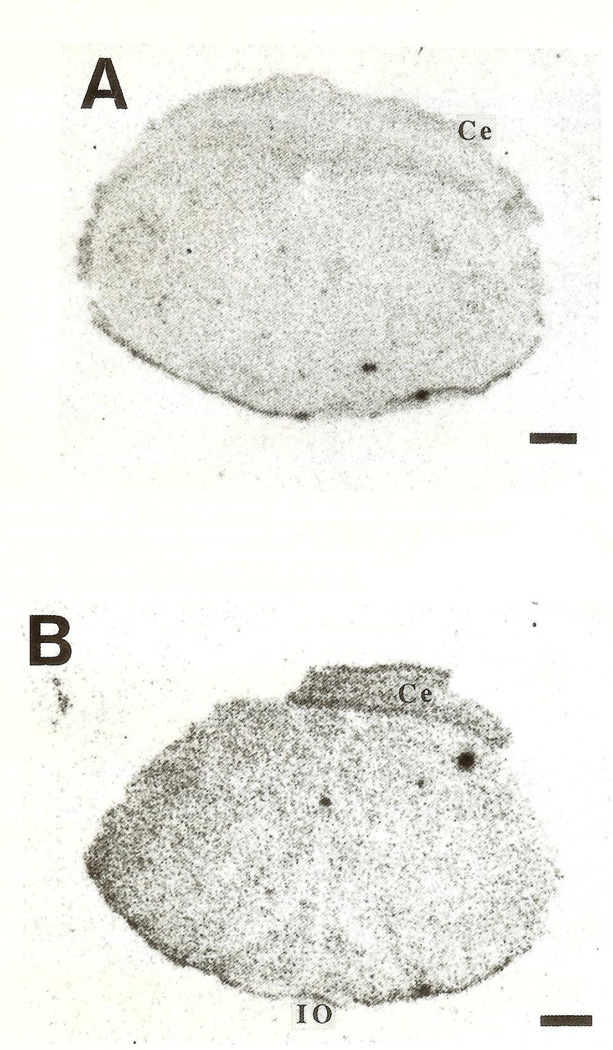
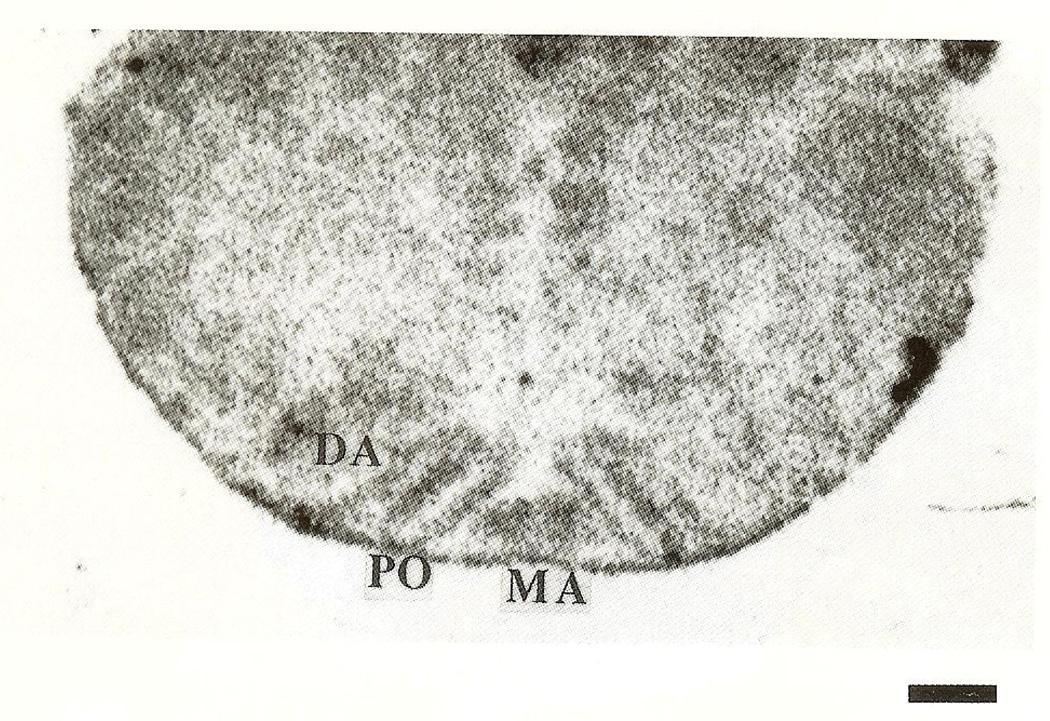
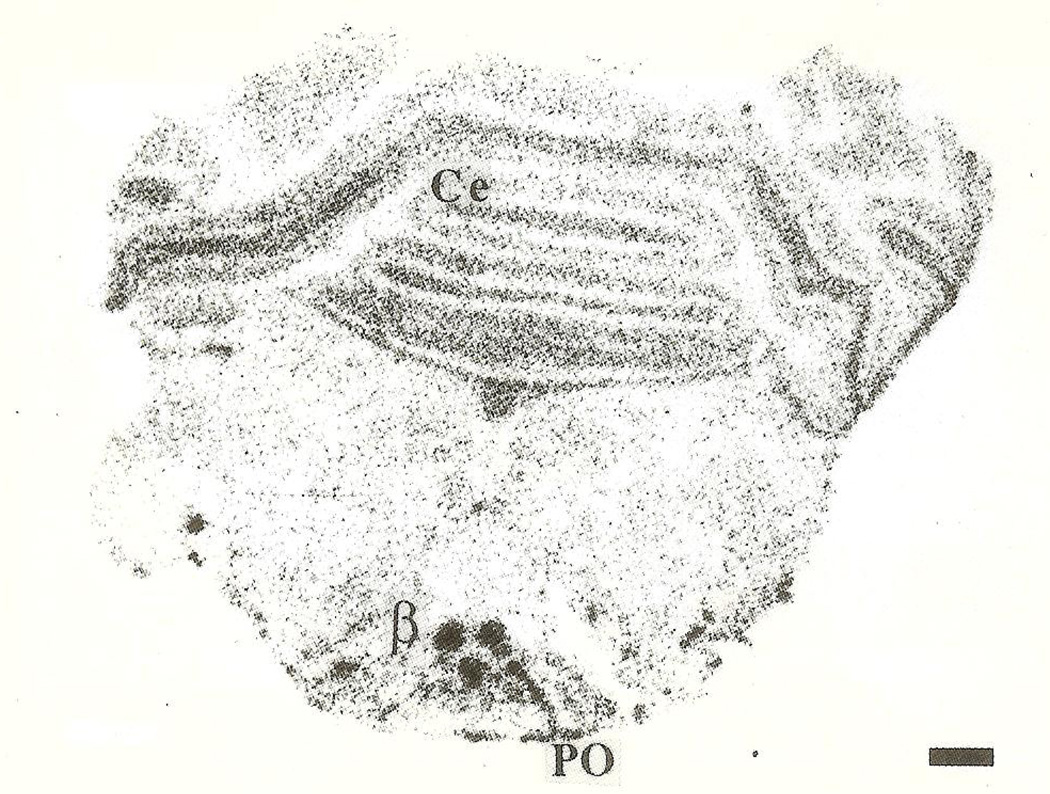
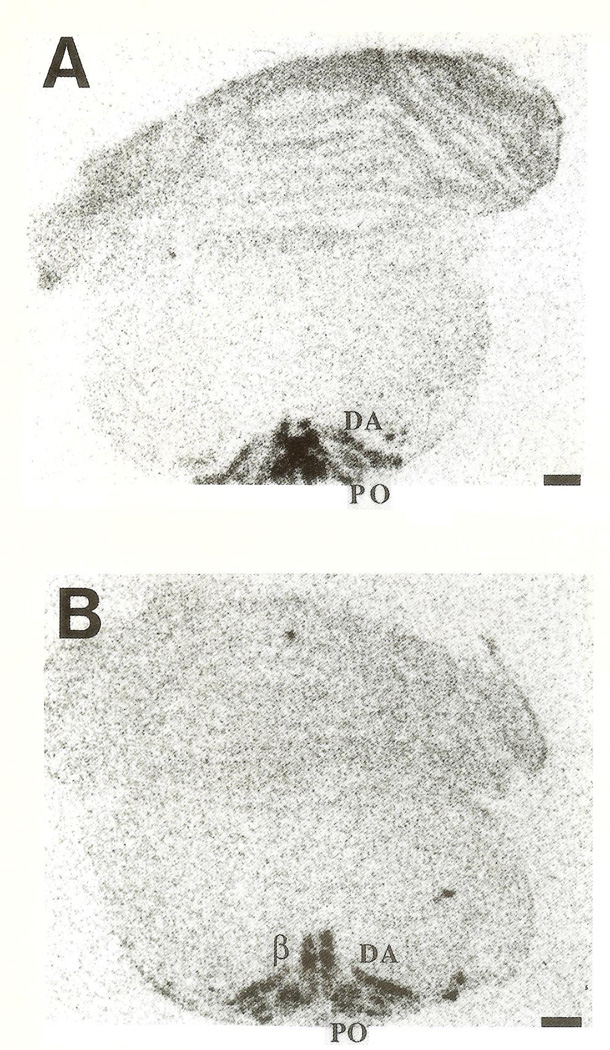
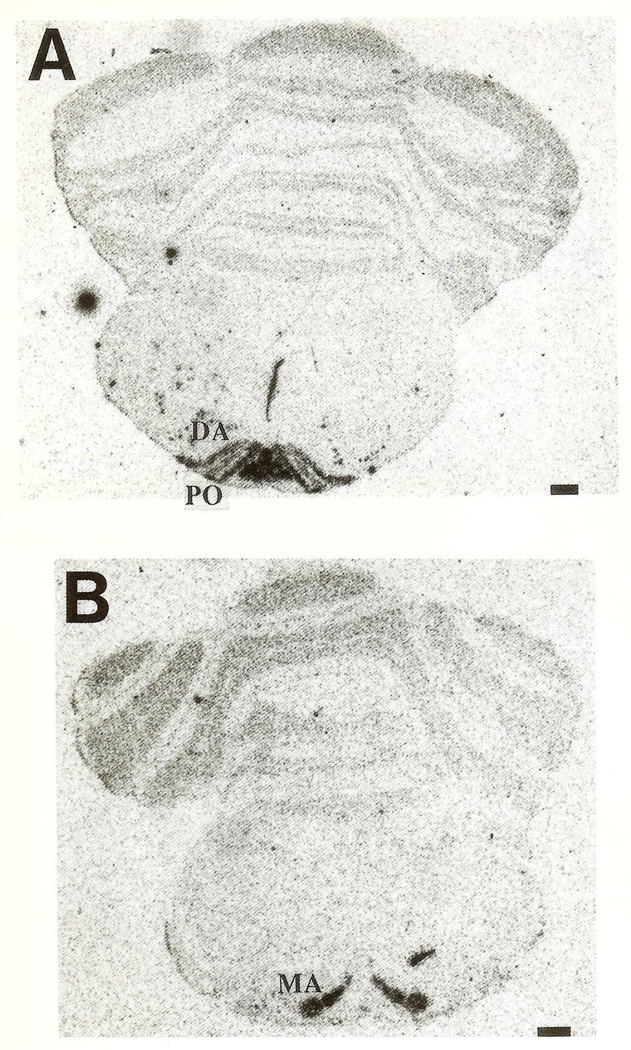
CRH-mRNA was not detectable in the inferior olive of rats aged one to three days (Fig. 1A, B). Pilot data revealed no hybridization to the CRH probe in prenatal olive and no signal was observed using the sense strand control (not shown). On the fifth postnatal day, a faint hybridization signal above background was observed in all olivary nuclei (Fig. 2), in two of four animals examined. CRH-mRNA abundance remained low through the end of the first postnatal week. Subsequently, abundance of CRH transcript increased with age, starting with the medial olivary nuclei: a section through the rostro-caudal middle-third of the inferior olive of a 10-day-old rat is shown (Fig. 3). Robust CRH gene expression was evident medially, in the beta subnucleus and dorsomedial cell column of the principal olivary nucleus (Table 1). By day 14, CRH-mRNA abundance was high in all major olivary complex components: dorsal accessory (DA) and medial accessory (MA) and the beta subnucleus of the principal olive (Fig. 4A, B). On postnatal day 18, maximal hybridization signal density occurred medially, over the beta subnucleus of the principal olivary nucleus. Dorsal accessory and medial accessory nuclei of the inferior olive contained CRH-mRNA, with a tendency to a medial–lateral gradient (Fig. 5A, B; Table 1). In our hands, this distribution pattern was also seen in the adult (not shown).
Fig. 1.
Coronal sections of rat medulla, at the level of the inferior olive (IO), subjected to in-situ hybridization with an oligonucleotide probe directed against CRH-RNA. On postnatal day 2 (A) and 3 (B) no hybridization is visible over the IO and the sections are identical to those hybridized with sense-strand controls (not shown). Ce=cerebellum. Bar=0.5 mm.
Fig. 2.
