Abstract
Spinal muscular atrophies (SMAs) are hereditary disorders characterized by degeneration of lower motor neurons. Different SMA types are clinically and genetically heterogeneous and many of them show significant phenotypic overlap. We recently described the clinical phenotype of a new disease in two Finnish families with a unique autosomal dominant late-onset lower motor neuronopathy. The studied families did not show linkage to any known locus of hereditary motor neuron disease and thus seemed to represent a new disease entity. For this study, we recruited two more family members and performed a more thorough genome-wide scan. We obtained significant linkage on chromosome 22q, maximum LOD score being 3.43 at marker D22S315. The linked area is defined by flanking markers D22S686 and D22S276, comprising 18.9 Mb. The region harbours 402 genes, none of which is previously known to be associated with SMAs. This study confirms that the disease in these two families is a genetically distinct entity and also provides evidence for a founder mutation segregating in both pedigrees.
Keywords: motor neuron disease, spinal muscular atrophy, linkage analysis
Introduction
Spinal muscular atrophies (SMAs) are hereditary disorders characterized by degeneration of lower motor neurons. Most SMA patients have an autosomal recessive disorder caused by mutations of SMN1 gene on 5q. Non-5q SMAs are rare and clinically and genetically heterogeneous.1, 2, 3 They are commonly classified by the inheritance pattern and distribution of weakness (distal, proximal or bulbar). However, because many separate disease entities show significant phenotypic overlap and, on the other hand, mutations in one gene may result in several allelic phenotypes, no final classification exists to fully categorize these disorders.4
Current understanding of the pathophysiology of motor neuron diseases is limited. Much information, however, has accumulated through characterization of disease-causing genes. The proteins encoded by these genes have miscellaneous cellular functions such as vesicle trafficking,5 copper transport,6 aminoacylation7 or RNA processing.8, 9 Nonetheless, it seems that many of them are involved in axonal transport and RNA metabolism.2 It has been hypothesized that large axons of motor neurons have so high metabolic requirements for maintenance, transport over long distances and precise connectivity that they are particularly vulnerable to defects in ubiquitously expressed proteins.1
For autosomal dominant non-5q SMAs, nine genes and four loci are known. Autosomal dominant forms of SMAs with predominant distal weakness result from mutations in HSPB8,10 HSPB1,11 HSPB3,12 GARS,7 DCTN15 and BSCL29 or show linkage to 2q1413 or 7q34-q36.14 Proximal adult-onset SMAs with autosomal dominant inheritance have been reported to be caused by mutations in VAPB,15 LMNA16 and TRPV417 or to be linked to 3q13.118 or 14q32.19 Mutations in many of these genes cause several allelic disorders. For example, mutations in TRPV4 are associated with scapuloperoneal SMA (OMIM 181405) or hereditary motor and sensory neuropathy type 2C (OMIM 606071),17 and mutations in GARS are known to cause Charcot-Marie-Tooth disease type 2D (OMIM 601472) and distal SMA type V (dSMAV, OMIM 600794).7 Moreover, dSMAV may also be caused by mutations in BSCL2 that on the other hand is also associated with spastic paraplegia 17 (OMIM 270685) and congenital generalized lipodystrophy type 2 (OMIM 269700).9
We recently described a new form of autosomal dominant lower motor neuron disease characterized by adult onset of painful cramps and slowly progressive very late-onset weakness and atrophy of proximal and distal muscles.20 The studied families did not show linkage to any known locus of hereditary motor neuron disease and thus seemed to represent a new disease entity. In this study, we show by a genome-wide scan and subsequent fine genetic mapping that both reported families have the same disease and that the disorder is a new genetically distinct entity linked to a novel SMA locus on chromosome 22q11.2-q13.2.
Subjects and methods
Subjects
We studied 18 members of two kindreds (F1 and F2) that were previously described.20 DNA samples of two new unaffected individuals (F1:III-22 and F2:IV-10) were collected for this study. Eleven individuals were affected by lower motor neuron disease, six were unaffected and the affection status of one family member (F1:III-30) was uncertain. The study was approved by the Institutional Review Board of the Northern Karelia Hospital District. Blood samples for molecular genetic studies were obtained after written informed consent.