Photomicrograph of a section of mid-inferior olive revealing the presence of low-abundance CRH-mRNA in the dorsal accessory (DA), medial accessory (MA) and principal (PO) olivary nuclei of a 5-day-old rat. Bar=0.5 mm.
Fig. 3.
Section through the IO of a 10-day-old rat. Robust CRH gene expression is evident medially, over the beta subnucleus and dorsal portion of the principal olive (PO). Bar=0.5 mm.
Table 1.
Ontogeny of CRH-mRNA in rat inferior olive
| Developmental age (postnatal day) | ||||||
|---|---|---|---|---|---|---|
| Region | 2–3 | 5 | 7 | 10 | 14 | 18 |
| Dorsal accessory | − | ± | + | + | +++ | ++ |
| Medical accessory | − | ± | + | + | ++ | +++ |
| Principal | − | ± | ++ | ++ | +++ | ++++ |
| Beta subnucleus | − | − | + | ++ | ++++ | ++++ |
Seen in two of four rats.
Fig. 4.
CRH-mRNA distribution in (A) rostral and (B) caudal IO olive complex of a 14-day-old rat. DA=dorsal accessory, PO=principal olivary nucleus; MA=medial accessory nucleus; β=beta olivary subnucleus. Bar=0.5 mm.
Fig. 5.
CRH-mRNA distribution in the rostral (A) and caudal (B) IO of an 18-day-old rat. MA = medial accessory, DA = dorsal accessory, PO=principal olivary nuclei. Bar=0.5 mm.
DISCUSSION
CRH gene expression has been demonstrated in the mature inferior olive of a number of species, including mouse,17 rat,22,28 rabbit,11 cat,19 monkey species12 and humans.28 In the rat, CRH was found in neurons in the principal, and medial and dorsal accessory nuclei. In the adult mouse, higher CRH-mRNA abundance was described in dorsal and medial accessory nuclei of the inferior olive vs. the beta subnucleus,17 but CRH-mRNA has not been quantified in the rat.
The ontogeny of CRH in the inferior olive has been described in the mouse17 and opossum.9 In the mouse, very low levels of CRH-mRNA, hardly above background signal, have been reported as early as fetal day 13.5 (see Fig. 3g in Reference 17). Significant levels of CRH-mRNA were demonstrated by postnatal day 7, and adult levels in all olivary divisions were found by postnatal day 14.17
In the opossum, Cummings el al.9 recently described CRH-mRNA in cells destined to migrate to the inferior olive.9 Olivary neurons thus expressed CRH prior to the time that CRH-containing axons appeared in the cerebellar anlage. The authors concluded that CRH might have a role in target recognition and synaptic organization of the developing olivo-cerebellar pathways.
In the rat, inferior olive neurons are born on fetal days 13 and 14.1 Neurons settling rostrally in the principal nucleus antedate those ending in the caudal medial accessory nucleus. Olivary neurons, fully differentiated, migrate from the neuroepithelium of the lateral recess of the fourth ventricle. Increases in cell volume and maturation of synaptic formation and of afferent innervation, however, occur mainly postnatally.8,14
We have previously demonstrated a post-mitotic, post-migrational onset of CRH gene expression in neurons of rat hypothalamic paraventricular nucleus.4,5 The current study suggests that, in the rat inferior olive, the onset of CRH synthesis follows migration of olivary neurons to their final destination. Though the sensitivity of our in-situ hybridization technique may limit earlier detection of very small quantities of CRH-mRNA, the identical method was capable of detecting robust CRH-gene expression in single paraventricular nucleus neurons on the 17th fetal day,4,5 in agreement with others.15
The role of CRH in the olivary–cerebellar pathway is currently a topic of research.6,7,9,13 Iontophoresis of CRH to Purkinje cells suppresses hyperpolarization13 following depolarization, increases spontaneous firing rate and blocks inhibitory effects of GABA and enkephalin.6,7 CRH has excitatory properties on most neurons in a number of species3,6,7,13 and has been implicated in age-specific myoclonic and limbic seizures.3 CRH induces far more severe seizures (status epilepticus) in neonatal rats as compared to adults.3 Thus, CRH may have species-, age- and site-specific roles in the central nervous system.
In conclusion, CRH-mRNA is first evident in the inferior olive of the rat postnatally, on days 5–7. CRH-gene expression increases linearly over the subsequent two weeks, and reaches adult levels and distribution by the 18th postnatal day. Species specificity may account for distinct and differing roles for CRH in the opossum, rat and mouse.