Genetic analysis
Genomic DNA was extracted from the peripheral blood leukocytes by standard methods. A genome-wide scan was done with 16 samples of the two kindreds using deCODE 536 microsatellite marker set at an average of 8 cM density.21 Linkage analysis on all the autosomes was run with Merlin22 using a parametric dominant model with a disease allele frequency of 0.0001 and absolute disease risk of 0.0001 for noncarriers and 0.9 for carriers. Merlin was also used to construct haplotypes. To confirm and refine the location of the disease-causing gene, seven additional markers, D22S264, D22S446, D22S306, D22S686, D22S926, D22S419 and D22S421, were analyzed. Fluorescently labeled PCR products were analyzed using ABI3130xl automatic DNA Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and Peak Scanner Software v1.0 (Applied Biosystems). DNA from additional two unaffected relatives (F1:III-22 and F2:IV-10) were included in these analyses.
Results
Clinical features
Our 2–3-year clinical follow-up of patients accords with previously reported findings (Table 1). The general features include onset of painful cramps in adulthood, followed by slowly progressive proximal and distal weakness, usually after 45 years of age and very late in some patients. All patients were still ambulant, at least for short distances, although one 63-year-old male patient uses a mobility scooter outdoors because of muscle weakness and poor balance. Respiratory, bulbar or facial involvement has not occurred in any patient. One female patient, aged 50, has developed neck muscle weakness with a tendency to head drooping. Mild muscle atrophy is observed only very late in the disease course.
Table 1. Follow-up data on clinical findings in five patients from the two families described previously.
| Patient | Sex | Age at examination (years) | Disease duration (years) | Neurogenic EMG in all limbs | Hypo/areflexia | Weakness | Walking ability preserved | Musculature (atrophy) |
|---|---|---|---|---|---|---|---|---|
| F1:III-23 | M | 57 | 17 | Yes | All limbs | All limbs and abd | Yes | Well formed (pseudohypertrophic calves) |
| F1:III-34 | F | 49 | 5 | Yes | All limbs | Right lower limb | Yes | Well formed but mild right-calf atrophy |
| F2:IV-8 | M | 63 | 23 | Yes | All limbs | All limbs and abd | Walking sticks or mobility scooter | Mild atrophy in lower limbs |
| F2:IV-3 | M | 63 | 18 | Yes | All limbs | All limbs and abd | Yes | No atrophy |
| F2:V-3 | M | 39 | 5 | Yes | All limbs | All limbs and abd | Yes | Calves atrophic |
Abbreviation: abd, abdominal muscles.
The follow-up on these tabulated patients ranged from 2 to 16 years at the neurological department.
Molecular genetic results
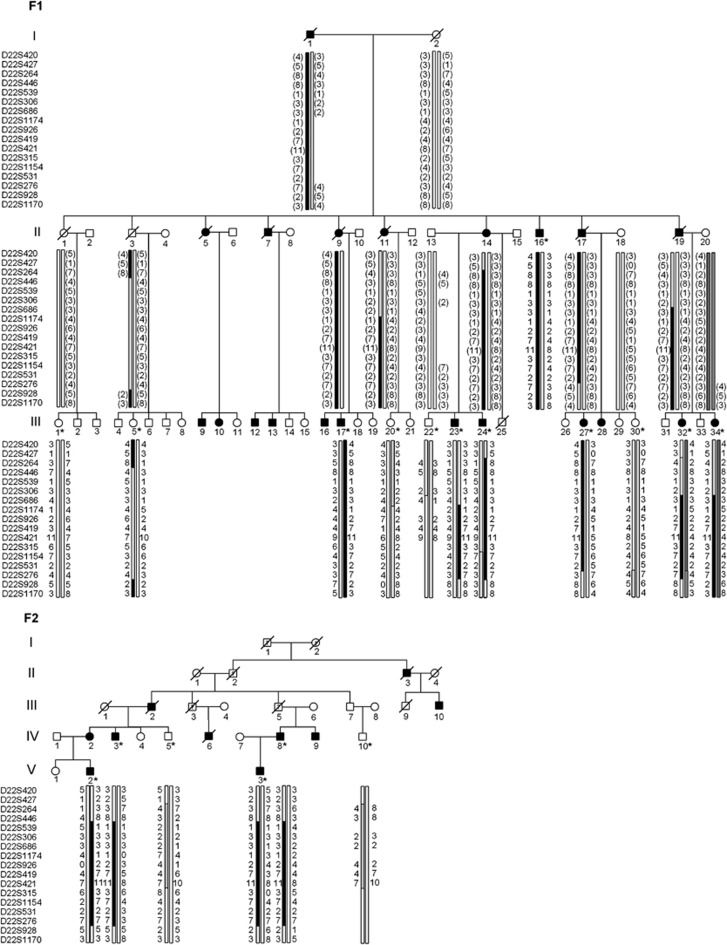