Acknowledgements
The technical assistance of Linda Schultz is appreciated. Supported by NS 01307 and NS 28912 (T. Z. B.) and CHLA and USC funding (D.C.).
Abbreviations
- CRH
corticotropin releasing hormone
- RNA
ribonucleic acid
- ISH
in-situ hybridization
REFERENCES
- 1.Altman J, Bayer S. Prenatal development of the cerebellar system in the rat. J. comp. Neural. 1978;179:49–76. doi: 10.1002/cne.901790105. [DOI] [PubMed] [Google Scholar]
- 2.Anderson MC, Chung E, Van Woert MH. Effect of inferior olive lesion on seizure threshold in the rat. Life Sci. 1987;40:2367–2375. doi: 10.1016/0024-3205(87)90511-x. [DOI] [PubMed] [Google Scholar]
- 3.Baram TZ, Hirsch E, Snead OC, III, Schultz L. CRH induced seizures in the infant brain originate in the amygdala. Ann. Neural. 1992;31:488–494. doi: 10.1002/ana.410310505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baram TZ, Lerner SP. Corticotropin releasing hormone—ontogeny of gene expression in rat hypothalamus. Int. J. Devl Neurosci. 1991;9:473–478. doi: 10.1016/0736-5748(91)90033-i. [DOI] [PubMed] [Google Scholar]
- 5.Baram TZ, Schultz L. CRH gene expression in the fetal rat is not increased after pharmacological adrenalectomy. Neurosci. Lett. 1992;142:215–218. doi: 10.1016/0304-3940(92)90376-i. [DOI] [PubMed] [Google Scholar]
- 6.Bishop GA. Neuromodulatory effects of corticotropin releasing factor on cerebellar Purkinje cells: an in vivo study in the cat. Neuroscience. 1990;39:251–257. doi: 10.1016/0306-4522(90)90238-y. [DOI] [PubMed] [Google Scholar]
- 7.Bishop GA, King JS. Differential modulation of Purkinje cell activity by enkephalin and corticotropin releasing factor. Neuropeptides. 1992;22:167–174. doi: 10.1016/0143-4179(92)90159-t. [DOI] [PubMed] [Google Scholar]
- 8.Bourrat F, Sotelo C. Postnatal development of the inferior olivary complex in the rat: morphometric analysis of volumetric growth and neuronal cell number. Devl Brain Res. 1984;16:241–251. doi: 10.1016/0165-3806(84)90029-4. [DOI] [PubMed] [Google Scholar]
- 9.Cummings SL, Young WS, III, King JS. Early development of cerebellar afferent systems that contain corticotropin releasing factor. J. comp. Neurol. 1994;350:534–540. doi: 10.1002/cne.903500403. [DOI] [PubMed] [Google Scholar]
- 10.Cummings SL, Young WS, III, Bishop GA, De Souza EB, King SJ. Distribution of CRF in the cerebellum and pre-cerebellar nuclei of the opposum. J. comp. Neurol. 1989;280:501–521. doi: 10.1002/cne.902800402. [DOI] [PubMed] [Google Scholar]
- 11.Errico P, Barmack NH. Origin of cerebellar mossy and climbing fibers immunoreactive for corticotropin-releasing factor in the rabbit. J. comp. Neurol. 1993;336:307–320. doi: 10.1002/cne.903360211. [DOI] [PubMed] [Google Scholar]
- 12.Foote SL, Cha CI. Distribution of corticotropin-releasing factor like immunoreactivity in brainstem of two monkey species: an immunohistochemical study. J. comp. Neurol. 1988;276:239–264. doi: 10.1002/cne.902760208. [DOI] [PubMed] [Google Scholar]
- 13.Fox EA, Gruol DL. CRF suppresses the after-hyperpolarization in cerebellar Purkinje neurons. Neurosci. Lett. 1993;149:103–107. doi: 10.1016/0304-3940(93)90358-r. [DOI] [PubMed] [Google Scholar]
- 14.Gotow T, Sotelo C. Postnatal development of the inferior olive complex in the rat: synaptogenesis of GABAergic afferents. J. comp. Neurol. 1987;263:526–552. doi: 10.1002/cne.902630406. [DOI] [PubMed] [Google Scholar]
- 15.Grino MW, Young WS, III, Burgunder JM. Ontogeny of expression of the CRF gene in the hypothalamic paraventricular nucleus and of the proopiomelanocortin gene in rat pituitary. Endocrinology. 1989;124:60–68. doi: 10.1210/endo-124-1-60. [DOI] [PubMed] [Google Scholar]