The disease occurred in both genders in successive generations and with male-to-male transmission, indicating autosomal dominant inheritance. In the genome-wide scan, significant evidence for linkage was obtained on chromosome 22 between markers D22S427 and D22S276, maximum parametric LOD score being 5.01 at marker D22S315. Refined linkage analysis was performed with seven additional markers, which defined the area further between markers D22S686 and D22S276, this time maximum parametric LOD score being 3.43 at marker D22S315 (Table 2). The disease-associated haplotype was identical in both families and was shared by all affected members of the two families (Figure 1). No unaffected individuals, including the one individual whose affection status was uncertain, carried the haplotype. Two recombination events, one in F1:III-23 and the other in F1:III-27, narrowed the linked region between markers D22S686 and D22S276, spanning 18.9 Mb (chromosome 22: 23 068 722–42 012 370). The linked genomic area harbours 402 genes, some of which are hypothetical or pseudogenes (http://www.ncbi.nlm.nih.gov/mapview).
Table 2. Parametric linkage LOD scores for both the studied families on chromosome 22.
| Marker | LOD | Location (Mb) |
|---|---|---|
| D22S420 | −3.57 | 17.86 |
| D22S427 | −4.92 | 18.59 |
| D22S264 | −5.98 | 20.77 |
| D22S446 | −4.55 | 22.02 |
| D22S539 | −4.59 | 22.26 |
| D22S306 | −4.72 | 22.56 |
| D22S686 | −0.59 | 23.07 |
| D22S1174 | 3.27 | 24.48 |
| D22S926 | 3.41 | 25.82 |
| D22S419 | 3.42 | 25.94 |
| D22S421 | 3.42 | 25.95 |
| D22S315 | 3.43 | 26.02 |
| D22S1154 | 3.35 | 26.62 |
| D22S531 | 3.13 | 30.70 |
| D22S276 | 1.71 | 42.01 |
| D22S928 | −6.46 | 45.48 |
| D22S1170 | −2.99 | 48.35 |
Markers inside the linked area are indicated in bold.
Figure 1.
Pedigrees and chromosome 22 haplotypes of members of families F1 and F2. DNA sample was available from individuals marked with an asterisk. Blackened symbols indicate individuals affected with lower motor neuron disease. The black haplotype represents the disease haplotype. Inferred haplotypes are presented in parentheses. Observed recombination events in F1:III-23 and F1:III-27 suggest that the minimum probable candidate interval is flanked by D22S686 and D22S276.
Discussion
We have recently reported two Finnish families with a slowly progressive motor neuron disease. The clinical phenotype of their disease differed from previously described autosomal dominant SMAs and all the known motor neuron disorder loci were excluded from being associated with the disease.20 In this study, a genome-wide linkage study identified significant linkage to chromosome 22q11.2-q13.2 with a unique haplotype segregating in the affected members of both of the families.
The most common first symptoms of this new entity of SMA are painful cramps and fasciculations presented in the fourth and fifth decade. Muscle atrophy and weakness develop gradually late in the disease course and no patient was wheelchair-dependent. The disease does not cause symptoms or signs from respiratory, bulbar or facial muscles or upper motor neurons. EMG shows mild to moderate, widespread chronic and active neurogenic changes. Neurogenic changes are also the main morphological findings on muscle biopsy. One of the subjects included in this study (F1:III-30) could not be definitely classified. In this study, she proved not to carry the disease haplotype, indicating that her EMG findings with polyphasic motor unit potentials and fibrillations were unrelated to the familial disease.
Locus 22q11.2-q13.2 has not been previously linked to SMAs. Most of the genes inside the linked area are poorly described and none of them is an obvious candidate gene. Because the genes known to cause motor neuron disorders have such variation in their functions, practically no gene can be excluded a priori. Nevertheless, in the linked region, there are altogether 36 immunoglobulin lambda variable genes that seem to be highly unlikely candidates.
The only known gene linked to motor neuron disease in chromosome 22 is NEFH, coding for human heavy neurofilament subunit protein. Neurofilaments have a role in axonal transport, and deletions in NEFH have been found in ALS patients. It could not be determined, however, whether the deletions segregated with the disease or not.23 The symptoms of the patients in our families differ from those of ALS, but otherwise NEFH is probably one of the strongest candidate genes in the linked region.
Of the other genes in the area that have been associated with nervous system, ADORA2A is probably the best described. Its product is one of the several receptor subtypes for adenosine. The activity of the encoded protein, a G-protein-coupled receptor family member, is mediated by G proteins, which activate adenylyl cyclase. The encoded protein is abundant in basal ganglia, vasculature and platelets and it is a major target of caffeine. It has been suggested to be involved in the pathogenesis of Huntington's disease.24 UPB1 encodes beta-ureidopropionase that catalyzes the last step in the pyrimidine degradation pathway. Ureidopropionase deficiencies are associated with N-carbamyl-beta-amino aciduria and may lead to abnormalities in central nervous functions. SGSM1 (small G-protein-signaling modulator 1) is poorly described. It is mainly expressed in brain, heart and testis, but it is known to be expressed also in spinal cord and peripheral nerves. SGSM proteins modulate small G-protein-mediated (RAP and RAB) neuronal signal transduction and the vesicular transportation pathways.25 One of these genes could be the disease-causing gene, but because the number of potential genes is over 400, selecting the right gene for sequencing is hardly conceivable.
Possible approaches for finding the disease-causing gene could be exome sequencing or high-throughput sequencing of the linked 18.9 Mb region. However, because the disease-associated haplotype is the same in these two families, all the possibly detected variations in the 18.9 Mb area will be common to all our patients and the number of potential disease-causing mutations would still be high. Another approach is to narrow down the linked area further and our next aim is to increase the patient material. With the linked haplotype identified in this study, it is now possible to try to find both other Finnish patients affected by this disease and new critical recombinants. Because the disease is dominantly inherited, it is likely that more patients can be found.
The finding that both families in the study share a common haplotype provides evidence of a common founder mutation. The families are not known to be related, but because they both originate from the same geographical region, they should have a common ancestor. Whether or not this new entity of spinal muscular neuronopathy is purely a Finnish peculiarity remains to be seen. With the linkage data provided, families with a comparable phenotype can now be checked by the molecular genetic methods. Any identified family in other populations linked to this new locus will be of high interest for determination of the causative gene.
Acknowledgments
This study was supported by Finska Läkaresällskapet grant.
The authors declare no conflict of interest.