- 16.Jingami H, Mizuno NT, Takahashi H, Shibahara S, Furutani Y, Imura H, Numa S. Cloning and sequence analysis of cDNA for rat CRF precursor. FEBS Lett. 1985;191:63–66. doi: 10.1016/0014-5793(85)80994-7. [DOI] [PubMed] [Google Scholar]
- 17.Keegan CE, Herman JP, Karolyi J, O’Shea S, Camper SA, Seasholtz AF. Differential expression of corticotropin releasing hormone in developing mouse embryos and adult brain. Endocrinology. 1994;134:2547–2555. doi: 10.1210/endo.134.6.8194481. [DOI] [PubMed] [Google Scholar]
- 18.King JS, Bishop GA. Ontogenesis of cerebellar afferents identified by cholecystokinin-like immunoreactivity. Devl Brain Res. 1992;65:237–252. doi: 10.1016/0165-3806(92)90185-y. [DOI] [PubMed] [Google Scholar]
- 19.Kitama K, Luppi PH, Tramu G, Sastre JP, Buda C, Jouvet M. Localization of CRH immunoreactive neurons in the cat medulla oblongata: their presence in the inferior olive. Cell Tissue Res. 1988;251:137–143. doi: 10.1007/BF00215458. [DOI] [PubMed] [Google Scholar]
- 20.Llinas R, Volkind RA. The olivocerebellar system: functional properties as revealed by harmaline-induced tremor. Exp. Brain Res. 1973;18:69–87. doi: 10.1007/BF00236557. [DOI] [PubMed] [Google Scholar]
- 21.Morara S, Provini L, Rosina A. CGRP expression in the rat olivocerebellar system during postnatal development. Brain Res. 1989;504:315–319. doi: 10.1016/0006-8993(89)91376-0. [DOI] [PubMed] [Google Scholar]
- 22.Palkovits M, Leranth C, Gorcs T, Young WS., III Corticotropin releasing factor in the olivocerebellar tract of rats: demonstration by light- and electron microscopy. Proc. natn. Acad. Sci. U.S.A. 1987;84:3911–3915. doi: 10.1073/pnas.84.11.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paradies MA, Eisenman LM. Evidence for early topographic organization in the embryonic olivocerebellar projection: a model system for study of pattern formation processes in the central nervous system. Dev. Dynam. 1993;197:125–145. doi: 10.1002/aja.1001970206. [DOI] [PubMed] [Google Scholar]
- 24.Pellegrino LJ, Pellegrino AS, Cushman AJ. A Stereotaxic Atlas of the Rat Brain. 2nd edn. New York: Plenum Press; 1981. [Google Scholar]
- 25.Walker JJ, King JS. Ontogenesis of enkephalinergic afferent systems in opossum cerebellum. Devl Brain Res. 1989;48:35–58. doi: 10.1016/0165-3806(89)90092-8. [DOI] [PubMed] [Google Scholar]
- 26.Wassef M, Cholley B, Hcizmann CW, Sotelo C. Development of the olivocerebellar projection in the rat. J. comp. Neurol. 1992;323:537–550. doi: 10.1002/cne.903230406. [DOI] [PubMed] [Google Scholar]
- 27.Watts AG, Swanson LW. Diurnal variation in the content of preprocorticotropin releasing hormone mRNA in the hypothalamic PVN of rats of both sexes as measured by in situ hybridization. Endocrinology. 1989;125:1734–1738. doi: 10.1210/endo-125-3-1734. [DOI] [PubMed] [Google Scholar]
- 28.Young WS, III, Walker LC, Powers RE. Corticotropin releasing factor mRNA is expressed in the inferior olives of rodents and primates. Mol. Brain Res. 1986;1:189–196. doi: 10.1016/0169-328x(86)90010-0. [DOI] [PubMed] [Google Scholar]
- 29.Yi SJ, Baram TZ. Corticotropin releasing factor mediates the response to cold stress in the neonatal rat, without compensatory enhancement of the peptide’s gene expression. Endocrinology. 1994;135:2364–2368. doi: 10.1210/endo.135.6.7988418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi SJ, Masters JN, Baram TZ. The effect of a specific glucocorticoid receptor antagonist on CRH gene expression in the neonatal rat hypothalamus. Devl Brain Res. 1993;73:253–259. doi: 10.1016/0165-3806(93)90145-z. [DOI] [PubMed] [Google Scholar]