References
- Irobi J, Dierick I, Jordanova A, Claeys KG, De Jonghe P, Timmerman V. Unraveling the genetics of distal hereditary motor neuronopathies. Neuromolecular Med. 2006;8:131–146. doi: 10.1385/nmm:8:1-2:131. [DOI] [PubMed] [Google Scholar]
- Van Den Bosch L, Timmerman V. Genetics of motor neuron disease. Curr Neurol Neurosci Rep. 2006;6:423–431. doi: 10.1007/s11910-996-0024-9. [DOI] [PubMed] [Google Scholar]
- Zerres K, Rudnik-Schoneborn S. 93rd ENMC international workshop: non-5q-spinal muscular atrophies (SMA)–clinical picture (6–8 April 2001, Naarden, The Netherlands) Neuromuscul Disord. 2003;13:179–183. doi: 10.1016/s0960-8966(02)00211-0. [DOI] [PubMed] [Google Scholar]
- Darras BT. Non-5q spinal muscular atrophies: the alphanumeric soup thickens. Neurology. 2011;77:312–314. doi: 10.1212/WNL.0b013e3182267bd8. [DOI] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Kennerson ML, Nicholson GA, Kaler SG, et al. Missense mutations in the copper transporter gene ATP7A cause X-linked distal hereditary motor neuropathy. Am J Hum Genet. 2010;86:343–352. doi: 10.1016/j.ajhg.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann K, Schuelke M, Diers A, et al. Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet. 2001;29:75–77. doi: 10.1038/ng703. [DOI] [PubMed] [Google Scholar]
- Windpassinger C, Auer-Grumbach M, Irobi J, et al. Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and silver syndrome. Nat Genet. 2004;36:271–276. doi: 10.1038/ng1313. [DOI] [PubMed] [Google Scholar]
- Irobi J, Van Impe K, Seeman P, et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet. 2004;36:597–601. doi: 10.1038/ng1328. [DOI] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova I, Irobi J, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- Kolb SJ, Snyder PJ, Poi EJ, et al. Mutant small heat shock protein B3 causes motor neuropathy: utility of a candidate gene approach. Neurology. 2010;74:502–506. doi: 10.1212/WNL.0b013e3181cef84a. [DOI] [PubMed] [Google Scholar]
- McEntagart M, Norton N, Williams H, et al. Localization of the gene for distal hereditary motor neuronopathy VII (dHMN-VII) to chromosome 2q14. Am J Hum Genet. 2001;68:1270–1276. doi: 10.1086/320122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath S, Blair IP, Kennerson ML, Durnall JC, Nicholson GA. A novel locus for distal motor neuron degeneration maps to chromosome 7q34-q36. Hum Genet. 2007;121:559–564. doi: 10.1007/s00439-007-0348-9. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnik-Schoneborn S, Botzenhart E, Eggermann T, et al. Mutations of the LMNA gene can mimic autosomal dominant proximal spinal muscular atrophy. Neurogenetics. 2007;8:137–142. doi: 10.1007/s10048-006-0070-0. [DOI] [PubMed] [Google Scholar]
- Deng HX, Klein CJ, Yan J, et al. Scapuloperoneal spinal muscular atrophy and CMT2C are allelic disorders caused by alterations in TRPV4. Nat Genet. 2010;42:165–169. doi: 10.1038/ng.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Nakagawa M, Suehara M, et al. Gene for hereditary motor and sensory neuropathy (proximal dominant form) mapped to 3q13.1. Neuromuscul Disord. 1999;9:368–371. doi: 10.1016/s0960-8966(99)00021-8. [DOI] [PubMed] [Google Scholar]
- Harms MB, Allred P, Gardner R, et al. Dominant spinal muscular atrophy with lower extremity predominance: linkage to 14q32. Neurology. 2010;75:539–546. doi: 10.1212/WNL.0b013e3181ec800c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokela M, Penttila S, Huovinen S, et al. Late-onset lower motor neuronopathy: a new autosomal dominant disorder. Neurology. 2011;77:334–340. doi: 10.1212/WNL.0b013e3182267b71. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Al-Chalabi A, Andersen PM, Nilsson P, et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet. 1999;8:157–164. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- Taherzadeh-Fard E, Saft C, Wieczorek S, Epplen JT, Arning L. Age at onset in Huntington's disease: replication study on the associations of ADORA2A, HAP1 and OGG1. Neurogenetics. 2010;11:435–439. doi: 10.1007/s10048-010-0248-3. [DOI] [PubMed] [Google Scholar]
- Yang H, Sasaki T, Minoshima S, Shimizu N. Identification of three novel proteins (SGSM1, 2, 3) which modulate small G protein (RAP and RAB)-mediated signaling pathway. Genomics. 2007;90:249–260. doi: 10.1016/j.ygeno.2007.03.013. [DOI] [PubMed] [Google Scholar]